Perspectives in Genome-Editing Techniques for Livestock
Simple Summary
Abstract
1. Introduction
2. Precision Genome-Editing Tools Successfully Used in Livestock
2.1. Zinc Finger Nucleases (ZFNs)
2.2. Transcription Activator-like Effector Nucleases (TALENs)
2.3. Clustered Regularly Interspaced Palindromic Repeats (CRISPR)
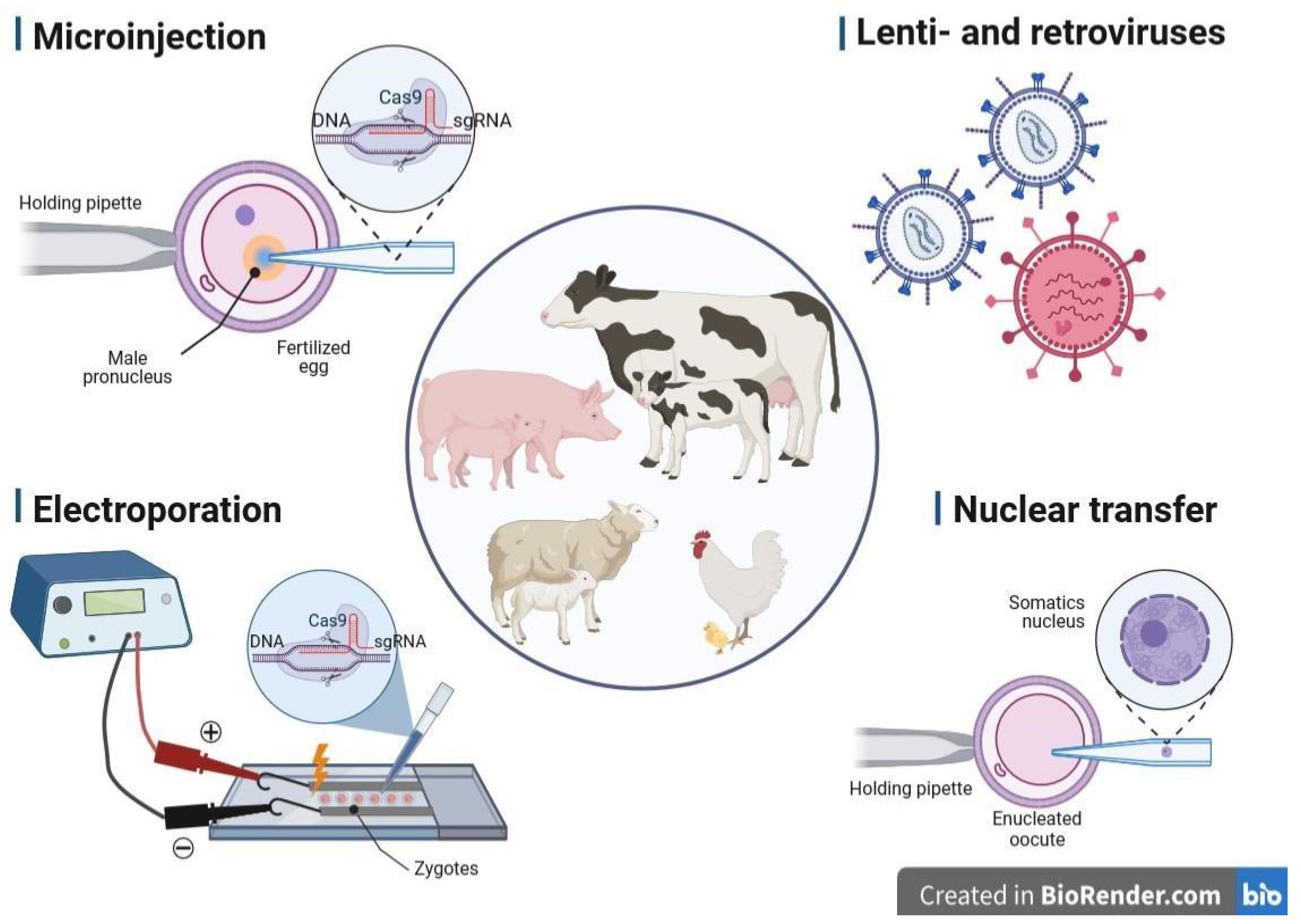
3. Methods of Gene Delivery Successfully Applied in Livestock Biotechnology
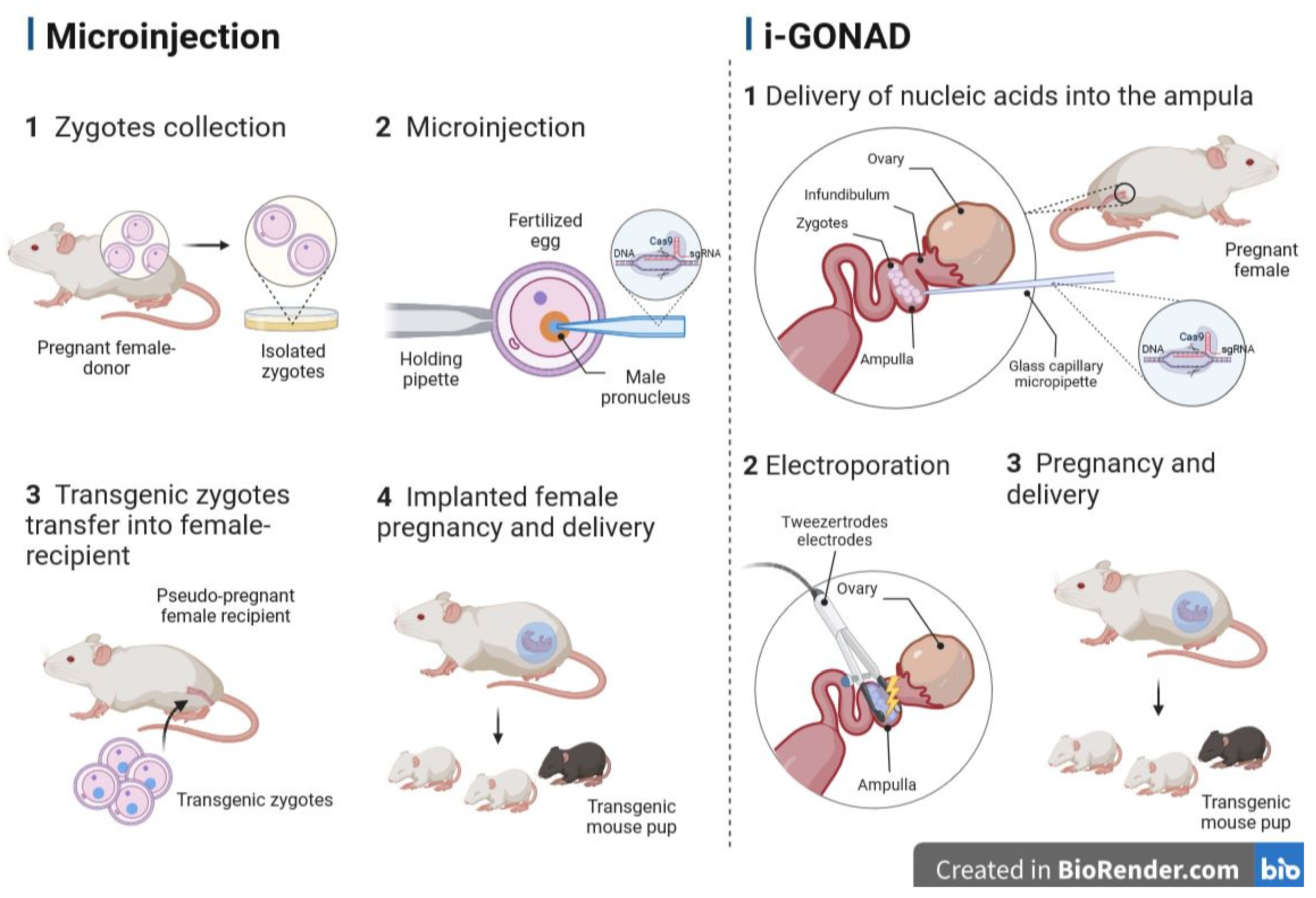
3.1. Microinjection into the Pronucleus
3.2. Retro- and Lentiviral Vectors
3.3. Electroporation of Zygotes
3.4. Transfer of Nucleus (Cloning, SCNT)
3.5. Sperm-Mediated Gene Transfer (SMGT)
4. Transgenic Livestock Successfully Approved for Industrial Use
5. Gene-Editing Systems and Delivery Methods Not yet Applied in Farm Animals or Applied with Little Success
5.1. Blastocyst Injection
5.2. Genome Editing via Oviductal Nucleic Acid Delivery (GONAD)
5.3. Transplacental Gene Delivery (TPGD)
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Van Eenennaam, A.L.; Young, A.E. Genetic Improvement of Food Animals: Past and Future. In Encyclopedia of Food Security and Sustainability; Elsevier: Amsterdam, The Netherlands, 2018; pp. 171–180. [Google Scholar] [CrossRef]
- Singh, P.; Ali, S.A. Impact of CRISPR-Cas9-Based Genome Engineering in Farm Animals. Vet. Sci. 2021, 8, 122. [Google Scholar] [CrossRef]
- McFarlane, G.R.; Salvesen, H.A.; Sternberg, A.; Lillico, S.G. On-Farm Livestock Genome Editing Using Cutting Edge Reproductive Technologies. Front. Sustain. Food Syst. 2019, 3, 106. [Google Scholar] [CrossRef]
- Turnbull, C.; Lillemo, M.; Hvoslef-Eide, T.A.K. Global Regulation of Genetically Modified Crops Amid the Gene Edited Crop Boom—A Review. Front. Plant Sci. 2021, 12, 630396. [Google Scholar] [CrossRef] [PubMed]
- Prakash, D.; Verma, S.; Bhatia, R.; Tiwary, B.N. Risks and Precautions of Genetically Modified Organisms. ISRN Ecol. 2011, 2011, 369573. [Google Scholar] [CrossRef]
- Hallerman, E.M.; Bredlau, J.P.; Camargo, L.S.A.; Dagli, M.L.Z.; Karembu, M.; Ngure, G.; Romero-Aldemita, R.; Rocha-Salavarrieta, P.J.; Tizard, M.; Walton, M.; et al. Towards progressive regulatory approaches for agricultural applications of animal biotechnology. Transgenic Res. 2022, 31, 167–199. [Google Scholar] [CrossRef]
- Wells, D.J. Genetically Modified Animals and Pharmacological Research. Comp. Vet. Pharmacol. 2010, 199, 213–226. [Google Scholar] [CrossRef]
- Niemann, H.; Kues, W.A. Transgenic farm animals: An update. Reprod. Fertil. Dev. 2007, 19, 762–770. [Google Scholar] [CrossRef]
- Eriksson, S.; Jonas, E.; Rydhmer, L.; Röcklinsberg, H. Invited review: Breeding and ethical perspectives on genetically modified and genome edited cattle. J. Dairy Sci. 2018, 101, 1–17. [Google Scholar] [CrossRef]
- Van Eenennaam, A.L. Application of genome editing in farm animals: Cattle. Transgenic Res. 2019, 28, 93–100. [Google Scholar] [CrossRef]
- Niemann, H.; Kues, W.; Carnwath, J.W. Transgenic Farm Animals: Current Status and Perspectives for Agriculture and Biomedicine; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Houdebine, L.-M. Production of pharmaceutical proteins by transgenic animals. Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 107–121. [Google Scholar] [CrossRef]
- Hata, T.; Uemoto, S.; Kobayashi, E. Transplantable liver production plan. Organogenesis 2013, 9, 235–238. [Google Scholar] [CrossRef]
- Hryhorowicz, M.; Zeyland, J.; Słomski, R.; Lipiński, D. Genetically Modified Pigs as Organ Donors for Xenotransplantation. Mol. Biotechnol. 2017, 59, 435–444. [Google Scholar] [CrossRef] [PubMed]
- West, J.; Gill, W.W. Genome Editing in Large Animals. J. Equine Vet. Sci. 2016, 41, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kubo, Y.; Yamashita, K.; Saito, T.; Tanaka, K.; Makino, T.; Takahashi, T.; Kurokawa, Y.; Yamasaki, M.; Eguchi, H.; Doki, Y.; et al. Heparinized swine models for better surgical/endoscopic training. DEN Open 2021, 2, e64. [Google Scholar] [CrossRef]
- Tamme, R.; Laing, D.; Steinmann, W.-D.; Bauer, T. Transgenic Livestock for Food Production, Introduction. In Encyclopedia of Sustainability Science and Technology; Elsevier: Amsterdam, The Netherlands, 2012; pp. 10812–10814. [Google Scholar] [CrossRef]
- Keiser, N.W.; Engelhardt, J.F. New animal models of cystic fibrosis: What are they teaching us? Curr. Opin. Pulm. Med. 2011, 17, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Noble, M.S.; Rodriguez-Zas, S.; Cook, J.B.; Bleck, G.T.; Hurley, W.L.; Wheeler, M.B. Lactational performance of first-parity transgenic gilts expressing bovine alpha-lactalbumin in their milk. J. Anim. Sci. 2002, 80, 1090–1096. [Google Scholar] [CrossRef]
- Carlson, D.F.; Lancto, C.A.; Zang, B.; Kim, E.-S.; Walton, M.; Oldeschulte, D.; Seabury, C.; Sonstegard, T.S.; Fahrenkrug, S.C. Production of hornless dairy cattle from genome-edited cell lines. Nat. Biotechnol. 2016, 34, 479–481. [Google Scholar] [CrossRef]
- Young, A.E.; Mansour, T.A.; McNabb, B.R.; Owen, J.R.; Trott, J.F.; Brown, C.T.; Van Eenennaam, A.L. Genomic and phenotypic analyses of six offspring of a genome-edited hornless bull. Nat. Biotechnol. 2019, 38, 225–232. [Google Scholar] [CrossRef]
- Niu, Y.; Zhao, X.; Zhou, J.; Li, Y.; Huang, Y.; Cai, B.; Liu, Y.; Ding, Q.; Zhou, S.; Zhao, J.; et al. Efficient generation of goats with defined point mutation (I397V) in GDF9 through CRISPR/Cas9. Reprod. Fertil. Dev. 2018, 30, 307. [Google Scholar] [CrossRef]
- Fabre, S.; Pierre, A.; Mulsant, P.; Bodin, L.; Di Pasquale, E.; Persani, L.; Monget, P.; Monniaux, D. Regulation of ovulation rate in mammals: Contribution of sheep genetic models. Reprod. Biol. Endocrinol. 2006, 4, 20. [Google Scholar] [CrossRef]
- Zheng, Q.; Lin, J.; Huang, J.; Zhang, H.; Zhang, R.; Zhang, X.; Cao, C.; Hambly, C.; Qin, G.; Yao, J.; et al. Reconstitution of UCP1 using CRISPR/Cas9 in the white adipose tissue of pigs decreases fat deposition and improves thermogenic capacity. Proc. Natl. Acad. Sci. USA 2017, 114, E9474–E9482. [Google Scholar] [CrossRef]
- Wang, X.; Yu, H.; Lei, A.; Zhou, J.; Zeng, W.; Zhu, H.; Dong, Z.; Niu, Y.; Shi, B.; Cai, B.; et al. Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Sci. Rep. 2015, 5, srep13878. [Google Scholar] [CrossRef] [PubMed]
- Oishi, I.; Yoshii, K.; Miyahara, D.; Kagami, H.; Tagami, T. Targeted mutagenesis in chicken using CRISPR/Cas9 system. Sci. Rep. 2016, 6, 23980. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.K.; Pinzón, C.; Huggins, S.; Pryor, J.H.; Falck, A.; Herman, F.; Oldeschulte, J.; Chavez, M.B.; Foster, B.L.; White, S.H.; et al. Genetic engineering a large animal model of human hypophosphatasia in sheep. Sci. Rep. 2018, 8, 16945. [Google Scholar] [CrossRef]
- Hu, S.; Wang, Z.; Polejaeva, I. 40 KNOCKOUT OF GOAT NUCLEOPORIN 155 (NUP155) GENE USING CRISPR/Cas9 SYSTEMS. Reprod. Fertil. Dev. 2014, 26, 134. [Google Scholar] [CrossRef]
- Ni, W.; Qiao, J.; Hu, S.; Zhao, X.; Regouski, M.; Yang, M.; Polejaeva, I.A.; Chen, C. Efficient Gene Knockout in Goats Using CRISPR/Cas9 System. PLoS ONE 2014, 9, e106718. [Google Scholar] [CrossRef] [PubMed]
- Program and Abstracts of the 14th Transgenic Technology Meeting (TT2017): Snowbird Resort, Salt Lake City, Utah, USA, 1–4 October 2017. Transgenic Res. 2017, 26, 1–45. [CrossRef] [PubMed]
- Fan, Z.; Yang, M.; Regouski, M.; Polejaeva, I.A. Gene Knockouts in Goats Using CRISPR/Cas9 System and Somatic Cell Nuclear Transfer. Methods Mol. Biol. 2019, 1874, 373–390. [Google Scholar] [CrossRef] [PubMed]
- Vilarino, M.; Rashid, S.T.; Suchy, F.P.; McNabb, B.R.; van der Meulen, T.; Fine, E.J.; Ahsan, S.D.; Mursaliyev, N.; Sebastiano, V.; Diab, S.S.; et al. CRISPR/Cas9 microinjection in oocytes disables pancreas development in sheep. Sci. Rep. 2017, 7, 17472. [Google Scholar] [CrossRef]
- Vilarino, M.; Suchy, F.P.; Rashid, S.T.; Lindsay, H.; Reyes, J.; McNabb, B.R.; van der Meulen, T.; Huising, M.O.; Nakauchi, H.; Ross, P.J. Mosaicism diminishes the value of pre-implantation embryo biopsies for detecting CRISPR/Cas9 induced mutations in sheep. Transgenic Res. 2018, 27, 525–537. [Google Scholar] [CrossRef]
- Carroll, D. Genome editing: Past, present, and future. Yale J. Biol. Med. 2017, 90, 653–659. [Google Scholar] [PubMed]
- Sun, Z.; Wang, M.; Han, S.; Ma, S.; Zou, Z.; Ding, F.; Li, X.; Li, L.; Tang, B.; Wang, H.; et al. Production of hypoallergenic milk from DNA-free beta-lactoglobulin (BLG) gene knockout cow using zinc-finger nucleases mRNA. Sci. Rep. 2018, 8, 15430. [Google Scholar] [CrossRef] [PubMed]
- Hauschild, J.; Petersen, B.; Santiago, Y.; Queisser, A.-L.; Carnwath, J.W.; Lucas-Hahn, A.; Zhang, L.; Meng, X.; Gregory, P.D.; Schwinzer, R.; et al. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 2011, 108, 12013–12017. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Y.; Tian, Y.; Yu, Y.; Gao, M.; Hu, G.; Su, F.; Pan, S.; Luo, Y.; Guo, Z.; et al. Generation of mastitis resistance in cows by targeting human lysozyme gene to β-casein locus using zinc-finger nucleases. Proc. R. Soc. B Boil. Sci. 2014, 281, 20133368. [Google Scholar] [CrossRef]
- Flisikowska, T.; Thorey, I.S.; Offner, S.; Ros, F.; Lifke, V.; Zeitler, B.; Rottmann, O.; Vincent, A.; Zhang, L.; Jenkins, S.; et al. Efficient Immunoglobulin Gene Disruption and Targeted Replacement in Rabbit Using Zinc Finger Nucleases. PLoS ONE 2011, 6, e21045. [Google Scholar] [CrossRef]
- Geurts, A.M.; Cost, G.J.; Freyvert, Y.; Zeitler, B.; Miller, J.C.; Choi, V.M.; Jenkins, S.S.; Wood, A.; Cui, X.; Meng, X.; et al. Knockout Rats via Embryo Microinjection of Zinc-Finger Nucleases. Science 2009, 325, 433. [Google Scholar] [CrossRef] [PubMed]
- Lillico, S.G.; Proudfoot, C.; King, T.J.; Tan, W.; Zhang, L.; Mardjuki, R.; Paschon, D.E.; Rebar, E.J.; Urnov, F.D.; Mileham, A.J.; et al. Mammalian interspecies substitution of immune modulatory alleles by genome editing. Sci. Rep. 2016, 6, 21645. [Google Scholar] [CrossRef] [PubMed]
- Bedell, V.M.; Wang, Y.; Campbell, J.M.; Poshusta, T.L.; Starker, C.G.; Krug, R.G., 2nd; Tan, W.; Penheiter, S.G.; Ma, A.C.; Leung, A.Y.H.; et al. In vivo genome editing using a high-efficiency TALEN system. Nature 2012, 491, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Sanjana, N.E.; Cong, L.; Zhou, Y.; Cunniff, M.M.; Feng, G.; Zhang, F. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 2012, 7, 171–192. [Google Scholar] [CrossRef]
- Moscou, M.J.; Bogdanove, A.J. A Simple Cipher Governs DNA Recognition by TAL Effectors. Science 2009, 326, 1501. [Google Scholar] [CrossRef]
- Method of the Year 2011. Nat. Methods 2011, 9, 1. [CrossRef]
- Moghaddassi, S.; Eyestone, W.; Bishop, C.E. TALEN-Mediated Modification of the Bovine Genome for Large-Scale Production of Human Serum Albumin. PLoS ONE 2014, 9, e89631. [Google Scholar] [CrossRef] [PubMed]
- Carlson, D.F.; Tan, W.; Lillico, S.G.; Stverakova, D.; Proudfoot, C.; Christian, M.; Voytas, D.F.; Long, C.R.; Whitelaw, C.B.A.; Fahrenkrug, S.C. Efficient TALEN-mediated gene knockout in livestock. Proc. Natl. Acad. Sci. USA 2012, 109, 17382–17387. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Mali, P.; Esvelt, K.M.; Church, G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods 2013, 10, 957–963. [Google Scholar] [CrossRef]
- Hao, F.; Yan, W.; Li, X.; Wang, H.; Wang, Y.; Hu, X.; Liu, X.; Liang, H.; Liu, D. Generation of Cashmere Goats Carrying an EDAR Gene Mutant Using CRISPR-Cas9-Mediated Genome Editing. Int. J. Biol. Sci. 2018, 14, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Kalds, P.; Zhou, S.; Cai, B.; Liu, J.; Wang, Y.; Petersen, B.; Sonstegard, T.; Wang, X.; Chen, Y. Sheep and Goat Genome Engineering: From Random Transgenesis to the CRISPR Era. Front. Genet. 2019, 10, 750. [Google Scholar] [CrossRef]
- Zinovieva, N.A.; Volkova, N.A.; Bagirov, V.A.; Brem, G. Transgenic farm animals: Status of the current researches and the future. Ecol. Genet. 2015, 13, 58–76. [Google Scholar] [CrossRef][Green Version]
- Hashimoto, M.; Yamashita, Y.; Takemoto, T. Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non-mosaic mutants in the mouse. Dev. Biol. 2016, 418, 1–9. [Google Scholar] [CrossRef]
- O’Neil, E.V.; Brooks, K.; Burns, G.W.; Ortega, M.S.; Denicol, A.C.; Aguiar, L.H.; Pedroza, G.H.; Benne, J.; Spencer, T.E. Prostaglandin-endoperoxide synthase 2 is not required for preimplantation ovine conceptus development in sheep. Mol. Reprod. Dev. 2020, 87, 142–151. [Google Scholar] [CrossRef]
- Heo, Y.T.; Quan, X.; Xu, Y.N.; Baek, S.; Choi, H.; Kim, N.-H.; Kim, J. CRISPR/Cas9 Nuclease-Mediated Gene Knock-In in Bovine-Induced Pluripotent Cells. Stem Cells Dev. 2015, 24, 393–402. [Google Scholar] [CrossRef]
- Tan, W.; Carlson, D.F.; Lancto, C.A.; Garbe, J.R.; Webster, D.A.; Hackett, P.B.; Fahrenkrug, S.C. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc. Natl. Acad. Sci. USA 2013, 110, 16526–16531. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Nie, J.; Tang, Y.; He, W.; Xiao, K.; Pang, C.; Liang, X.; Lu, Y.; Zhang, M. Generation of Transgenic Cloned Buffalo Embryos Harboring the EGFP Gene in the Y Chromosome Using CRISPR/Cas9-Mediated Targeted Integration. Front. Vet. Sci. 2020, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Hongbing, H.; Yonghe, M.; Tao, W.; Ling, L.; Xiuzhi, T.; Rui, H.; Shoulong, D.; Kongpan, L.; Feng, W.; Ning, L.; et al. One-step generation of myostatin gene knockout sheep via the CRISPR/Cas9 system. Front. Agric. Sci. Eng. 2014, 1, 2–5. [Google Scholar] [CrossRef]
- Crispo, M.; Mulet, A.P.; Tesson, L.; Barrera, N.; Cuadro, F.; dos Santos-Neto, P.C.; Nguyen, T.H.; Crénéguy, A.; Brusselle, L.; Anegón, I.; et al. Efficient Generation of Myostatin Knock-Out Sheep Using CRISPR/Cas9 Technology and Microinjection into Zygotes. PLoS ONE 2015, 10, e0136690. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Y.; Zhou, J.; Yu, H.; Kou, Q.; Lei, A.; Zhao, X.; Yan, H.; Cai, B.; Shen, Q.; et al. Multiplex gene editing via CRISPR/Cas9 exhibits desirable muscle hypertrophy without detectable off-target effects in sheep. Sci. Rep. 2016, 6, 32271. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.; Niu, Y.; Li, Y.; Zhou, S.; Li, C.; Ma, B.; Kou, Q.; Petersen, B.; Sonstegard, T.; et al. Low incidence of SNVs and indels in trio genomes of Cas9-mediated multiplex edited sheep. BMC Genom. 2018, 19, 397. [Google Scholar] [CrossRef]
- Zhou, S.; Cai, B.; He, C.; Wang, Y.; Ding, Q.; Liu, J.; Liu, Y.; Ding, Y.; Zhao, X.; Li, G.; et al. Programmable Base Editing of the Sheep Genome Revealed No Genome-Wide Off-Target Mutations. Front. Genet. 2019, 10, 215. [Google Scholar] [CrossRef] [PubMed]
- Stern, C. The chick model system: A distinguished past and a great future. Int. J. Dev. Biol. 2018, 62, 1–4. [Google Scholar] [CrossRef]
- Park, T.S.; Lee, H.J.; Kim, K.H.; Kim, J.-S.; Han, J.Y. Targeted gene knockout in chickens mediated by TALENs. Proc. Natl. Acad. Sci. USA 2014, 111, 12716–12721. [Google Scholar] [CrossRef] [PubMed]
- Park, T.S.; Han, J.Y. piggyBac transposition into primordial germ cells is an efficient tool for transgenesis in chickens. Proc. Natl. Acad. Sci. USA 2012, 109, 9337–9341. [Google Scholar] [CrossRef] [PubMed]
- Koslová, A.; Kučerová, D.; Reinišová, M.; Geryk, J.; Trefil, P.; Hejnar, J. Genetic Resistance to Avian Leukosis Viruses Induced by CRISPR/Cas9 Editing of Specific Receptor Genes in Chicken Cells. Viruses 2018, 10, 605. [Google Scholar] [CrossRef]
- Kim, G.-D.; Lee, J.H.; Song, S.; Kim, S.W.; Han, J.S.; Shin, S.P.; Park, B.-C.; Park, T.S. Generation of myostatin-knockout chickens mediated by D10A-Cas9 nickase. FASEB J. 2020, 34, 5688–5696. [Google Scholar] [CrossRef]
- Lee, J.; Kim, D.-H.; Lee, K. Muscle Hyperplasia in Japanese Quail by Single Amino Acid Deletion in MSTN Propeptide. Int. J. Mol. Sci. 2020, 21, 1504. [Google Scholar] [CrossRef]
- Lee, J.; Ma, J.; Lee, K. Direct delivery of adenoviral CRISPR/Cas9 vector into the blastoderm for generation of targeted gene knockout in quail. Proc. Natl. Acad. Sci. USA 2019, 116, 13288–13292. [Google Scholar] [CrossRef]
- Lee, J.; Kim, D.-H.; Karolak, M.C.; Shin, S.; Lee, K. Generation of genome-edited chicken and duck lines by adenovirus-mediated in vivo genome editing. Proc. Natl. Acad. Sci. USA 2022, 119, e2214344119. [Google Scholar] [CrossRef]
- Park, T.S.; Park, J.; Lee, J.H.; Park, J.-W.; Park, B.-C. Disruption of G0/G1 switch gene 2 ( G0S2 ) reduced abdominal fat deposition and altered fatty acid composition in chicken. FASEB J. 2018, 33, 1188–1198. [Google Scholar] [CrossRef]
- Park, T.S. —Invited Review—Gene-editing techniques and their applications in livestock and beyond. Anim. Biosci. 2023, 36, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.R. Gene Therapy: The Potential Applicability of Gene Transfer Technology to the Human Germline. Int. J. Med. Sci. 2004, 1, 76–91. [Google Scholar] [CrossRef][Green Version]
- Rexroad, C.; Vallet, J.; Matukumalli, L.K.; Reecy, J.; Bickhart, D.; Blackburn, H.; Boggess, M.; Cheng, H.; Clutter, A.; Cockett, N.; et al. Genome to Phenome: Improving Animal Health, Production, and Well-Being—A New USDA Blueprint for Animal Genome Research 2018–2027. Front. Genet. 2019, 10, 327. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, G.; Hao, Z.; Zhang, G.; Qing, Y.; Liu, S.; Qing, L.; Pan, W.; Chen, L.; Liu, G.; et al. Generation of biallelic knock-out sheep via gene-editing and somatic cell nuclear transfer. Sci. Rep. 2016, 6, srep33675. [Google Scholar] [CrossRef] [PubMed]
- Samiec, M.; Skrzyszowska, M. Transgenic mammalian species, generated by somatic cell cloning, in biomedicine, biopharmaceutical industry and human nutrition/dietetics—Recent achievements. Pol. J. Vet. Sci. 2011, 14, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Preisinger, D.; Winogrodzki, T.; Klinger, B.; Schnieke, A.; Rieblinger, B. Genome Editing in Pigs. Methods Mol. Biol. 2023, 2631, 393–417. [Google Scholar] [CrossRef] [PubMed]
- Skrzyszowska, M.; Samiec, M. Generating Cloned Goats by Somatic Cell Nuclear Transfer—Molecular Determinants and Application to Transgenics and Biomedicine. Int. J. Mol. Sci. 2021, 22, 7490. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Proudfoot, C.; Lillico, S.G.; Whitelaw, C.B.A. Gene targeting, genome editing: From Dolly to editors. Transgenic Res. 2016, 25, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Wiater, J.; Samiec, M.; Wartalski, K.; Smorąg, Z.; Jura, J.; Słomski, R.; Skrzyszowska, M.; Romek, M. Characterization of Mono- and Bi-Transgenic Pig-Derived Epidermal Keratinocytes Expressing Human FUT2 and GLA Genes—In Vitro Studies. Int. J. Mol. Sci. 2021, 22, 9683. [Google Scholar] [CrossRef]
- Skrzyszowska, M.; Smorąg, Z.; Słomski, R.; Kątska-Książkiewicz, L.; Kalak, R.; Michalak, E.; Wielgus, K.; Lehmann, J.; Lipiński, D.; Szalata, M.; et al. Generation of Transgenic Rabbits by the Novel Technique of Chimeric Somatic Cell Cloning. Biol. Reprod. 2006, 74, 1114–1120. [Google Scholar] [CrossRef]
- Wu, H.; Zhou, W.; Liu, H.; Cui, X.; Ma, W.; Wu, H.; Li, G.; Wang, L.; Zhang, J.; Zhang, X.; et al. Whole-genome methylation analysis reveals epigenetic variation between wild-type and nontransgenic cloned, ASMT transgenic cloned dairy goats generated by the somatic cell nuclear transfer. J. Anim. Sci. Biotechnol. 2022, 13, 145. [Google Scholar] [CrossRef]
- Gordon, J.W.; Scangos, G.A.; Plotkin, D.J.; A Barbosa, J.; Ruddle, F.H. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc. Natl. Acad. Sci. USA 1980, 77, 7380–7384. [Google Scholar] [CrossRef]
- Isola, L.M.; Gordon, J.W. Transgenic animals: A new era in developmental biology and medicine. Biotechnology 1991, 16, 3–20. [Google Scholar] [PubMed]
- Palmiter, R.D.; Brinster, R.L.; Hammer, R.E.; Trumbauer, M.E.; Rosenfeld, M.G.; Birnberg, N.C.; Evans, R.M. Dramatic growth of mice that develop from eggs microinjected with metallothionein–growth hormone fusion genes. Nature 1982, 300, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Pursel, V.; Rexroad, C.; Bolt, D.; Miller, K.; Wall, R.; Hammer, R.; Pinkert, C.; Palmiter, R.; Brinster, R. Progress on gene transfer in farm animals. Vet. Immunol. Immunopathol. 1987, 17, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Eyestone, W. Challenges and progress in the production of transgenic cattle. Reprod. Fertil. Dev. 1994, 6, 647–652. [Google Scholar] [CrossRef]
- Su, F.; Wang, Y.; Liu, G.; Ru, K.; Liu, X.; Yu, Y.; Liu, J.; Wu, Y.; Quan, F.; Guo, Z.; et al. Generation of transgenic cattle expressing human β-defensin 3 as an approach to reducing susceptibility to Mycobacterium bovis infection. FEBS J. 2016, 283, 776–790. [Google Scholar] [CrossRef][Green Version]
- Hofmann, A.; Kessler, B.; Ewerling, S.; Kabermann, A.; Brem, G.; Wolf, E.; Pfeifer, A. Epigenetic Regulation of Lentiviral Transgene Vectors in a Large Animal Model. Mol. Ther. 2006, 13, 59–66. [Google Scholar] [CrossRef]
- Van Eenennaam, A. The contribution of transgenic and genome-edited animals to agricultural and industrial applications. Rev. Sci. et Tech. de l’OIE 2018, 37, 97–112. [Google Scholar] [CrossRef]
- Mashiko, D.; Fujihara, Y.; Satouh, Y.; Miyata, H.; Isotani, A.; Ikawa, M. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci. Rep. 2013, 3, 3355. [Google Scholar] [CrossRef]
- Li, D.; Qiu, Z.; Shao, Y.; Chen, Y.; Guan, Y.; Liu, M.; Li, Y.; Gao, N.; Wang, L.; Lu, X.; et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 681–683. [Google Scholar] [CrossRef]
- Niu, Y.Y.; Shen, B.; Cui, Y.Q.; Chen, Y.C.; Wang, J.Y.; Wang, L.; Kang, Y.; Zhao, X.Y.; Si, W.; Li, W.; et al. Generation of Gene-Modified Cynomolgus Monkey via Cas9/RNA-Mediated Gene Targeting in One-Cell Embryos. Cell 2014, 156, 836–843. [Google Scholar] [CrossRef]
- Hammer, R.E.; Pursel, V.G.; Rexroad, C.E.; Wall, R.J.; Bolt, D.J.; Ebert, K.M.; Palmiter, R.D.; Brinster, R.L. Production of transgenic rabbits, sheep and pigs by microinjection. Nature 1985, 315, 680–683. [Google Scholar] [CrossRef] [PubMed]
- Houdebine, L.-M. The methods to generate transgenic animals and to control transgene expression. J. Biotechnol. 2002, 98, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Chrenek, P.; Makarevich, A.V.; Pivko, J.; Bulla, J. Transgenic farm animal production and application. Slovak. J. Anim. Sci. 2010, 43, 45–49. [Google Scholar]
- Horii, T.; Arai, Y.; Yamazaki, M.; Morita, S.; Kimura, M.; Itoh, M.; Abe, Y.; Hatada, I. Validation of microinjection methods for generating knockout mice by CRISPR/Cas-mediated genome engineering. Sci. Rep. 2014, 4, 4513. [Google Scholar] [CrossRef]
- Serov, O.L. Transgenic animals: Fundamental and applied aspects. Russ. J. Genet. Appl. Res. 2014, 4, 200–207. [Google Scholar] [CrossRef]
- Wall, R.J. Pronuclear microinjection. Cloning Stem Cells 2001, 3, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Wolf, E.; Schernthaner, W.; Zakhartchenko, V.; Prelle, K.; Stojkovic, M.; Brem, G. Transgenic Technology in Farm Animals—Progress and Perspectives. Exp. Physiol. 2000, 85, 615–625. [Google Scholar] [CrossRef]
- Maga, E.A.; Sargent, R.G.; Zeng, H.; Pati, S.; Zarling, D.A.; Oppenheim, S.M.; Collette, N.M.; Moyer, A.L.; Conrad-Brink, J.S.; Rowe, J.D.; et al. Increased efficiency of transgenic livestock production. Transgenic Res. 2003, 12, 485–496. [Google Scholar] [CrossRef]
- Chrenek, P.; Vasicek, D.; Makarevich, A.V.; Jurcik, R.; Suvegova, K.; Parkanyi, V.; Bauer, M.; Rafay, J.; Batorova, A.; Paleyanda, R.K. Increased transgene integration efficiency upon microinjection of DNA into both pronuclei of rabbit embryos. Transgenic Res. 2005, 14, 417–428. [Google Scholar] [CrossRef]
- Sumiyama, K.; Kawakami, K.; Yagita, K. A simple and highly efficient transgenesis method in mice with the Tol2 transposon system and cytoplasmic microinjection. Genomics 2010, 95, 306–311. [Google Scholar] [CrossRef]
- Wang, Y.; Du, Y.; Shen, B.; Zhou, X.; Li, J.; Liu, Y.; Wang, J.; Zhou, J.; Hu, B.; Kang, N.; et al. Efficient generation of gene-modified pigs via injection of zygote with Cas9/sgRNA. Sci. Rep. 2015, 5, 8256. [Google Scholar] [CrossRef] [PubMed]
- Tanihara, F.; Hirata, M.; Nguyen, N.T.; LE, Q.A.; Hirano, T.; Otoi, T. Effects of concentration of CRISPR/Cas9 components on genetic mosaicism in cytoplasmic microinjected porcine embryos. J. Reprod. Dev. 2019, 65, 209–214. [Google Scholar] [CrossRef]
- Su, X.; Chen, W.; Cai, Q.; Liang, P.; Chen, Y.; Cong, P.; Huang, J. Production of non-mosaic genome edited porcine embryos by injection of CRISPR/Cas9 into germinal vesicle oocytes. J. Genet. Genom. 2019, 46, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Hai, T.; Teng, F.; Guo, R.; Li, W.; Zhou, Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014, 24, 372–375. [Google Scholar] [CrossRef]
- Whitworth, K.M.; Lee, K.; Benne, J.A.; Beaton, B.P.; Spate, L.D.; Murphy, S.L.; Samuel, M.S.; Mao, J.; O’Gorman, C.; Walters, E.M.; et al. Use of the CRISPR/Cas9 System to Produce Genetically Engineered Pigs from In Vitro-Derived Oocytes and Embryos1. Biol. Reprod. 2014, 91, 78. [Google Scholar] [CrossRef]
- Chan, A.W.S.; Homan, E.J.; Ballou, L.U.; Burns, J.C.; Bremel, R.D. Transgenic cattle produced by reverse-transcribed gene transfer in oocytes. Proc. Natl. Acad. Sci. USA 1998, 95, 14028–14033. [Google Scholar] [CrossRef]
- Palmiter, R.D.; Sandgren, E.P.; Avarbock, M.R.; Allen, D.D.; Brinster, R.L. Heterologous introns can enhance expression of transgenes in mice. Proc. Natl. Acad. Sci. USA 1991, 88, 478–482. [Google Scholar] [CrossRef]
- Haskell, R.E.; Bowen, R.A. Efficient production of transgenic cattle by retroviral infection of early embryos. Mol. Reprod. Dev. 1995, 40, 386–390. [Google Scholar] [CrossRef]
- Hofmann, A.; Kessler, B.; Ewerling, S.; Weppert, M.; Vogg, B.; Ludwig, H.; Stojkovic, M.; Boelhauve, M.; Brem, G.; Wolf, E.; et al. Efficient transgenesis in farm animals by lentiviral vectors. EMBO Rep. 2003, 4, 1054–1058. [Google Scholar] [CrossRef]
- Whitelaw, C.A.; Radcliffe, P.A.; Ritchie, W.A.; Carlisle, A.; Ellard, F.M.; Pena, R.N.; Rowe, J.; Clark, A.; King, T.J.; A Mitrophanous, K. Efficient generation of transgenic pigs using equine infectious anaemia virus (EIAV) derived vector. FEBS Lett. 2004, 571, 233–236. [Google Scholar] [CrossRef] [PubMed]
- McGrew, M.J.; Sherman, A.; Ellard, F.M.; Lillico, S.G.; Gilhooley, H.J.; Kingsman, A.J.; Mitrophanous, K.A.; Sang, H. Efficient production of germline transgenic chickens using lentiviral vectors. EMBO Rep. 2004, 5, 728–733. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Mashimo, T. Simple Genome Editing of Rodent Intact Embryos by Electroporation. PLoS ONE 2015, 10, e0142755. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T. Reproductive technologies for the generation and maintenance of valuable animal strains. J. Reprod. Dev. 2018, 64, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Tanihara, F.; Hirata, M.; Nguyen, N.T.; Le, Q.A.; Wittayarat, M.; Fahrudin, M.; Hirano, T.; Otoi, T. Generation of CD163-edited pig via electroporation of the CRISPR/Cas9 system into porcine in vitro-fertilized zygotes. Anim. Biotechnol. 2019, 32, 147–154. [Google Scholar] [CrossRef]
- Lin, J.C.; Van Eenennaam, A.L. Electroporation-Mediated Genome Editing of Livestock Zygotes. Front. Genet. 2021, 12, 648482. [Google Scholar] [CrossRef]
- Modzelewski, A.J.; Chen, S.; Willis, B.J.; Lloyd, K.C.K.; Wood, J.A.; He, L. Efficient mouse genome engineering by CRISPR-EZ technology. Nat. Protoc. 2018, 13, 1253–1274. [Google Scholar] [CrossRef]
- Teixeira, M.; Py, B.F.; Bosc, C.; Laubreton, D.; Moutin, M.-J.; Marvel, J.; Flamant, F.; Markossian, S. Electroporation of mice zygotes with dual guide RNA/Cas9 complexes for simple and efficient cloning-free genome editing. Sci. Rep. 2018, 8, 474. [Google Scholar] [CrossRef]
- Chen, S.; Lee, B.; Lee, A.Y.-F.; Modzelewski, A.J.; He, L. Highly Efficient Mouse Genome Editing by CRISPR Ribonucleoprotein Electroporation of Zygotes. J. Biol. Chem. 2016, 291, 14457–14467. [Google Scholar] [CrossRef]
- Peng, H.; Wu, Y.; Zhang, Y. Efficient Delivery of DNA and Morpholinos into Mouse Preimplantation Embryos by Electroporation. PLoS ONE 2012, 7, e43748. [Google Scholar] [CrossRef]
- Tröder, S.E.; Ebert, L.K.; Butt, L.; Assenmacher, S.; Schermer, B.; Zevnik, B. An optimized electroporation approach for efficient CRISPR/Cas9 genome editing in murine zygotes. PLoS ONE 2018, 13, e0196891. [Google Scholar] [CrossRef]
- Nishio, K.; Tanihara, F.; Nguyen, T.-V.; Kunihara, T.; Nii, M.; Hirata, M.; Takemoto, T.; Otoi, T. Effects of voltage strength during electroporation on the development and quality of in vitro-produced porcine embryos. Reprod. Domest. Anim. 2018, 53, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Tanihara, F.; Hirata, M.; Nguyen, N.T.; Le, Q.A.; Hirano, T.; Takemoto, T.; Nakai, M.; Fuchimoto, D.; Otoi, T. Generation of PDX -1 mutant porcine blastocysts by introducing CRISPR /Cas9-system into porcine zygotes via electroporation. Anim. Sci. J. 2018, 90, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T. Genome Editing in Mouse and Rat by Electroporation. Methods Mol. Biol. 2017, 1630, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Takemoto, T. Electroporation enables the efficient mRNA delivery into the mouse zygotes and facilitates CRISPR/Cas9-based genome editing. Sci. Rep. 2015, 5, 11315. [Google Scholar] [CrossRef]
- Tanihara, F.; Hirata, M.; Nguyen, N.T.; Le, Q.A.; Hirano, T.; Takemoto, T.; Nakai, M.; Fuchimoto, D.-I.; Otoi, T. Generation of a TP53-modified porcine cancer model by CRISPR/Cas9-mediated gene modification in porcine zygotes via electroporation. PLoS ONE 2018, 13, e0206360. [Google Scholar] [CrossRef]
- Tanihara, F.; Takemoto, T.; Kitagawa, E.; Rao, S.; Do, L.T.K.; Onishi, A.; Yamashita, Y.; Kosugi, C.; Suzuki, H.; Sembon, S.; et al. Somatic cell reprogramming-free generation of genetically modified pigs. Sci. Adv. 2016, 2, e1600803. [Google Scholar] [CrossRef]
- Miao, D.; Giassetti, M.I.; Ciccarelli, M.; Lopez-Biladeau, B.; Oatley, J.M. Simplified pipelines for genetic engineering of mammalian embryos by CRISPR-Cas9 electroporation†. Biol. Reprod. 2019, 101, 177–187. [Google Scholar] [CrossRef]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef]
- Tian, X.C.; Kubota, C.; Enright, B.; Yang, X. Cloning animals by somatic cell nuclear transfer—Biological factors. Reprod. Biol. Endocrinol. 2003, 1, 98. [Google Scholar] [CrossRef]
- Willadsen, S.M. A method for culture of micromanipulated sheep embryos and its use to produce monozygotic twins. Nature 1979, 277, 298–300. [Google Scholar] [CrossRef]
- Willadsen, S.M. The development capacity of blastomeres from 4- and 8-cell sheep embryos. J. Embryol. Exp. Morphol. 1981, 65, 165–172. [Google Scholar]
- Willadsen, S.; Polge, C. Attempts to produce monozygotic quadruplets in cattle by blastomere separation. Vet. Rec. 1981, 108, 211–213. [Google Scholar] [CrossRef]
- McGrath, J.; Solter, D. Nuclear transplantation in mouse embryos. J. Exp. Zool. 1983, 228, 355–362. [Google Scholar] [CrossRef]
- Robl, J.M.; First, N.L. Manipulation of gametes and embryos in the pig. J. Reprod. Fertil. Suppl. 1985, 33, 101–114. [Google Scholar]
- Willadsen, S.M. Nuclear transplantation in sheep embryos. Nature 1986, 320, 63–65. [Google Scholar] [CrossRef] [PubMed]
- Willadsen, S.M. Cloning of sheep and cow embryos. Genome 1989, 31, 956–962. [Google Scholar] [CrossRef] [PubMed]
- Prather, R.S.; Barnes, F.L.; Sims, M.M.; Robl, J.M.; Eyestone, W.H.; First, N.L. Nuclear Transplantation in the Bovine Embryo: Assessment of Donor Nuclei and Recipient Oocyt14. Biol. Reprod. 1987, 37, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Stice, S.L.; Keefer, C.L. Multiple Generational Bovine Embryo Cloning. Biol. Reprod. 1993, 48, 715–719. [Google Scholar] [CrossRef]
- Collas, P.; Barnes, F.L. Nuclear transplantation by microinjection of inner cell mass and granulosa cell nuclei. Mol. Reprod. Dev. 1994, 38, 264–267. [Google Scholar] [CrossRef]
- Keefer, C.L.; Stice, S.L.; Matthews, D.L. Bovine Inner Cell Mass Cells as Donor Nuclei in the Production of Nuclear Transfer Embryos and Calves1. Biol. Reprod. 1994, 50, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Peura, T.T.; Lane, M.W.; Lewis, I.M.; Trounson, A.O. Development of bovine embryo-derived clones after increasing rounds of nuclear recycling. Mol. Reprod. Dev. 2001, 58, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Keefer, C.L. Artificial cloning of domestic animals. Proc. Natl. Acad. Sci. USA 2015, 112, 8874–8878. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.H.S.; McWhir, J.; Ritchie, W.A.; Wilmut, I. Sheep cloned by nuclear transfer from a cultured cell line. Nature 1996, 380, 64–66. [Google Scholar] [CrossRef] [PubMed]
- Schnieke, A.E.; Kind, A.J.; Ritchie, W.A.; Mycock, K.; Scott, A.R.; Ritchie, M.; Wilmut, I.; Colman, A.; Campbell, K.H.S. Human Factor IX Transgenic Sheep Produced by Transfer of Nuclei from Transfected Fetal Fibroblasts. Science 1997, 278, 2130–2133. [Google Scholar] [CrossRef]
- Cibelli, J.B.; Stice, S.L.; Golueke, P.J.; Kane, J.J.; Jerry, J.; Blackwell, C.; de León, F.A.P.; Robl, J.M. Cloned Transgenic Calves Produced from Nonquiescent Fetal Fibroblasts. Science 1998, 280, 1256–1258. [Google Scholar] [CrossRef]
- Baguisi, A.; Behboodi, E.; Melican, D.T.; Pollock, J.S.; Destrempes, M.M.; Cammuso, C.; Williams, J.L.; Nims, S.D.; Porter, C.A.; Midura, P.; et al. Production of goats by somatic cell nuclear transfer. Nat. Biotechnol. 1999, 17, 456–461. [Google Scholar] [CrossRef]
- Onishi, A.; Iwamoto, M.; Akita, T.; Mikawa, S.; Takeda, K.; Awata, T.; Hanada, H.; Perry, A.C.F. Pig Cloning by Microinjection of Fetal Fibroblast Nuclei. Science 2000, 289, 1188–1190. [Google Scholar] [CrossRef]
- Polejaeva, I.A.; Chen, S.-H.; Vaught, T.D.; Page, R.L.; Mullins, J.; Ball, S.; Dai, Y.; Boone, J.; Walker, S.; Ayares, D.L.; et al. Cloned pigs produced by nuclear transfer from adult somatic cells. Nature 2000, 407, 86–90. [Google Scholar] [CrossRef]
- Ruan, Z.; Zhao, X.; Li, Z.; Qin, X.; Shao, Q.; Ruan, Q.; Deng, Y.; Jiang, J.; Huang, B.; Lu, F.; et al. Effect of sex differences in donor foetal fibroblast on the early development and DNA methylation status of buffalo (Bubalus bubalis) nuclear transfer embryos. Reprod. Domest. Anim. 2018, 54, 11–22. [Google Scholar] [CrossRef]
- Li, Z.; Ruan, Z.; Zhao, X.; Qin, X.; Zhang, J.; Feng, Y.; Lu, J.; Shi, D.; Lu, F. RNAi-mediated knockdown of Xist improves development of the female buffalo (Bubalus bubalis) nuclear transfer embryos. Theriogenology 2022, 187, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Dixon-McDougall, T.; Brown, C. The making of a Barr body: The mosaic of factors that eXIST on the mammalian inactive X chromosome. Biochem. Cell Biol. 2016, 94, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Augui, S.; Nora, E.P.; Heard, E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 2011, 12, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Su, J.-M.; Yang, B.; Wang, Y.-S.; Li, Y.-Y.; Xiong, X.-R.; Wang, L.-J.; Guo, Z.-K.; Zhang, Y. Expression and methylation status of imprinted genes in placentas of deceased and live cloned transgenic calves. Theriogenology 2011, 75, 1346–1359. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Z.; Zhao, X.; Qin, X.; Luo, C.; Liu, X.; Deng, Y.; Zhu, P.; Li, Z.; Huang, B.; Shi, D.; et al. DNA methylation and expression of imprinted genes are associated with the viability of different sexual cloned buffaloes. Reprod. Domest. Anim. 2017, 53, 203–212. [Google Scholar] [CrossRef]
- Inoue, K.; Kohda, T.; Sugimoto, M.; Sado, T.; Ogonuki, N.; Matoba, S.; Shiura, H.; Ikeda, R.; Mochida, K.; Fujii, T.; et al. Impeding Xist Expression from the Active X Chromosome Improves Mouse Somatic Cell Nuclear Transfer. Science 2010, 330, 496–499. [Google Scholar] [CrossRef]
- Matoba, S.; Inoue, K.; Kohda, T.; Sugimoto, M.; Mizutani, E.; Ogonuki, N.; Nakamura, T.; Abe, K.; Nakano, T.; Ishino, F.; et al. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc. Natl. Acad. Sci. USA 2011, 108, 20621–20626. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Huang, Z.; Yuan, Y.; Shi, J.; Cai, G.; Liu, D.; Wu, Z.; Li, Z. Effects of RNAi-mediated knockdown of Xist on the developmental efficiency of cloned male porcine embryos. J. Reprod. Dev. 2016, 62, 591–597. [Google Scholar] [CrossRef]
- Janke, C.; Montagnac, G. Causes and Consequences of Microtubule Acetylation. Curr. Biol. 2017, 27, R1287–R1292. [Google Scholar] [CrossRef]
- Qu, P.; Zuo, Z.; Liu, Z.; Niu, Z.; Du, Y.; Ma, X.; Qiao, F.; Wang, M.; Zhang, Y.; Qing, S.; et al. Sperm-borne small RNAs regulate α-tubulin acetylation and epigenetic modification of early bovine somatic cell nuclear transfer embryos. Mol. Hum. Reprod. 2019, 25, 471–482. [Google Scholar] [CrossRef]
- Wang, M.; Du, Y.; Gao, S.; Wang, Z.; Qu, P.; Gao, Y.; Wang, J.; Liu, Z.; Zhang, J.; Zhang, Y.; et al. Sperm-borne miR-202 targets SEPT7 and regulates first cleavage of bovine embryos via cytoskeletal remodeling. Development 2021, 148, dev189670. [Google Scholar] [CrossRef]
- Gao, S.; Wang, Z.; Ma, J.; Wang, J.; Xing, X.; Shen, C.; Niu, Z.; Li, H.; Zhang, S.; Zhang, K.; et al. Effect of ACY-1215 on cytoskeletal remodeling and histone acetylation in bovine somatic cell nuclear transfer embryos. Theriogenology 2022, 183, 98–107. [Google Scholar] [CrossRef]
- Ogura, A.; Inoue, K.; Wakayama, T. Recent advancements in cloning by somatic cell nuclear transfer. Philos. Trans. R. Soc. B: Biol. Sci. 2013, 368, 20110329. [Google Scholar] [CrossRef]
- Loi, P.; Iuso, D.; Czernik, M.; Ogura, A. A New, Dynamic Era for Somatic Cell Nuclear Transfer? Trends Biotechnol. 2016, 34, 791–797. [Google Scholar] [CrossRef]
- Fulka, J.; Miyashita, N.; Nagai, T.; Ogura, A. Do cloned mammals skip a reprogramming step? Nat. Biotechnol. 2004, 22, 25–26. [Google Scholar] [CrossRef] [PubMed]
- Tamashiro, K.L.K.; Wakayama, T.; Akutsu, H.; Yamazaki, Y.; Lachey, J.L.; Wortman, M.D.; Seeley, R.J.; D’Alessio, D.A.; Woods, S.C.; Yanagimachi, R.; et al. Cloned mice have an obese phenotype not transmitted to their offspring. Nat. Med. 2002, 8, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Wakayama, S.; Kohda, T.; Obokata, H.; Tokoro, M.; Li, C.; Terashita, Y.; Mizutani, E.; Nguyen, V.T.; Kishigami, S.; Ishino, F.; et al. Successful Serial Recloning in the Mouse over Multiple Generations. Cell Stem Cell 2013, 12, 293–297. [Google Scholar] [CrossRef]
- Klymiuk, N.; Aigner, B.; Brem, G.; Wolf, E. Genetic modification of pigs as organ donors for xenotransplantation. Mol. Reprod. Dev. 2009, 77, 209–221. [Google Scholar] [CrossRef]
- Cooper, D.K.C.; Satyananda, V.; Ekser, B.; van der Windt, D.J.; Hara, H.; Ezzelarab, M.B.; Schuurman, H.-J. Progress in pig-to-non-human primate transplantation models (1998–2013): A comprehensive review of the literature. Xenotransplantation 2014, 21, 397–419. [Google Scholar] [CrossRef]
- Polejaeva, I.A.; Rutigliano, H.M.; Wells, K.D. Livestock in biomedical research: History, current status and future prospective. Reprod. Fertil. Dev. 2016, 28, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Vaught, T.D.; Boone, J.; Chen, S.-H.; Phelps, C.J.; Ball, S.; Monahan, J.A.; Jobst, P.M.; McCreath, K.J.; Lamborn, A.E.; et al. Targeted disruption of the α1,3-galactosyltransferase gene in cloned pigs. Nat. Biotechnol. 2002, 20, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.; Kolber-Simonds, D.; Park, K.-W.; Cheong, H.-T.; Greenstein, J.L.; Im, G.-S.; Samuel, M.; Bonk, A.; Rieke, A.; Day, B.N.; et al. Production of α-1,3-Galactosyltransferase Knockout Pigs by Nuclear Transfer Cloning. Science 2002, 295, 1089–1092. [Google Scholar] [CrossRef] [PubMed]
- Phelps, C.J.; Koike, C.; Vaught, T.D.; Boone, J.; Wells, K.D.; Chen, S.-H.; Ball, S.; Specht, S.M.; Polejaeva, I.A.; Monahan, J.A.; et al. Production of α1,3-Galactosyltransferase-Deficient Pigs. Science 2003, 299, 411–414. [Google Scholar] [CrossRef] [PubMed]
- McCreath, K.J.; Howcroft, J.; Campbell, K.H.S.; Colman, N.A.; Schnieke, A.E.; Kind, A.J. Production of gene-targeted sheep by nuclear transfer from cultured somatic cells. Nature 2000, 405, 1066–1069. [Google Scholar] [CrossRef] [PubMed]
- Kuroiwa, Y.; Kasinathan, P.; Matsushita, H.; Sathiyaselan, J.; Sullivan, E.J.; Kakitani, M.; Tomizuka, K.; Ishida, I.; Robl, J.M. Sequential targeting of the genes encoding immunoglobulin-μ and prion protein in cattle. Nat. Genet. 2004, 36, 775–780. [Google Scholar] [CrossRef][Green Version]
- Kues, W. The contribution of farm animals to human health. Trends Biotechnol. 2004, 22, 286–294. [Google Scholar] [CrossRef]
- Brackett, B.G.; Baranska, W.; Sawicki, W.; Koprowski, H. Uptake of Heterologous Genome by Mammalian Spermatozoa and Its Transfer to Ova through Fertilization. Proc. Natl. Acad. Sci. USA 1971, 68, 353–357. [Google Scholar] [CrossRef]
- Lavitrano, M.; Camaioni, A.; Fazio, V.M.; Dolci, S.; Farace, M.G.; Spadafora, C. Sperm cells as vectors for introducing foreign DNA into eggs: Genetic transformation of mice. Cell 1989, 57, 717–723. [Google Scholar] [CrossRef]
- Maione, B.; Lavitrano, M.; Spadafora, C.; Kiessling, A.A. Sperm-mediated gene transfer in mice. Mol. Reprod. Dev. 1998, 50, 406–409. [Google Scholar] [CrossRef]
- Lavitrano, M.; Forni, M.; Varzi, V.; Pucci, L.; Bacci, M.; Di Stefano, C.; Fioretti, D.; Zoraqi, G.; Moioli, B.; Rossi, M.; et al. Sperm-mediated gene transfer: Production of pigs transgenic for a human regulator of complement activation. Transplant. Proc. 1997, 29, 3508–3509. [Google Scholar] [CrossRef]
- Lavitrano, M.; Bacci, M.L.; Forni, M.; Lazzereschi, D.; Di Stefano, C.; Fioretti, D.; Giancotti, P.; Marfé, G.; Pucci, L.; Renzi, L.; et al. Efficient production by sperm-mediated gene transfer of human decay accelerating factor (hDAF) transgenic pigs for xenotransplantation. Proc. Natl. Acad. Sci. USA 2002, 99, 14230–14235. [Google Scholar] [CrossRef]
- Lavitrano, M.; Forni, M.; Bacci, M.L.; Di Stefano, C.; Varzi, V.; Wang, H.; Seren, E. Sperm mediated gene transfer in pig: Selection of donor boars and optimization of DNA uptake. Mol. Reprod. Dev. 2003, 64, 284–291. [Google Scholar] [CrossRef]
- Schellander, K.; Peli, J.; Schmoll, F.; Brem, G. Artificial insemination in cattle with DNA-treated sperm. Anim. Biotechnol. 1995, 6, 41–50. [Google Scholar] [CrossRef]
- Sperandio, S.; Lulli, V.; Bacci, M.; Forni, M.; Maione, B.; Spadafora, C.; Lavitrano, M. Sperm-mediated DNA transfer in bovine and swine species. Anim. Biotechnol. 1996, 7, 59–77. [Google Scholar] [CrossRef]
- Lavitrano, M.; Busnelli, M.; Cerrito, M.G.; Giovannoni, R.; Manzini, S.; Vargiolu, A. Sperm-mediated gene transfer. Reprod. Fertil. Dev. 2006, 18, 19–23. [Google Scholar] [CrossRef]
- Niemann, H.; Kues, W.A. Application of transgenesis in livestock for agriculture and biomedicine. Anim. Reprod. Sci. 2003, 79, 291–317. [Google Scholar] [CrossRef]
- Smith, K.R. Sperm-mediated gene transfer: Concepts and controversies. In Sperm-Mediated Gene Transfer: Concepts and Controversies; Bentham Science Publishers: Sharjah, United Arab Emirates, 2012. [Google Scholar] [CrossRef]
- Pereyra-Bonnet, F.; Fernández-Martín, R.; Olivera, R.; Jarazo, J.; Vichera, G.; Gibbons, A.; Salamone, D. A unique method to produce transgenic embryos in ovine, porcine, feline, bovine and equine species. Reprod. Fertil. Dev. 2008, 20, 741–749. [Google Scholar] [CrossRef]
- Shadanloo, F.; Najafi, M.H.; Hosseini, S.M.; Hajian, M.; Forouzanfar, M.; Ghaedi, K.; Abedi, P.; Ostadhosseini, S.; Hosseini, L.; Eskandari-Nasab, M.-P.; et al. Sperm status and DNA dose play key roles in sperm/ICSI-mediated gene transfer in caprine. Mol. Reprod. Dev. 2010, 77, 868–875. [Google Scholar] [CrossRef]
- Pereyra-Bonnet, F.; Gibbons, A.; Cueto, M.; Sipowicz, P.; Fernández-Martín, R.; Salamone, D. Efficiency of Sperm-Mediated Gene Transfer in the Ovine by Laparoscopic Insemination, In Vitro Fertilization and ICSI. J. Reprod. Dev. 2011, 57, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Pramod, R.K.; Kumar, R.; Mitra, A. Transgenic expression of green fluorescent protein in caprine embryos produced through electroporation-aided sperm-mediated gene transfer. Gene 2016, 576, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wei, H.; Wang, Y.; Wang, L.; Yu, M.; Fan, J.; Zheng, S.; Zhao, C. Production of Transgenic Goats by Sperm-mediated Exogenous DNA Transfer Method. Asian-Australas. J. Anim. Sci. 2009, 23, 33–40. [Google Scholar] [CrossRef]
- Rieth, A.; Pothier, F.; Sirard, M. Electroporation of bovine spermatozoa to carry DNA containing highly repetitive sequences into oocytes and detection of homologous recombination events. Mol. Reprod. Dev. 2000, 57, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Ebert, K.M.; Selgrath, J.P.; DiTullio, P.; Denman, J.; Smith, T.E.; Memon, M.A.; Schindler, J.E.; Monastersky, G.M.; Vitale, J.A.; Gordon, K. Transgenic Production of a Variant of Human Tissue-Type Plasminogen Activator in Goat Milk: Generation of Transgenic Goats and Analysis of Expression. Nat. Biotechnol. 1991, 9, 835–838. [Google Scholar] [CrossRef] [PubMed]
- Meade, H.; Echelard, Y.; Ziomek, C.; Young, M.; Harvey, M.; Cole, E.; Groet, S.; Smith, T.; Curling, J. Expression of Recombinant Proteins in the Milk of Transgenic Animals. Gene Expr. Syst. 1999, 399–427. [Google Scholar] [CrossRef]
- Sánchez-Villalba, E.; Arias, M.E.; Loren, P.; Fuentes, F.; Pereyra-Bonnet, F.; Salamone, D.; Felmer, R. Improved expression of green fluorescent protein in cattle embryos produced by ICSI-mediated gene transfer with spermatozoa treated with streptolysin-O. Anim. Reprod. Sci. 2018, 196, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Liang, S. Progress in gene transfer by germ cells in mammals. J. Genet. Genom. 2008, 35, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Felgner, P.L.; Gadek, T.R.; Holm, M.; Roman, R.; Chan, H.W.; Wenz, M.; Northrop, J.P.; Ringold, G.M.; Danielsen, M. Lipofection: A highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA 1987, 84, 7413–7417. [Google Scholar] [CrossRef]
- Rubessa, M.; Lotti, S.N.; Kandel, M.E.; Popescu, G.; Wheeler, M.B. SLIM microscopy allows for visualization of DNA-containing liposomes designed for sperm-mediated gene transfer in cattle. Mol. Biol. Rep. 2018, 46, 695–703. [Google Scholar] [CrossRef]
- Chang, K.; Qian, J.; Jiang, M.; Liu, Y.-H.; Wu, M.-C.; Chen, C.-D.; Lai, C.-K.; Lo, H.-L.; Hsiao, C.-T.; Brown, L.; et al. Effective generation of transgenic pigs and mice by linker based sperm-mediated gene transfer. BMC Biotechnol. 2002, 2, 5. [Google Scholar] [CrossRef]
- Van Eenennaam, A.L.; Silva, F.D.F.; Trott, J.F.; Zilberman, D. Genetic Engineering of Livestock: The Opportunity Cost of Regulatory Delay. Annu. Rev. Anim. Biosci. 2021, 9, 453–478. [Google Scholar] [CrossRef]
- Du, S.J.; Gong, Z.; Fletcher, G.L.; Shears, M.A.; King, M.J.; Idler, D.R.; Hew, C.L. Growth Enhancement in Transgenic Atlantic Salmon by the Use of an “All Fish” Chimeric Growth Hormone Gene Construct. Nat. Biotechnol. 1992, 10, 176–181. [Google Scholar] [CrossRef]
- FDA. Approves First-of-Its-Kind Intentional Genomic Alteration in Line of Domestic Pigs for Both Human Food, Potential Therapeutic Uses|FDA. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-its-kind-intentional-genomic-alteration-line-domestic-pigs-both-human-food (accessed on 26 January 2023).
- Huai, G.; Qi, P.; Yang, H.; Wang, Y. Characteristics of α-Gal epitope, anti-Gal antibody, α1,3 galactosyltransferase and its clinical exploitation (Review). Int. J. Mol. Med. 2015, 37, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Hilger, C.; Fischer, J.; Wölbing, F.; Biedermann, T. Role and Mechanism of Galactose-Alpha-1,3-Galactose in the Elicitation of Delayed Anaphylactic Reactions to Red Meat. Curr. Allergy Asthma Rep. 2019, 19, 3. [Google Scholar] [CrossRef] [PubMed]
- FDA. CVM Risk Assessment Summary-V-006378 PRLR-SLICK Cattle Executive Summary. Available online: https://www.fda.gov/media/155706/download (accessed on 26 January 2023).
- Littlejohn, M.D.; Henty, K.M.; Tiplady, K.; Johnson, T.; Harland, C.; Lopdell, T.; Sherlock, R.G.; Li, W.; Lukefahr, S.D.; Shanks, B.C.; et al. Functionally reciprocal mutations of the prolactin signalling pathway define hairy and slick cattle. Nat. Commun. 2014, 5, 5861. [Google Scholar] [CrossRef] [PubMed]
- Dikmen, S.; Khan, F.A.; Huson, H.J.; Sonstegard, T.S.; Moss, J.I.; Dahl, G.E.; Hansen, P.J. The SLICK hair locus derived from Senepol cattle confers thermotolerance to intensively managed lactating Holstein cows. J. Dairy Sci. 2014, 97, 5508–5520. [Google Scholar] [CrossRef]
- Whitworth, K.M.; Rowland, R.R.; Ewen, C.L.; Trible, B.R.; Kerrigan, M.A.; Cino-Ozuna, A.G.; Samuel, M.S.; Lightner, J.E.; McLaren, D.G.; Mileham, A.J.; et al. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 2016, 34, 20–22. [Google Scholar] [CrossRef]
- Kling, J. First US approval for a transgenic animal drug. Nat. Biotechnol. 2009, 27, 302–303. [Google Scholar] [CrossRef]
- van Veen, H.A.; Koiter, J.; Vogelezang, C.J.; van Wessel, N.; van Dam, T.; Velterop, I.; van Houdt, K.; Kupers, L.; Horbach, D.; Salaheddine, M.; et al. Characterization of recombinant human C1 inhibitor secreted in milk of transgenic rabbits. J. Biotechnol. 2012, 162, 319–326. [Google Scholar] [CrossRef]
- Rabbit milk Ruconest for hereditary angioedema. Nat. Biotechnol. 2014, 32, 849. [CrossRef]
- Shirley, M. Sebelipase Alfa: First Global Approval. Drugs 2015, 75, 1935–1940. [Google Scholar] [CrossRef]
- Becker, R. US government approves transgenic chicken. Nature 2015, 1038, 18985. [Google Scholar] [CrossRef]
- Gong, Z.; Wan, H.; Tay, T.L.; Wang, H.; Chen, M.; Yan, T. Development of transgenic fish for ornamental and bioreactor by strong expression of fluorescent proteins in the skeletal muscle. Biochem. Biophys. Res. Commun. 2003, 308, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Larson, M.A. Blastocyst Microinjection with Embryonic Stem Cells. Methods Mol. Biol. 2019, 2066, 83–88. [Google Scholar] [CrossRef]
- Gardner, R.L. The Relationship Between Cell Lineage and Differentiation in the Early Mouse Embryo. Results Probl. Cell Differ. 1978, 9, 205–241. [Google Scholar] [CrossRef]
- Bai, Q.; Desprat, R.; Klein, B.; Lemaitre, J.-M.; De Vos, J. Embryonic stem cells or induced pluripotent stem cells? A DNA integrity perspective. Curr. Gene Ther. 2013, 13, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Rashid, T.; Kobayashi, T.; Nakauchi, H. Revisiting the Flight of Icarus: Making Human Organs from PSCs with Large Animal Chimeras. Cell Stem Cell 2014, 15, 406–409. [Google Scholar] [CrossRef]
- Gardner, R.L. Clonal analysis of early mammalian development. Philos. Trans. R. Soc. London. B, Biol. Sci. 1985, 312, 163–178. [Google Scholar] [CrossRef]
- Beddington, R.S.P.; Robertson, E.J. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development 1989, 105, 733–737. [Google Scholar] [CrossRef]
- George, S.H.L.; Gertsenstein, M.; Vintersten, K.; Korets-Smith, E.; Murphy, J.; Stevens, M.E.; Haigh, J.J.; Nagy, A. Developmental and adult phenotyping directly from mutant embryonic stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 4455–4460. [Google Scholar] [CrossRef]
- MacKay, G.E.; West, J.D. Fate of tetraploid cells in 4n↔2n chimeric mouse blastocysts. Mech. Dev. 2005, 122, 1266–1281. [Google Scholar] [CrossRef]
- Eakin, G.S.; Hadjantonakis, A.-K.; Papaioannou, V.E.; Behringer, R.R. Developmental potential and behavior of tetraploid cells in the mouse embryo. Dev. Biol. 2005, 288, 150–159. [Google Scholar] [CrossRef]
- Misra, R.P.; Bronson, S.K.; Xiao, Q.; Garrison, W.; Li, J.; Zhao, R.; Duncan, S.A. Generation of single-copy transgenic mouse embryos directly from ES cells by tetraploid embryo complementation. BMC Biotechnol. 2001, 1, 12. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Talluri, T.R.; Selokar, N.L.; Hyder, I.; Kues, W.A. Perspectives of pluripotent stem cells in livestock. World J. Stem Cells 2021, 13, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shiue, Y.; Bertolini, L.; Medrano, J.; BonDurant, R.; Anderson, G. Establishment of pluripotent cell lines from porcine preimplantation embryos. Theriogenology 1999, 52, 195–212. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, D.; Hou, Y.; Jiao, L.; Zheng, X.; Wang, W.-H. Isolation and culture of embryonic stem cells from porcine blastocysts. Mol. Reprod. Dev. 2003, 65, 429–434. [Google Scholar] [CrossRef]
- First, N.; Sims, M.; Park, S.; Kent-First, M. Systems for production of calves from cultured bovine embryonic cells. Reprod. Fertil. Dev. 1994, 6, 553–562. [Google Scholar] [CrossRef]
- Mitalipova, M.; Beyhan, Z.; First, N.L.; Hou, D.-R.; Jin, Y.; Nie, X.-W.; Zhang, M.-L.; Ta, N.; Zhao, L.-H.; Yang, N.; et al. Pluripotency of Bovine Embryonic Cell Line Derived from Precompacting Embryos. Cloning 2001, 3, 59–67. [Google Scholar] [CrossRef]
- Yadav, P.S.; Kues, W.A.; Herrmann, D.; Carnwath, J.W.; Niemann, H. Bovine ICM derived cells express theOct4 ortholog. Mol. Reprod. Dev. 2005, 72, 182–190. [Google Scholar] [CrossRef]
- Zhu, S.-X.; Sun, Z.; Zhang, J.-P. Ovine (Ovis aries) blastula from an in vitro production system and isolation of primary embryonic stem cells. Zygote 2007, 15, 35–41. [Google Scholar] [CrossRef]
- Behboodi, E.; Bondareva, A.; Begin, I.; Rao, K.; Neveu, N.; Pierson, J.; Wylie, C.; Piero, F.; Huang, Y.; Zeng, W.; et al. Establishment of goat embryonic stem cells from in vivo produced blastocyst-stage embryos. Mol. Reprod. Dev. 2011, 78, 202–211. [Google Scholar] [CrossRef]
- De, A.K.; Malakar, D.; Akshey, Y.S.; Jena, M.K.; Dutta, R. Isolation and Characterization of Embryonic Stem Cell-Like Cells From in vitro Produced Goat (Capra hircus) Embryos. Anim. Biotechnol. 2011, 22, 181–196. [Google Scholar] [CrossRef]
- Saito, S.; Ugai, H.; Sawai, K.; Yamamoto, Y.; Minamihashi, A.; Kurosaka, K.; Kobayashi, Y.; Murata, T.; Obata, Y.; Yokoyama, K. Isolation of embryonic stem-like cells from equine blastocysts and their differentiation in vitro1. FEBS Lett. 2002, 531, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; George, A.; Kamble, N.M.; Singh, K.P.; Chauhan, M.S.; Singla, S.K.; Palta, P.; Sood, T.J.; Lagah, S.V.; Sharma, A.; et al. Optimization of Culture Conditions to Support Long-Term Self-Renewal of Buffalo (Bubalus bubalis) Embryonic Stem Cell-Like Cells. Cell. Reprogramming 2011, 13, 539–549. [Google Scholar] [CrossRef]
- Koyama, S.; Kimura, T.; Ogita, K.; Nakamura, H.; Tabata, C.; Ali, K.M.A.H.N.; Temma-Asano, K.; Shimoya, K.; Tsutsui, T.; Koyama, M.; et al. Simple and highly efficient method for transient in vivo gene transfer to mid-late pregnant mouse uterus. J. Reprod. Immunol. 2006, 70, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, G.; Gurumurthy, C.B.; Wada, K.; Miura, H.; Sato, M.; Ohtsuka, M. GONAD: Genome-editing via Oviductal Nucleic Acids Delivery system: A novel microinjection independent genome engineering method in mice. Sci. Rep. 2015, 5, 11406. [Google Scholar] [CrossRef]
- Ohtsuka, M.; Sato, M.; Miura, H.; Takabayashi, S.; Matsuyama, M.; Koyano, T.; Arifin, N.; Nakamura, S.; Wada, K.; Gurumurthy, C.B. i-GONAD: A robust method for in situ germline genome engineering using CRISPR nucleases. Genome Biol. 2018, 19, 25. [Google Scholar] [CrossRef] [PubMed]
- Takabayashi, S.; Aoshima, T.; Kabashima, K.; Aoto, K.; Ohtsuka, M.; Sato, M. i-GONAD (improved genome-editing via oviductal nucleic acids delivery), a convenient in vivo tool to produce genome-edited rats. Sci. Rep. 2018, 8, 12059. [Google Scholar] [CrossRef]
- Hirose, M.; Honda, A.; Fulka, H.; Tamura-Nakano, M.; Matoba, S.; Tomishima, T.; Mochida, K.; Hasegawa, A.; Nagashima, K.; Inoue, K.; et al. Acrosin is essential for sperm penetration through the zona pellucida in hamsters. Proc. Natl. Acad. Sci. USA 2020, 117, 2513–2518. [Google Scholar] [CrossRef]
- Tsukamoto, M.; Ochiya, T.; Yoshida, S.; Sugimura, T.; Terada, M. Gene transfer and expression in progeny after intravenous DNA injection into pregnant mice. Nat. Genet. 1995, 9, 243–248. [Google Scholar] [CrossRef]
- Nakamura, S.; Watanabe, S.; Ando, N.; Ishihara, M.; Sato, M. Transplacental Gene Delivery (TPGD) as a Noninvasive Tool for Fetal Gene Manipulation in Mice. Int. J. Mol. Sci. 2019, 20, 5926. [Google Scholar] [CrossRef]
- Nakamura, S.; Ishihara, M.; Ando, N.; Watanabe, S.; Sakurai, T.; Sato, M. Transplacental delivery of genome editing components causes mutations in embryonic cardiomyocytes of mid-gestational murine fetuses. IUBMB Life 2019, 71, 835–844. [Google Scholar] [CrossRef]
- Nakamura, S.; Ando, N.; Watanabe, S.; Akasaka, E.; Ishihara, M.; Sato, M. Hydrodynamics-Based Transplacental Delivery as a Useful Noninvasive Tool for Manipulating Fetal Genome. Cells 2020, 9, 1744. [Google Scholar] [CrossRef] [PubMed]
- Madsen, E.M.; Lindegaard, M.L.; Andersen, C.B.; Damm, P.; Nielsen, L.B. Human Placenta Secretes Apolipoprotein B-100-containing Lipoproteins. J. Biol. Chem. 2004, 279, 55271–55276. [Google Scholar] [CrossRef] [PubMed]
- Beckman, D.A.; Lloyd, J.B.; Brent, R.L. Investigations into mechanisms of amino acid supply to the rat embryo using whole-embryo culture. Int. J. Dev. Biol. 1997, 41, 315–318. [Google Scholar] [PubMed]
- Rugh, R. The Mouse: Its Reproduction and Development; Oxford University Press: Oxford, UK, 1990; Volume 438. [Google Scholar]
- Kikuchi, N.; Nakamura, S.; Ohtsuka, M.; Kimura, M.; Sato, M. Possible mechanism of gene transfer into early to mid-gestational mouse fetuses by tail vein injection. Gene Ther. 2002, 9, 1529–1541. [Google Scholar] [CrossRef]
- Yamashita, M.S.; Melo, E.O. Animal Transgenesis and Cloning: Combined Development and Future Perspectives. Methods Mol. Biol. 2023, 2647, 121–149. [Google Scholar] [CrossRef]
- Galli, C.; Lazzari, G. Current applications of SCNT in advanced breeding and genome editing in livestock. Reproduction 2021, 162, F23–F32. [Google Scholar] [CrossRef]
- Morita, K.; Honda, A.; Asano, M. A Simple and Efficient Method for Generating KO Rats Using In Vitro Fertilized Oocytes. Methods Mol. Biol. 2023, 2637, 233–246. [Google Scholar] [CrossRef]
- Namula, Z.; Le, Q.A.; Wittayarat, M.; Lin, Q.; Takebayashi, K.; Hirata, M.; Do, L.T.K.; Tanihara, F.; Otoi, T. Triple gene editing in porcine embryos using electroporation alone or in combination with microinjection. Vet. World 2022, 15, 496–501. [Google Scholar] [CrossRef]
- Bevacqua, R.; Fernandez-Martín, R.; Savy, V.; Canel, N.; Gismondi, M.; Kues, W.; Carlson, D.; Fahrenkrug, S.; Niemann, H.; Taboga, O.; et al. Efficient edition of the bovine PRNP prion gene in somatic cells and IVF embryos using the CRISPR/Cas9 system. Theriogenology 2016, 86, 1886–1896.e1. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Kaneko, T. Rapid and efficient production of genome-edited animals by electroporation into oocytes injected with frozen or freeze-dried sperm. Cryobiology 2019, 90, 71–74. [Google Scholar] [CrossRef]
- Mizushima, S.; Sasanami, T.; Ono, T.; Kuroiwa, A. Current Approaches to and the Application of Intracytoplasmic Sperm Injection (ICSI) for Avian Genome Editing. Genes 2023, 14, 757. [Google Scholar] [CrossRef] [PubMed]
- Lotti, S.N.; Polkoff, K.M.; Rubessa, M.; Wheeler, M.B. Modification of the Genome of Domestic Animals. Anim. Biotechnol. 2017, 28, 198–210. [Google Scholar] [CrossRef] [PubMed]
| Species | Gene | Trait | Effect | Commercial Name | References |
|---|---|---|---|---|---|
| fish | gh1 | Production trait | fast-growing salmon: 2- to 6-fold as compared to the wild-type fish | AquAdvantage | [204] |
| pig | GGTA1 | Medical use: xenotransplantation | reduces the risk of transplant rejection due to no alpha-gal sugar on cell surfaces | GalSafe | [205,206,207] |
| cattle | SLICK | Breed quality | substantially increases thermotolerance and thermoregulatory ability | PRLR-SLICK cattle | [208] |
| goat | ATryn1 | Medical use: drug production | the human ATryn1 (antithrombin-III) expressed by goats in milk | ATryn | [212] |
| rabbit | C1INH | Medical use: drug production | producing recombinant human C1 esterase inhibitor (Rhucin) in milk | Ruconest | [213,214] |
| chicken | LIPA | Medical use: drug production | express lipase A, lysosomal acid type, in eggs for long-term enzyme replacement therapy | Kanuma | [215] |
| fish | mylz2 | Fluorescent protein overexpression | overexpress GFP, YFP and RFP under a strong muscle-specific mylz2 promoter | GloFish | [217] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popova, J.; Bets, V.; Kozhevnikova, E. Perspectives in Genome-Editing Techniques for Livestock. Animals 2023, 13, 2580. https://doi.org/10.3390/ani13162580
Popova J, Bets V, Kozhevnikova E. Perspectives in Genome-Editing Techniques for Livestock. Animals. 2023; 13(16):2580. https://doi.org/10.3390/ani13162580
Chicago/Turabian StylePopova, Julia, Victoria Bets, and Elena Kozhevnikova. 2023. "Perspectives in Genome-Editing Techniques for Livestock" Animals 13, no. 16: 2580. https://doi.org/10.3390/ani13162580
APA StylePopova, J., Bets, V., & Kozhevnikova, E. (2023). Perspectives in Genome-Editing Techniques for Livestock. Animals, 13(16), 2580. https://doi.org/10.3390/ani13162580






