InDel and CNV within the AKAP13 Gene Revealing Strong Associations with Growth Traits in Goat
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Samples Collection
2.2. Genomic DNA Isolation
2.3. Primer Design and Genotype Detection
2.4. InDel and CNV Genotyping of AKAP13 Gene
2.5. Statistical Analyses
2.6. Genetic Linkage Analysis
3. Results
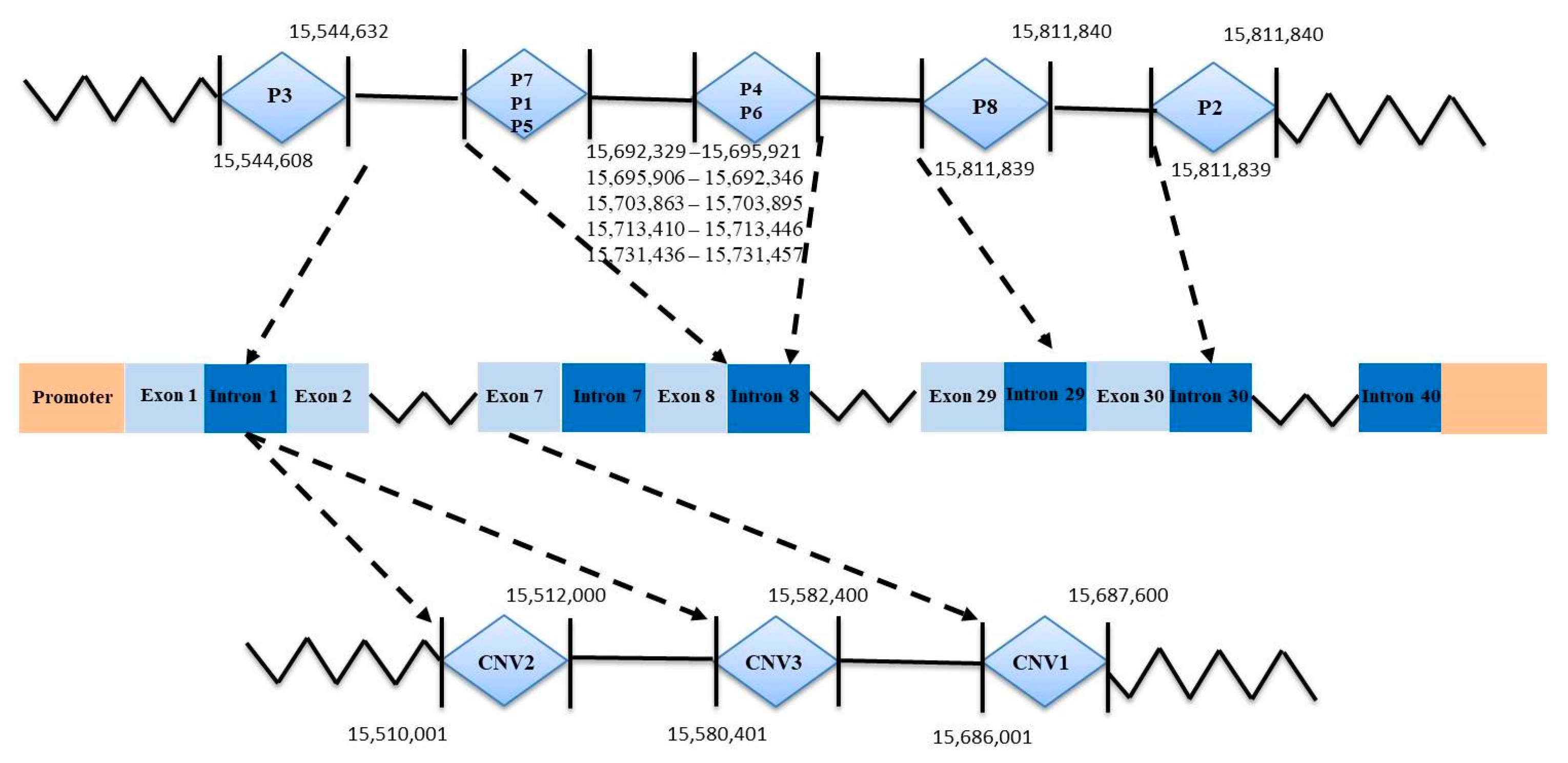
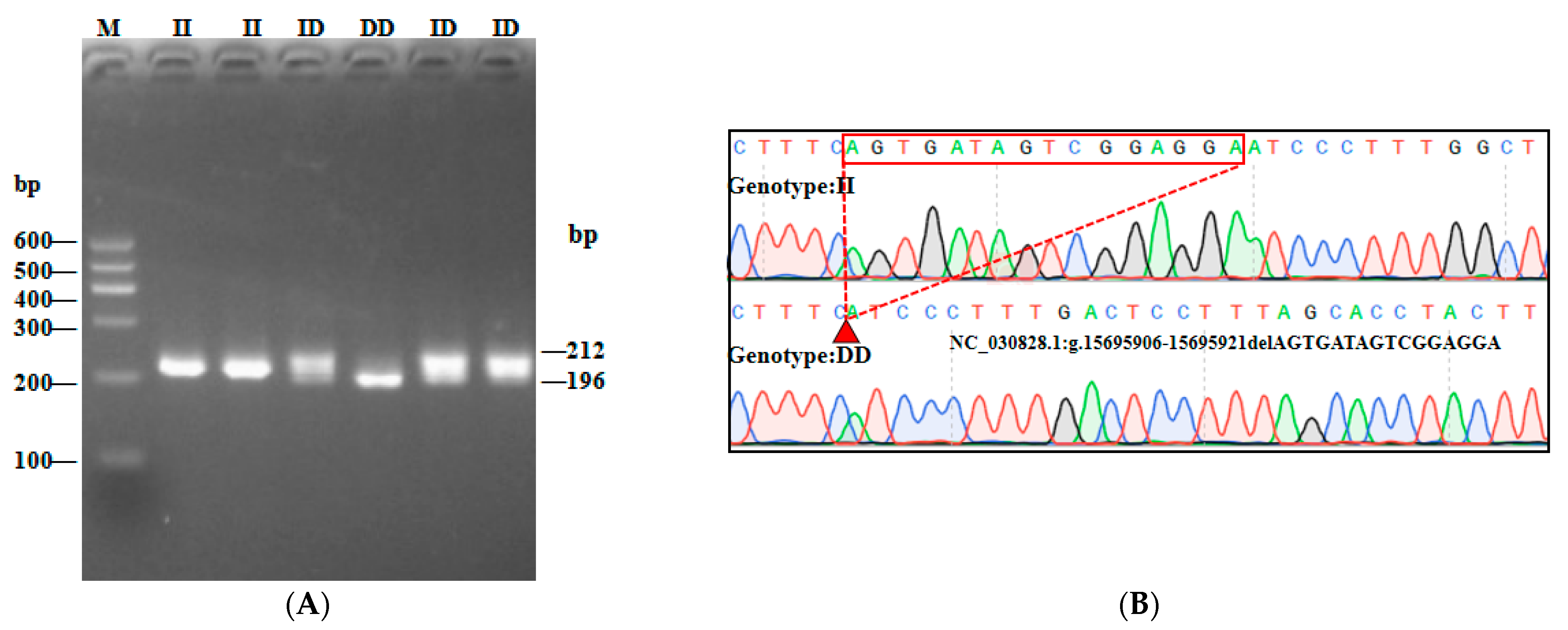
3.1. Characterization of Three InDel and CNV Loci in the AKAP13 Gene of SBWC Goats
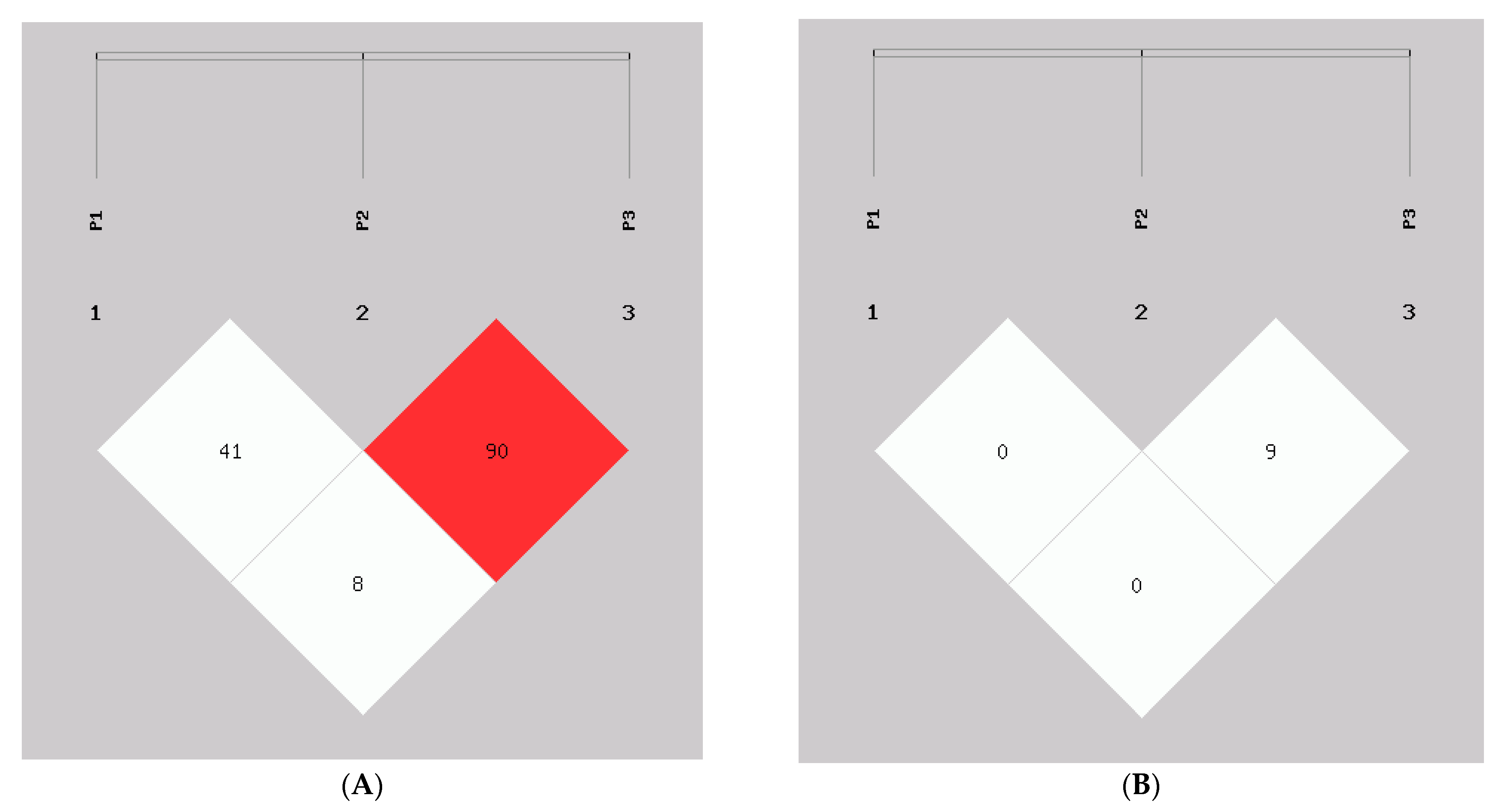
3.2. InDel Detection: Genotype Frequency, Linkage Disequilibrium, and Haplotype Analyses of the Goat AKAP13 Gene
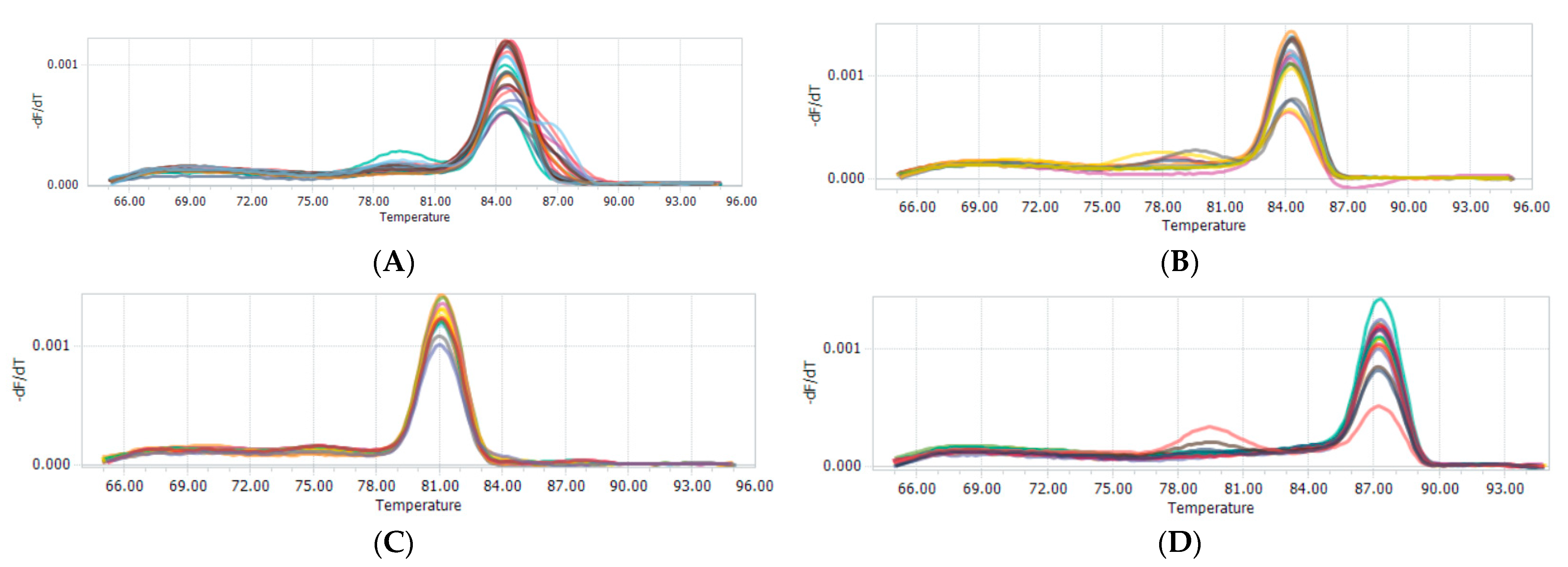
3.3. CNV Detection: Frequency of the Goat AKAP13 Gene Genotypes
3.4. Association Analysis of Mutations with Growth Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, X.; Yang, Q.; Zhang, S.; Zhang, X.; Pan, C.; Chen, H.; Zhu, H.; Lan, X. Genetic Effects of Single Nucleotide Polymorphisms in the Goat GDF9 Gene on Prolificacy: True or False Positive? Animals 2019, 9, 886. [Google Scholar] [CrossRef]
- Zhang, S.; Jiang, E.; Kang, Z.; Bi, Y.; Liu, H.; Xu, H.; Wang, Z.; Lei, C.; Chen, H.; Lan, X. CircRNA Profiling Reveals an Abundant circBDP1 that Regulates Bovine Fat Development by Sponging miR-181b/miR-204 Targeting Sirt1/TRARG1. J. Agric. Food Chem. 2022, 70, 14312–14328. [Google Scholar] [CrossRef]
- Zhao, W.; Zhong, T.; Wang, L.J.; Li, L.; Zhang, H.P. Extensive female-mediated gene flow and low phylogeography among seventeen goat breeds in southwest China. Biochem. Genet. 2014, 52, 355–364. [Google Scholar] [CrossRef]
- Fantazi, K.; Tolone, M.; Amato, B.; Sahraoui, H.; Vincenzo di Marco, L.P.; Gaouar, S.B.S.; Vitale, M. Characterization of morphological traits in Algerian indigenous goats by multivariate analyses. Genet. Biodivers. J. 2017, 1, 20–30. [Google Scholar] [CrossRef]
- Benyoub, K.; AmeurAmeur, A.; Gaouar, S.B.S. Phenotypic characterization of local gaots populations in western algerian: Morphometric measurements and milk quality. Genet. Biodivers. J. 2018, 2, 73–80. [Google Scholar]
- Tefiel, H.; Ata, N.; Chahbar, M.; Benyarou, M.; Fantazi, K.; Yilmaz, O.; Cemal, I.; Karaca, O.; Boudouma, D.; Gaouar, S.B.S. Genetic characterization of four Algerian goat breeds assessed by microsatellite markers. Rumin 2018, 160, 65–71. [Google Scholar] [CrossRef]
- Jiang, E.; Kang, Z.; Wang, X.; Liu, Y.; Liu, X.; Wang, Z.; Li, X.; Lan, X. Detection of insertions/deletions (InDels) within the goat Runx2 gene and their association with litter size and growth traits. Anim. Biotechnol. 2021, 32, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Fantazi, K.; Migliore, S.; Kdidi, S.; Racinaro, L.; Tefiel, H.; Boukhari, R.; Federico, G.; Di Marco Lo Presti, V.; Gaouar, S.B.S.; Vitale, M. Analysis of differences in prion protein gene (PRNP) polymorphisms between Algerian and Southern Italy’s goats. Ital. J. Anim. Sci. 2018, 17, 578–585. [Google Scholar] [CrossRef]
- Sahraoui, H.; Madani, T.; Khaled, F.; Khouane Asma, C.; Dettori, M.L. Genetic variability in the A microsatellite at SLC11A1 gene and possible implications with innate resistance against brucellosis in Algerian native goats. Biodivers. J. Biol. Divers. 2020, 21, 5630–5636. [Google Scholar] [CrossRef]
- Tang, Q.; Zhang, X.; Wang, X.; Wang, K.; Yan, H.; Zhu, H.; Lan, X.; Lei, Q.; Pan, C. Detection of two insertion/deletions (InDels) within the ADAMTS9 gene and their associations with growth traits in goat. Rumin 2019, 180, 9–14. [Google Scholar] [CrossRef]
- Wang, K.; Cui, Y.; Wang, Z.; Yan, H.; Meng, Z.; Zhu, H.; Qu, L.; Lan, X.; Pan, C. One 16 bp insertion/deletion (InDel) within the KDM6A gene revealing strong associations with growth traits in goat. Gene 2019, 686, 16–20. [Google Scholar] [CrossRef]
- Chen, N.; Fu, W.; Zhao, J.; Shen, J.; Chen, Q.; Zheng, Z.; Chen, H.; Sonstegard, T.S.; Lei, C.; Jiang, Y. BGVD: An integrated database for bovine sequencing variations and selective signatures. Genom. Proteom. Bioinform. 2020, 18, 186–193. [Google Scholar] [CrossRef]
- Fu, W.; Wang, R.; Yu, J.; Hu, D.; Cai, Y.; Shao, J.; Jiang, Y. GGVD: A goat genome variation database for tracking the dynamic evolutionary process of selective signatures and ancient introgressions. J. Genet. Genom. 2021, 48, 248–256. [Google Scholar] [CrossRef]
- Fu, W.; Wang, R.; Nanaei, H.A.; Wang, J.; Hu, D.; Jiang, Y. RGD v2.0: A major update of the ruminant functional and evolutionary genomics database. Nucleic Acids Res. 2022, 50, D1091–D1099. [Google Scholar] [CrossRef]
- Minden, A.; Lin, A.; Claret, F.X.; Abo, A.; Karin, M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell 1995, 81, 1147–1157. [Google Scholar] [CrossRef]
- Rubino, D.; Driggers, P.; Arbit, D.; Kemp, L.; Miller, B.; Coso, O.; Pagliai, K.; Gray, K.; Gutkind, S.; Segars, J. Characterization of Brx, a novel Dbl family member that modulates estrogen receptor action. Oncogene 1998, 16, 2513–2526. [Google Scholar] [CrossRef][Green Version]
- Melick, C.H.; Meng, D.; Jewell, J.L. A-kinase anchoring protein 8L interacts with mTORC1 and promotes cell growth. J. Biol. Chem. 2020, 295, 8096–8105. [Google Scholar] [CrossRef]
- Deng, Z.; Li, X.; Blanca Ramirez, M.; Purtell, K.; Choi, I.; Lu, J.H.; Yu, Q.; Yue, Z. Selective autophagy of AKAP11 activates cAMP/PKA to fuel mitochondrial metabolism and tumor cell growth. Proc. Natl. Acad. Sci. USA 2021, 118, e2020215118. [Google Scholar] [CrossRef]
- Ng, S.S.M.; Jorge, S.; Malik, M.; Britten, J.; Su, S.C.; Armstrong, C.R.; Brennan, J.T.; Chang, S.; Baig, K.M.; Driggers, P.H.; et al. A-Kinase Anchoring Protein 13 (AKAP13) Augments Progesterone Signaling in Uterine Fibroid Cells. J. Clin. Endocrinol. Metab. 2019, 104, 970–980. [Google Scholar] [CrossRef]
- Diviani, D.; Reggi, E.; Arambasic, M.; Caso, S.; Maric, D. Emerging roles of A-kinase anchoring proteins in cardiovascular pathophysiology. Biochim. Biophys. Acta 2016, 1863, 1926–1936. [Google Scholar] [CrossRef]
- Welch, E.J.; Jones, B.W.; Scott, J.D. Networking with AKAPs: Context-dependent regulation of anchored enzymes. Mol. Interv. 2010, 10, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Azeez, K.R.A.; Knapp, S.; Fernandes, J.M.; Klussmann, E.; Elkins, J.M. The crystal structure of the RhoA-AKAP-Lbc DH-PH domain complex. Biochem. J. 2014, 464, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Mayers, C.M.; Wadell, J.; McLean, K.; Venere, M.; Malik, M.; Shibata, T.; Driggers, P.H.; Kino, T.; Guo, X.C.; Koide, H.; et al. The Rho guanine nucleotide exchange factor AKAP13 (BRX) is essential for cardiac development in mice. J. Biol. Chem. 2010, 285, 12344–12354. [Google Scholar] [CrossRef] [PubMed]
- Koide, H.; Holmbeck, K.; Lui, J.C.; Guo, X.C.; Driggers, P.; Chu, T.; Tatsuno, I.; Quaglieri, C.; Kino, T.; Baron, J.; et al. Mice deficient in AKAP13 (BRX) are osteoporotic and have impaired osteogenesis. J. Bone Miner. Res. 2015, 30, 1887–1895. [Google Scholar] [CrossRef] [PubMed]
- Maravet Baig, K.; Su, S.C.; Mumford, S.L.; Giuliani, E.; Ng, S.S.M.; Armstrong, C.; Keil, M.F.; Vaught, K.C.; Olsen, N.; Pettiford, E.; et al. Mice deficient in AKAP13 (BRX) develop compulsive-like behavior and increased body weight. Brain Res. Bull. 2018, 140, 72–79. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, H.; Liu, X.; Yang, Q.; Pan, C.; Lei, C.; Dang, R.; Chen, H.; Lan, X. The muscle development transcriptome landscape of ovariectomized goat. R. Soc. Open Sci. 2017, 4, 171415. [Google Scholar] [CrossRef]
- Zhang, S.; Jiang, E.; Wang, K.; Zhang, Y.; Yan, H.; Qu, L.; Chen, H.; Lan, X.; Pan, C. Two Insertion/Deletion Variants within SPAG17 Gene Are Associated with Goat Body Measurement Traits. Animals 2019, 9, 379. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Yan, H.; Wang, K.; Xu, H.; Zhang, X.; Zhu, H.; Liu, J.; Qu, L.; Lan, X.; Pan, C. Insertion/Deletion within the KDM6A Gene Is Significantly Associated with Litter Size in Goat. Front. Genet. 2018, 9, 91. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Cui, W.; Chen, M.; Zhang, X.; Song, X.; Pan, C. A 21-bp InDel within the LLGL1 gene is significantly associated with litter size in goat. Anim. Biotechnol. 2021, 32, 213–218. [Google Scholar] [CrossRef]
- Wang, Q.; Bi, Y.; Wang, Z.; Zhu, H.; Liu, M.; Wu, X.; Pan, C. Goat SNX29: mRNA expression, InDel and CNV detection, and their associations with litter size. Front. Vet. Sci. 2022, 9, 981315. [Google Scholar] [CrossRef]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Z.; He, Z.; Tang, W.; Li, T.; Zeng, Z.; He, L.; Shi, Y. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: Update of the SHEsis (http://analysis.bio-x.cn). Cell Res. 2009, 19, 519–523. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, R.; Bai, Y.; Yang, Y.; Song, X.; Lan, X.; Pan, C. A deletion mutation within the goat AKAP13 gene is significantly associated with litter size. Anim. Biotechnol. 2023, 34, 350–356. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, X.; Jiang, E.; Yan, H.; Zhu, H.; Chen, H.; Liu, J.; Qu, L.; Pan, C.; Lan, X. InDels within caprine IGF2BP1 intron 2 and the 3’-untranslated regions are associated with goat growth traits. Anim. Genet. 2020, 51, 117–121. [Google Scholar] [CrossRef]
- Liu, S.; He, S.; Chen, L.; Li, W.; Di, J.; Liu, M. Estimates of linkage disequilibrium and effective population sizes in Chinese Merino (Xinjiang type) sheep by genome wide SNPs. Genes Genom. 2017, 39, 733–745. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zheng, Z.; Cai, Y.; Chen, T.; Li, C.; Fu, W.; Jiang, Y. CNVcaller: Highly efficient and widely applicable software for detecting copy number variations in large populations. Gigascience 2017, 6, gix115. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Fu, W.; Su, R.; Tian, X.; Du, D.; Zhao, Y.; Zheng, Z.; Chen, Q.; Gao, S.; Cai, Y.; et al. Towards the complete goat pan-genome by recovering missing genomic segments from the reference genome. Front. Genet. 2019, 10, 1169. [Google Scholar] [CrossRef]
- Rogers, R.; Norian, J.; Malik, M.; Christman, G.; Abu-Asab, M.; Chen, F.; Korecki, C.; Iatridis, J.; Catherino, W.H.; Tuan, R.S.; et al. Mechanical homeostasis is altered in uterine leiomyoma. Am. J. Obstet. Gynecol. 2008, 198, 474.e1–474.e11. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, C.N.C.; Islam, M.S.; Afrin, S.; Brennan, J.; Psoter, K.J.; Segars, J.H. Mechanical stiffness augments ligand-dependent progesterone receptor B activation via MEK 1/2 and Rho/ROCK-dependent signaling pathways in uterine fibroid cells. Fertil. Steril. 2021, 116, 255–265. [Google Scholar] [CrossRef]
- Kim, J.M.; Choi, J.S.; Kim, Y.H.; Jin, S.H.; Lim, S.; Jang, H.J.; Kim, K.T.; Ryu, S.H.; Suh, P.G. An activator of the cAMP/PKA/CREB pathway promotes osteogenesis from human mesenchymal stem cells. J. Cell. Physiol. 2013, 228, 617–626. [Google Scholar] [CrossRef]
- Arnsdorf, E.J.; Tummala, P.; Kwon, R.Y.; Jacobs, C.R. Mechanically induced osteogenic differentiation—The role of RhoA, ROCKII and cytoskeletal dynamics. J. Cell Sci. 2009, 122 Pt 4, 546–553. [Google Scholar] [CrossRef]
- Wang, N.; Robaye, B.; Agrawal, A.; Skerry, T.M.; Boeynaems, J.M.; Gartland, A. Reduced bone turnover in mice lacking the P2Y13 receptor of ADP. Mol. Endocrinol. 2012, 26, 142–152. [Google Scholar] [CrossRef]
- Lee, K.S.; Kim, H.J.; Li, Q.L.; Chi, X.Z.; Ueta, C.; Komori, T.; Wozney, J.M.; Kim, E.G.; Choi, J.Y.; Ryoo, H.M.; et al. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol. Cell. Biol. 2000, 20, 8783–8792. [Google Scholar] [CrossRef]
- Ling, W.; Rager, K.; Richardson, K.K.; Warren, A.D.; Ponte, F.; Aykin-Burns, N.; Manolagas, S.C.; Almeida, M.; Kim, H.N. Mitochondrial Sirt3 contributes to the bone loss caused by aging or estrogen deficiency. JCI Insight 2021, 6, e146728. [Google Scholar] [CrossRef] [PubMed]
- Subaran, R.L.; Odgerel, Z.; Swaminathan, R.; Glatt, C.E.; Weissman, M.M. Novel variants in ZNF34 and other brain-expressed transcription factors are shared among early-onset MDD relatives. Am. J. Med. Genet. 2016, 171, 333–341. [Google Scholar] [CrossRef]
- Mose, L.E.; Hayes, D.N.; Perou, C.M.; Parker, J.S. Improved InDel detection in RNA-seq data via assembly based re-alignment reveals expressed Epidermal Growth Factor Receptor InDels in Lung Adenocarcinoma. Cancer Res. 2017, 77 (Suppl. S13), 3592. [Google Scholar] [CrossRef]
- Guan, B.; Gao, M.; Wu, C.H.; Wang, T.L.; Shih, I.M. Functional analysis of in-frame InDel ARID1A mutations reveals new regulatory mechanisms of its tumor suppressor functions. Neoplasia 2012, 14, 986–993. [Google Scholar] [CrossRef]
- Minor, E.A.; Dubovy, S.; Wang, G. AMD-associated variants at the chromosome 10q26 locus and the stability of ARMS2 transcripts. Invest. Ophthalmol. Vis. Sci. 2013, 54, 5913–5919. [Google Scholar] [CrossRef]
- Liu, X.; Ma, L.; Wang, M.; Wang, K.; Li, J.; Yan, H.; Zhu, H.; Lan, X. Two InDel variants of prolactin receptor (PRLR) gene are associated with growth traits in goat. Anim. Biotechnol. 2020, 31, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Hui, Y.; Zhang, S.; Wang, M.; Yan, H.; Zhu, H.; Qu, L.; Lan, X.; Pan, C. A deletion mutation within the ATBF1 gene is strongly associated with goat litter size. Anim. Biotechnol. 2020, 31, 174–180. [Google Scholar] [CrossRef] [PubMed]


| Loci | Primer Sequence (5′–3′) | Region | Size (bp) | Tm (°C) |
|---|---|---|---|---|
| P1-16 bp | F: ACAGCCCTGAATGATGGATACAC | Intron | 196/212 | TD-PCR |
| R: TTCAGCAAAAGCAAACTCTCTGG | ||||
| P2-15 bp | F: GTTCAGGGCAGAGTGTGCTT | Intron | 225/240 | TD-PCR |
| R: CACCCAGTAGCACCAAAGGG | ||||
| P3-25 bp | F: TGGAATTGGGATGTTTTGTGTG | Intron | 218/243 | TD-PCR |
| R: AGCCACAGATTCCGAGCTTA | ||||
| P4-37 bp | F: CAGAGATAACCAGAGGAGGTGG | Intron | 159/196 | TD-PCR |
| R: ACGCTGGGAAAAAGTCAGGT | ||||
| P5-33 bp | F: TCTGGTTTGGGTGCCATACT | Intron | 175/208 | TD-PCR |
| R: TCATGTTGAAAGGGCATGCATTA | ||||
| P6-22 bp | F: GTGTCTTGTAATAACCTATAATGGGA | Intron | 201/223 | TD-PCR |
| R: GCTGCTCTTACCTGTTTTGATG | ||||
| P7-18 bp | F: GTATTATTTCAAAGTTCATCCATG | Intron | 214/232 | TD-PCR |
| R: TATAGAATTACCACATGAGCCA | ||||
| P8-15 bp | F: CCCCCAGCTGAGGAATTGAG | Intron | 138/153 | TD-PCR |
| R: GAGGATGAACAACAGGGAGACA |
| Primers | Sequences (5′–3′) | Sizes (bp) |
|---|---|---|
| CNV1 | F: CTTGAGCAGTGCTTTGCTGG | 110 |
| R: CCCTCAAACGGTTGCTTGTG | ||
| CNV2 | F: TTTCCAGCCTGTTGACTCCG | 117 |
| R: ACCAAGCCACTTCACCCAAT | ||
| CNV3 | F: ACGAACCTCTGCTTTCAACC | 120 |
| R: TAGGATCGAAGGTGTCCTGG | ||
| MC1R | F: GGCCTGAGAGGGGAATCACA | 126 |
| R: AGTGGGTCTCTGGATGGAGG |
| Primers | Chromosome | Start | End | Length | Location |
|---|---|---|---|---|---|
| CNV1 | 21 | 15,686,001 | 15,687,600 | 1599 | exonic |
| CNV2 | 21 | 15,510,001 | 15,512,000 | 1999 | intron |
| CNV3 | 21 | 15,580,401 | 15,582,400 | 1999 | intron |
| Loci | Frequencies | Ho | He | Ne | PIC | HWE p-Value | |||
|---|---|---|---|---|---|---|---|---|---|
| Genotypes | Alleles | ||||||||
| P1-16 bp (n = 408) | II | 0.802 (n = 327) | I | 0.884 | 0.164 | 0.206 | 1.259 | 0.185 | 0.00025 |
| ID | 0.164 (n = 67) | D | 0.116 | ||||||
| DD | 0.034 (n = 14) | ||||||||
| P2-15 bp (n = 400) | II | 0.025 (n = 10) | I | 0.169 | 0.288 | 0.281 | 1.390 | 0.241 | 0.8844 |
| ID | 0.2875 (n = 115) | D | 0.831 | ||||||
| DD | 0.6875 (n = 275) | ||||||||
| P3-25 bp (n = 235) | II | 0.089 (n = 21) | I | 0.347 | 0.515 | 0.453 | 1.828 | 0.350 | 0.1121 |
| ID | 0.515 (n = 121) | D | 0.653 | ||||||
| DD | 0.396 (n = 93) | ||||||||
| Loci | D′ | r2 | ||
|---|---|---|---|---|
| P2 | P3 | P2 | P3 | |
| P1 | 0.417 | 0.084 | 0.009 | 0.003 |
| P2 | 0.904 | 0.096 | ||
| Haplotypic Names | Haplotypic Types | Haplotypic Frequencies |
|---|---|---|
| Haplotype1 | IP1DP2IP3 | 0.004 |
| Haplotype2 | IP1DP2DP3 | 0.014 |
| Haplotype3 | IP1IP2IP3 | 0.069 |
| Haplotype4 | IP1IP2DP3 | 0.105 |
| Haplotype5 | DP1DP2DP2 | 0.161 |
| Haplotype6 | DP1IP2IP2 | 0.277 |
| Haplotype7 | DP1IP2DP2 | 0.369 |
| Body Measurement Traits | Combined Genotypes (Mean ± SE)/(Frequencies) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| IP1IP1–IP2IP2 (0.139) | IP1IP1–IP2DP2 (0.182) | IP1IP1–DP2DP2 (0.248) | IP1DP1–IP2IP2 (0.032) | IP1DP1–IP2DP2 (0.076) | IP1DP1–DP2DP2 (0.141) | DP1DP1–IP2IP2 (0.010) | DP1DP1–IP2DP2 (0.053) | DP1DP1–DP2DP2 (0.119) | |
| Body height (cm) | 56.31 ± 0.25 (n = 332) | 56.48 ± 0.20 (n = 435) | 56.24 ± 0.18 (n = 595) | 56.68 ± 0.43 (n = 77) | 56.88 ± 0.27 (n = 180) | 56.26 ± 0.23 (n = 340) | 56.69 ± 0.85 (n = 24) | 56.97 ± 0.32 (n = 127) | 56.19 ± 0.26 (n = 287) |
| Body length (cm) | 65.07 ± 0.38 bce (n = 332) | 65.76 ± 0.33 bce (n = 415) | 65.55 ± 0.28 bce (n = 574) | 67.02 ± 0.66 ae (n = 75) | 67.80 ± 0.42 ad (n = 158) | 66.40 ± 0.33 bcde (n = 317) | 68.34 ± 1.50 ac (n = 22) | 68.48 ± 0.52 a (n = 105) | 66.38 ± 0.38 bcde (n = 264) |
| Height at hip cross (cm) | 59.95 ± 0.33 (n = 164) | 60.15 ± 0.26 (n = 235) | 59.85 ± 0.23 (n = 319) | 60.43 ± 0.49 (n = 56) | 60.54 ± 0.31 (n = 127) | 59.92 ± 0.27 (n = 211) | 61.20 ± 0.92 (n = 15) | 60.72 ± 0.36 (n = 86) | 59.87 ± 0.30 (n = 170) |
| Chest circumference (cm) | 90.08 ± 2.03 (n = 334) | 91.61 ± 2.14 (n = 438) | 90.30 ± 1.16 (n = 596) | 90.31 ± 0.94 (n = 77) | 93.88 ± 3.60 (n = 181) | 90.52 ± 0.46 (n = 339) | 89.33 ± 1.74 (n = 24) | 95.17 ± 5.08 (n = 128) | 90.48 ± 0.51 (n = 286) |
| Chest depth (cm) | 27.07 ± 0.16 (n = 331) | 27.18 ± 0.14 (n = 427) | 27.14 ± 0.12 (n = 537) | 26.94 ± 0.44 (n = 74) | 27.28 ± 0.27 (n = 170) | 27.16 ± 0.18 (n = 280) | 28.45 ± 0.46 (n = 21) | 27.71 ± 0.29 (n = 117) | 27.36 ± 0.17 (n = 227) |
| Chest width (cm) | 17.48 ± 0.14 (n = 331) | 17.58 ± 0.13 (n = 427) | 17.55 ± 0.11 (n = 538) | 18.11 ± 0.24 (n = 74) | 17.99 ± 0.18 (n = 170) | 17.77 ± 0.14 (n = 281) | 18.31 ± 0.40 (n = 21) | 17.97 ± 0.22 (n = 117) | 17.71 ± 0.16 (n = 228) |
| Cannon circumference (cm) | 8.02 ± 0.05 (n = 334) | 8.08 ± 0.05 (n = 438) | 8.10 ± 0.04 (n = 596) | 8.23 ± 0.09 (n = 77) | 8.25 ± 0.06 (n = 181) | 8.21 ± 0.04 (n = 339) | 8.17 ± 0.15 (n = 24) | 8.24 ± 0.07 (n = 128) | 8.21 ± 0.05 (n = 286) |
| Hip width (cm) | 20.73 ± 0.18 (n = 159) | 20.80 ± 0.14 (n = 229) | 20.70 ± 0.13 (n = 311) | 20.76 ± 0.23 (n = 55) | 20.86 ± 0.15 (n = 125) | 20.70 ± 0.14 (n = 207) | 20.90 ± 0.39 (n = 15) | 20.94 ± 0.18 (n = 85) | 20.69 ± 0.17 (n = 167) |
| Loci | Size | Genotypic Frequencies | ||
|---|---|---|---|---|
| Gain | Medium | Loss | ||
| CNV1 | 79 | 0.886 (n = 70) | 0.079 (n = 6) | 0.038 (n = 3) |
| CNV2 | 78 | 0.692 (n = 54) | 0.090 (n = 7) | 0.218 (n = 17) |
| CNV3 | 79 | 1.000 (n = 79) | - | - |
| Locus | Body Measurement Traits | Genotype (Mean ± SE) | p-Values | ||
|---|---|---|---|---|---|
| II | ID | DD | |||
| P1-16 bp InDel | Body height | 55.29 ± 0.25 (n = 322) | 56.64 ± 0.44 (n = 67) | 56.50 ± 1.06 (n = 14) | 0.827 |
| Body length | 65.02 ± 0.39 a (n = 324) | 67.02 ± 0.65 b (n = 67) | 69.11 ± 1.61 b (n = 14) | 0.010 * | |
| Height at hip cross | 59.86 ± 0.34 (n = 159) | 60.22 ± 0.51 (n = 51) | 60.50 ± 1.04 (n = 10) | 0.794 | |
| Chest circumference | 90.11 ± 2.09 (n = 324) | 90.47 ± 0.99 (n = 67) | 89.43 ± 2.12 (n = 14) | 0.994 | |
| Chest depth | 27.02 ± 0.16 (n = 324) | 26.71 ± 0.48 (n = 67) | 28.11 ± 0.60 (n = 14) | 0.286 | |
| Chest width | 17.45 ± 0.14 (n = 324) | 18.03 ± 0.25 (n = 67) | 18.04 ± 0.44 (n = 14) | 0.177 | |
| Cannon circumference | 8.02 ± 0.05 (n = 324) | 8.27 ± 0.10 (n = 67) | 8.32 ± 0.17 (n = 14) | 0.094 | |
| Hip width | 20.72 ± 0.18 (n = 154) | 20.75 ± 0.24 (n = 50) | 20.95 ± 0.44 (n = 10) | 0.947 | |
| P2-15 bp InDel | Body height | 56.95 ± 1.47 (n = 10) | 57.03 ± 0.33 (n = 113) | 56.17 ± 0.26 (n = 273) | 0.169 |
| Body length | 67.00 ± 3.09 AB (n = 8) | 68.38 ± 0.56 A (n = 91) | 66.23 ± 0.39 B (n = 250) | 0.014 * | |
| Height at hip cross | 62.60 ± 1.81 (n = 5) | 60.75 ± 0.39 (n = 76) | 59.83 ± 0.31 (n = 160) | 0.080 | |
| Chest circumference | 89.20 ± 3.08 (n = 10) | 95.88 ± 5.69 (n = 114) | 90.53 ± 0.52 (n = 272) | 0.345 | |
| Chest depth | 29.14 ± 0.67 (n = 7) | 27.65 ± 0.32 (n = 103) | 27.31 ± 0.18 (n = 213) | 0.175 | |
| Chest width | 18.86 ± 0.86 (n = 7) | 17.97 ± 0.25 (n = 103) | 17.69 ± 0.17 (n = 214) | 0.337 | |
| Cannon circumference | 7.96 ± 0.28 (n = 10) | 8.23 ± 0.08 (n = 114) | 8.20 ± 0.05 (n = 272) | 0.600 | |
| Hip width | 20.80 ± 0.86 (n = 5) | 20.94 ± 0.19 (n = 75) | 20.68 ± 0.17 (n = 157) | 0.651 | |
| P3-25 bp InDel | Body height | 57.71 ± 0.75 (n = 21) | 56.45 ± 0.34 (n = 121) | 57.13 ± 0.39 (n = 93) | 0.218 |
| Body length | 70.29 ± 0.65 (n = 21) | 69.31 ± 0.39 (n = 121) | 70.39 ± 0.42 (n = 92) | 0.146 | |
| Height at hip cross | 61.33 ± 0.58 (n = 21) | 59.78 ± 0.36 (n = 121) | 60.78 ± 0.38 (n = 93) | 0.062 | |
| Chest circumference | 94.03 ± 1.17 (n = 21) | 92.12 ± 0.65 (n = 121) | 92.12 ± 0.61 (n = 93) | 0.440 | |
| Chest depth | 29.05 ± 0.35 (n = 21) | 28.10 ± 0.21 (n = 120) | 28.09 ± 0.20 (n = 93) | 0.146 | |
| Chest width | 17.90 ± 0.42 (n = 21) | 18.29 ± 0.21 (n = 121) | 18.24 ± 0.20 (n = 93) | 0.746 | |
| Cannon circumference | 8.70 ± 0.13 (n = 21) | 8.55 ± 0.05 (n = 121) | 8.63 ± 0.05 (n = 93) | 0.382 | |
| Hip width | 21.69 ± 0.38 (n = 21) | 20.82 ± 0.20 (n = 117) | 20.67 ± 0.18 (n = 92) | 0.100 | |
| Mutations | Growth Traits | Genotype (Mean ± SE) | p-Values | ||
|---|---|---|---|---|---|
| Gain | Medium | Loss | |||
| CNV1 | Body height | 60.00 a ± 0.44 (n = 70) | 56.83 b ± 1.28 (n = 6) | 56.67 ab ± 0.33 (n = 3) | 0.045 |
| Body length | 64.86 a ± 0.51 (n = 70) | 60.58 b ± 1.61 (n = 6) | 62.17 ab ± 3.20 (n = 3) | 0.046 | |
| Height at hip cross | 61.80 ± 0.43 (n = 69) | 60.83 ± 0.94 (n = 6) | 59.83 ± 1.17 (n = 3) | 0.532 | |
| Chest width | 14.81 ± 0.22 (n = 70) | 14.50 ± 0.67 (n = 6) | 14.00 ± 1.04 (n = 3) | 0.701 | |
| Chest depth | 27.14 ± 0.28 (n = 69) | 26.25 ± 0.40 (n = 6) | 25.67 ± 0.33 (n = 3) | 0.374 | |
| Chest circumference | 78.27 ± 0.72 (n = 70) | 75.88 ± 0.97 (n = 6) | 74.77 ± 1.63 (n = 3) | 0.390 | |
| Cannon circumference | 7.52 ± 0.07 (n = 70) | 7.25 ± 0.01 (n = 6) | 7.43 ± 0.24 (n = 3) | 0.493 | |
| CNV2 | Body height | 59.48 ± 0.51 (n = 54) | 58.43 ± 1.19 (n = 7) | 60.64 ± 0.89 (n = 17) | 0.354 |
| Body length | 64.76 ± 0.62 (n = 54) | 62.64 ± 1.66 (n = 7) | 64.41 ± 0.92 (n = 17) | 0.493 | |
| Height at hip cross | 62.17 ± 0.51 (n = 53) | 60.21 ± 1.08 (n = 7) | 60.79 ± 0.64 (n = 17) | 0.186 | |
| Chest width | 15.04 ± 0.23 (n = 54) | 15.07 ± 0.72 (n = 7) | 14.00 ± 0.42 (n = 17) | 0.092 | |
| Chest depth | 27.57 a ± 0.29 (n = 53) | 26.36 ab ± 0.52 (n = 7) | 25.82 b ± 0.58 (n = 17) | 0.011 | |
| Chest circumference | 79.24 a ± 0.77 (n = 54) | 75.57 ab ± 0.95 (n = 7) | 75.52 b ± 1.36 (n = 17) | 0.026 | |
| Cannon circumference | 7.56 a ± 0.05 (n = 54) | 7.79 ab ± 0.46 (n = 7) | 7.20 b ± 0.11 (n = 17) | 0.014 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, X.; Bai, Y.; Yuan, R.; Zhu, H.; Lan, X.; Qu, L. InDel and CNV within the AKAP13 Gene Revealing Strong Associations with Growth Traits in Goat. Animals 2023, 13, 2746. https://doi.org/10.3390/ani13172746
Song X, Bai Y, Yuan R, Zhu H, Lan X, Qu L. InDel and CNV within the AKAP13 Gene Revealing Strong Associations with Growth Traits in Goat. Animals. 2023; 13(17):2746. https://doi.org/10.3390/ani13172746
Chicago/Turabian StyleSong, Xiaoyue, Yangyang Bai, Rongrong Yuan, Haijing Zhu, Xianyong Lan, and Lei Qu. 2023. "InDel and CNV within the AKAP13 Gene Revealing Strong Associations with Growth Traits in Goat" Animals 13, no. 17: 2746. https://doi.org/10.3390/ani13172746
APA StyleSong, X., Bai, Y., Yuan, R., Zhu, H., Lan, X., & Qu, L. (2023). InDel and CNV within the AKAP13 Gene Revealing Strong Associations with Growth Traits in Goat. Animals, 13(17), 2746. https://doi.org/10.3390/ani13172746




