Investigating the Mechanism of Low-Salinity Environmental Adaptation in Sepia esculenta Larvae through Transcriptome Profiling
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Samples and Procedures
2.2. Detection of Sample RNA
2.3. Analysis of DEGs
2.4. qRT-PCR Validation
3. Results
3.1. Sequencing Results
3.2. Analysis of DEGs
3.3. Analysis Using GO and KEGG
3.4. Analysis of PPI Network Results
3.5. Analysis by qRT-PCR
4. Discussion
4.1. Enrichment Analysis of DEGs
4.2. Combined Analysis of KEGG and PPI
4.2.1. Analysis of Inflammatory and Immune Responses
4.2.2. Abnormalities in Metabolism
4.2.3. Signal Transduction was Enhanced by Low Salinity
4.3. Analysis of Two Hub Genes
4.4. Exploring the Functions of Other Key DEGs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aura, C.M.; Saitoh, S.I.; Liu, Y.; Hirawake, T.; Baba, K.; Yoshida, T. Implications of marine environment change on Japanese scallop (Mizuhopecten yessoensis) aquaculture suitability: A comparative study in Funka and Mutsu Bays, Japan. Aquac. Res. 2016, 47, 2164–2182. [Google Scholar] [CrossRef]
- Zhou, C.; Wong, K.; Tsou, J.Y.; Zhang, Y. Detection and Statistics of Offshore Aquaculture Rafts in Coastal Waters. J. Mar. Sci. Eng. 2022, 10, 781. [Google Scholar] [CrossRef]
- Hunt, J.D.; Byers, E. Reducing sea level rise with submerged barriers and dams in Greenland. Mitig. Adapt. Strateg. Glob. Chang. 2019, 24, 779–794. [Google Scholar] [CrossRef]
- Gregory, J.; Oerlemans, J. Simulated future sea-level rise due to glacier melt based on regionally and seasonally resolved temperature changes. Nature 1998, 391, 474–476. [Google Scholar] [CrossRef]
- Maynard, A.; Bible, J.M.; Pespeni, M.H.; Sanford, E.; Evans, T.G. Transcriptomic responses to extreme low salinity among locally adapted populations of Olympia oyster (Ostrea lurida). Mol. Ecol. 2018, 27, 4225–4240. [Google Scholar] [CrossRef]
- Knowles, G.; Handlinger, J.; Jones, B.; Moltschaniwskyj, N. Hemolymph chemistry and histopathological changes in Pacific oysters (Crassostrea gigas) in response to low salinity stress. J. Invertebr. Pathol. 2014, 121, 78–84. [Google Scholar] [CrossRef]
- Ren, Z.; Zhao, Y.; Song, W.; Wang, C.; Mu, C.; Li, R. Transcriptome analysis of the Sepia pharaonis: Identification of low salinity stress-related information and microsatellite markers. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 35, 100705. [Google Scholar] [CrossRef]
- Morozova, O.; Hirst, M.; Marra, M.A. Applications of new sequencing technologies for transcriptome analysis. Annu. Rev. Genom. Hum. Genet. 2009, 10, 135–151. [Google Scholar] [CrossRef]
- Qian, X.; Ba, Y.; Zhuang, Q.; Zhong, G. RNA-Seq technology and its application in fish transcriptomics. OMICS 2014, 18, 98–110. [Google Scholar] [CrossRef]
- Bao, X.; Wang, W.; Chen, X.; Feng, Y.; Xu, X.; Sun, G.; Li, B.; Liu, X.; Li, Z.; Yang, J. Exploration of immune response mechanisms in cadmium and copper co-exposed juvenile golden cuttlefish (Sepia esculenta) based on transcriptome profiling. Front. Immunol. 2022, 13, 963931. [Google Scholar] [CrossRef]
- Boamah, G.A.; Huang, Z.; Shen, Y.; Lu, Y.; Wang, Z.; Su, Y.; Xu, C.; Luo, X.; Ke, C.; You, W. Transcriptome analysis reveals fluid shear stress (FSS) and atherosclerosis pathway as a candidate molecular mechanism of short-term low salinity stress tolerance in abalone. BMC Genom. 2022, 23, 392. [Google Scholar] [CrossRef]
- Bao, X.; Wang, W.; Yuan, T.; Li, Y.; Chen, X.; Liu, X.; Xu, X.; Sun, G.; Li, B.; Yang, J.; et al. Transcriptome profiling based on larvae at different time points after hatching provides a core set of gene resource for understanding the immune response mechanisms of the egg-protecting behavior against Vibrio anguillarum infection in Amphioctopus fangsiao. Fish Shellfish Immunol. 2022, 124, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhang, D.; Wang, L.; Li, W.; He, P.; Näslund, J.; Zhang, X. Sperm competition in golden cuttlefish Sepia esculenta: The impact of mating order and male size. Aquaculture 2021, 530, 735929. [Google Scholar] [CrossRef]
- Wang, Y.; Bao, X.; Wang, W.; Xu, X.; Liu, X.; Li, Z.; Yang, J.; Yuan, T. Exploration of anti-stress mechanisms in high temperature exposed juvenile golden cuttlefish (Sepia esculenta) based on transcriptome profiling. Front. Physiol. 2023, 14, 1189375. [Google Scholar] [CrossRef]
- Gong, J.; Li, Q.; Yu, H.; Liu, S.; Kong, L. Effects of low salinity on hemolymph osmolality and transcriptome of the Iwagaki oyster, Crassostrea nippona. Fish Shellfish Immunol. 2022, 126, 211–216. [Google Scholar] [CrossRef]
- Masotti, A.; Preckel, T. Analysis of small RNAs with the Agilent 2100 Bioanalyzer. Nat. Methods 2006, 3, 658. [Google Scholar] [CrossRef]
- de Sena Brandine, G.; Smith, A.D. Falco: High-speed FastQC emulation for quality control of sequencing data. F1000Res 2019, 8, 1874. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zheng, X.; Ma, Y.; Bai, Y.; Huang, T.; Lv, X.; Deng, J.; Wang, Z.; Lian, W.; Tong, Y.; Zhang, X.; et al. Identification and validation of immunotherapy for four novel clusters of colorectal cancer based on the tumor microenvironment. Front. Immunol. 2022, 13, 984480. [Google Scholar] [CrossRef]
- Jiao, X.; Sherman, B.T.; Huang, D.W.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics 2012, 28, 1805–1806. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Zhu, B.; Zhang, Y.; Wang, H.; Li, C.; Su, Y.; Ba, C. The research of applying primer premier 5.0 to design PCR primer. J. Jinzhou. Med. Coll. 2004, 25, 43–46. [Google Scholar] [CrossRef]
- Eker, Ç.; İnan, H.C.; Çelebi, A.; Gözen, E.D.; Karaman, E. Investigation of Toll-like Receptor-2, -3 and -4 Gene Expressions in Larynx Squamous Cell Carcinoma. Turk. Arch. Otorhinolaryngol. 2022, 60, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Jones, H.R.; Johnson, K.M.; Kelly, M.W. Synergistic Effects of Temperature and Salinity on the Gene Expression and Physiology of Crassostrea virginica. Integr. Comp. Biol. 2019, 59, 306–319. [Google Scholar] [CrossRef]
- Ming, Z.; Pang, Y.; Liu, J. Mechanical Deformation Mediated Transmembrane Transport. Macromol. Rapid Commun. 2020, 41, e1900518. [Google Scholar] [CrossRef]
- Schendzielorz, A.B.; Bragoszewski, P.; Naumenko, N.; Gomkale, R.; Schulz, C.; Guiard, B.; Chacinska, A.; Rehling, P. Motor recruitment to the TIM23 channel’s lateral gate restricts polypeptide release into the inner membrane. Nat. Commun. 2018, 9, 4028. [Google Scholar] [CrossRef]
- Zhong, J.; Zhao, X. Transcriptomic Analysis of Viable but Non-Culturable Escherichia coli O157:H7 Formation Induced by Low Temperature. Microorganisms 2019, 7, 634. [Google Scholar] [CrossRef]
- McNamara, J.C.; Freire, C.A. Strategies of Invertebrate Osmoregulation: An Evolutionary Blueprint for Transmuting into Fresh Water from the Sea. Integr. Comp. Biol. 2022, 62, 376–387. [Google Scholar] [CrossRef]
- Wright-LaGreca, M.; Mackenzie, C.; Green, T.J. Ocean Acidification Alters Developmental Timing and Gene Expression of Ion Transport Proteins during Larval Development in Resilient and Susceptible Lineages of the Pacific Oyster (Crassostrea gigas). Mar Biotechnol. 2022, 24, 116–124. [Google Scholar] [CrossRef]
- Hu, M.Y.; Sucré, E.; Charmantier-Daures, M.; Charmantier, G.; Lucassen, M.; Himmerkus, N.; Melzner, F. Localization of ion-regulatory epithelia in embryos and hatchlings of two cephalopods. Cell Tissue Res. 2010, 339, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Liu, H.; Fu, S.; Dong, W.; Pan, B.; Bu, W. Transcriptome-wide identification and characterization of toll-like receptors response to Vibrio anguillarum infection in Manila clam (Ruditapes philippinarum). Fish Shellfish Immunol. 2021, 111, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Xu, X.; Xiao, X.; Wang, Y.; Yang, H.; Zhang, Z. Genome-wide identification and characterization of Toll-like receptors (TLR) genes in Haliotis discus hannai, H. rufescens, and H. laevigata. Fish Shellfish Immunol. 2023, 137, 108728. [Google Scholar] [CrossRef]
- Dong, X.; Kim, Y.S.; Kim, E.K.; Shin, W.B.; Park, J.S.; Kim, S.J.; Go, E.A.; Park, P.J.; Kwon, S.C. Scallop Extracts Inhibited LPS-Induced Inflammation by Suppressing MAPK and NF-κB Activation in RAW264.7 Macrophages. Adv. Exp. Med. Biol. 2019, 1155, 1069–1081. [Google Scholar] [CrossRef] [PubMed]
- Portillo-López, A.; Gould, M.C.; Stephano, J.L. MAPK is involved in metaphase I arrest in oyster and mussel oocytes. Biol. Cell. 2003, 95, 275–282. [Google Scholar] [CrossRef]
- Hatano, K.; Yoshida, M.A.; Hirayama, J.; Kitani, Y.; Hattori, A.; Ogiso, S.; Watabe, Y.; Sekiguchi, T.; Tabuchi, Y.; Urata, M.; et al. Deep ocean water alters the cholesterol and mineral metabolism of squid Todarodes pacificus and suppresses its weight loss. Sci. Rep. 2023, 13, 7591. [Google Scholar] [CrossRef]
- Fiorini, R.; Ventrella, V.; Trombetti, F.; Fabbri, M.; Pagliarani, A.; Nesci, S. Lipid-protein interactions in mitochondrial membranes from bivalve mollusks: Molecular strategies in different species. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2019, 227, 12–20. [Google Scholar] [CrossRef]
- Ahmad, I.; Ahmed, I.; Fatma, S.; Peres, H. Role of branched-chain amino acids on growth, physiology and metabolism of different fish species: A review. Aquacult. Nutr. 2021, 27, 1270–1289. [Google Scholar] [CrossRef]
- Chandhini, S.; Trumboo, B.; Jose, S.; Varghese, T.; Rajesh, M.; Kumar, V.J.R. Insulin-like growth factor signalling and its significance as a biomarker in fish and shellfish research. Fish. Physiol. Biochem. 2021, 47, 1011–1031. [Google Scholar] [CrossRef]
- Li, Z.; Bao, X.; Liu, X.; Li, Y.; Cui, M.; Liu, X.; Li, B.; Feng, Y.; Xu, X.; Sun, G.; et al. Transcriptome profiling based on protein-protein interaction networks provides a set of core genes for understanding the immune response mechanisms of the egg-protecting behavior in Octopus ocellatus. Fish Shellfish Immunol. 2021, 117, 113–123. [Google Scholar] [CrossRef]
- Wu, Y.; Si, X.; Qiu, L.; Chen, X.; Fu, P.; Buttino, I.; Guo, B.; Liao, Z.; Yan, X.; Qi, P. Regulation of innate immunity in marine mussel Mytilus coruscus: MicroRNA Mc-novel_miR_196 targets McTLR-like1 molecule to inhibit inflammatory response and apoptosis. Fish Shellfish Immunol. 2023, 138, 108868. [Google Scholar] [CrossRef] [PubMed]
- Canesi, L.; Auguste, M.; Balbi, T.; Prochazkova, P. Soluble mediators of innate immunity in annelids and bivalve mollusks: A mini-review. Front. Immunol. 2022, 13, 1051155. [Google Scholar] [CrossRef] [PubMed]
- Tomar, N.; De, R.K. A brief outline of the immune system. Methods Mol. Biol. 2014, 1184, 3–12. [Google Scholar] [CrossRef]
- Parkin, J.; Cohen, B. An overview of the immune system. Lancet 2001, 357, 1777–1789. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S. Inflammation, metaflammation and immunometabolic disorders. Nature 2017, 542, 177–185. [Google Scholar] [CrossRef]
- Shastri, A.; Bonifati, D.M.; Kishore, U. Innate immunity and neuroinflammation. Mediat. Inflamm. 2013, 2013, 342931. [Google Scholar] [CrossRef]
- Rahtes, A.; Geng, S.; Lee, C.; Li, L. Cellular and molecular mechanisms involved in the resolution of innate leukocyte inflammation. J. Leukoc. Biol. 2018, 104, 535–541. [Google Scholar] [CrossRef]
- Germolec, D.R.; Shipkowski, K.A.; Frawley, R.P.; Evans, E. Markers of Inflammation. Methods Mol. Biol. 2018, 1803, 57–79. [Google Scholar] [CrossRef]
- Ottaviani, E.; Franchini, A.; Malagoli, D. Inflammatory response in molluscs: Cross-taxa and evolutionary considerations. Curr. Pharm. Des. 2010, 16, 4160–4165. [Google Scholar] [CrossRef]
- Jiravanichpaisal, P.; Söderhäll, K.; Söderhäll, I. Inflammation in arthropods. Curr. Pharm. Des. 2010, 16, 4166–4174. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, M.; Sun, Y.; Wang, Y.; Zhang, Z. Transcriptomic analysis and discovery of genes involving in enhanced immune protection of Pacific abalone (Haliotis discus hannai) in response to the re-infection of Vibrio parahaemolyticus. Fish Shellfish Immunol. 2022, 125, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Hao, G.; Li, J.; Peng, W.; Geng, X.; Sun, J. Comparative analysis of dual specificity protein phosphatase genes 1, 2 and 5 in response to immune challenges in Japanese flounder Paralichthys olivaceus. Fish Shellfish Immunol. 2017, 68, 368–376. [Google Scholar] [CrossRef]
- Sun, Y.; Chu, Q.; Zhao, X.; Zhou, Z.; Bi, D.; Xu, T. microRNA-375 modulates the NF-κB pathway in miiuy croaker by targeting DUSP1 gene. Dev. Comp. Immunol. 2018, 86, 196–202. [Google Scholar] [CrossRef]
- Li, Q.; Verma, I.M. NF-kappaB regulation in the immune system. Nat. Rev. Immunol. 2002, 2, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Chakravarti, B.; Akhtar Siddiqui, J.; Anthony Sinha, R.; Raza, S. Targeting autophagy and lipid metabolism in cancer stem cells. Biochem. Pharmacol. 2023, 212, 115550. [Google Scholar] [CrossRef]
- Qu, M.; Zhou, X.; Wang, X.; Li, H. Lipid-induced S-palmitoylation as a Vital Regulator of Cell Signaling and Disease Development. Int. J. Biol. Sci. 2021, 17, 4223–4237. [Google Scholar] [CrossRef]
- Zhang, J.M.; Wang, X.H.; Hao, L.H.; Wang, H.; Zhang, X.Y.; Muhammad, I.; Qi, Y.; Li, G.L.; Sun, X.Q. Nrf2 is crucial for the down-regulation of Cyp7a1 induced by arachidonic acid in Hepg2 cells. Environ. Toxicol. Pharmacol. 2017, 52, 21–26. [Google Scholar] [CrossRef]
- Iqbal, J.; Sun, L.; Zaidi, M. Complexity in signal transduction. Ann. N. Y. Acad. Sci. 2010, 1192, 238–244. [Google Scholar] [CrossRef]
- Lafrenie, R.M.; Yamada, K.M. Integrin-dependent signal transduction. J. Cell Biochem. 1996, 61, 543–553. [Google Scholar] [CrossRef]
- Kang, Y.P.; Falzone, A.; Liu, M.; González-Sánchez, P.; Choi, B.H.; Coloff, J.L.; Saller, J.J.; Karreth, F.A.; DeNicola, G.M. PHGDH supports liver ceramide synthesis and sustains lipid homeostasis. Cancer Metab. 2020, 8, 6. [Google Scholar] [CrossRef]
- Sinkus, M.L.; Graw, S.; Freedman, R.; Ross, R.G.; Lester, H.A.; Leonard, S. The human CHRNA7 and CHRFAM7A genes: A review of the genetics, regulation, and function. Neuropharmacology 2015, 96 Pt B, 274–288. [Google Scholar] [CrossRef]
- Zhang, S.; Xue, R.; Geng, Y.; Wang, H.; Li, W. Fisetin Prevents HT22 Cells from High Glucose-Induced Neurotoxicity via PI3K/Akt/CREB Signaling Pathway. Front. Neurosci. 2020, 14, 241. [Google Scholar] [CrossRef] [PubMed]
- Alderton, W.K.; Cooper, C.E.; Knowles, R.G. Nitric oxide synthases: Structure, function and inhibition. Biochem. J. 2001, 357 Pt 3, 593–615. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yin, X.; Zhao, X.; Wu, Z.; Xiao, Y.; Di, Q.; Sun, P.; Tang, H.; Quan, J.; Chen, W. HDAC11 negatively regulates antifungal immunity by inhibiting Nos2 expression via binding with transcriptional repressor STAT3. Redox Biol. 2022, 56, 102461. [Google Scholar] [CrossRef]
- Alaupovic, P. Apolipoprotein composition as the basis for classifying plasma lipoproteins. Characterization of ApoA- and ApoB-containing lipoprotein families. Prog. Lipid Res. 1991, 30, 105–138. [Google Scholar] [CrossRef] [PubMed]
- Shaulian, E. AP-1—The Jun proteins: Oncogenes or tumor suppressors in disguise? Cell Signal. 2010, 22, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Hartl, M.; Bader, A.G.; Bister, K. Molecular targets of the oncogenic transcription factor jun. Curr. Cancer Drug Targets 2003, 3, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.C. Regulation of glutathione synthesis. Mol. Aspects Med. 2009, 30, 42–59. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Gari, A.; Qu, C.; Chen, J. Pan-Cancer Analysis of PARP1 Alterations as Biomarkers in the Prediction of Immunotherapeutic Effects and the Association of Its Expression Levels and Immunotherapy Signatures. Front. Immunol. 2021, 12, 721030. [Google Scholar] [CrossRef]
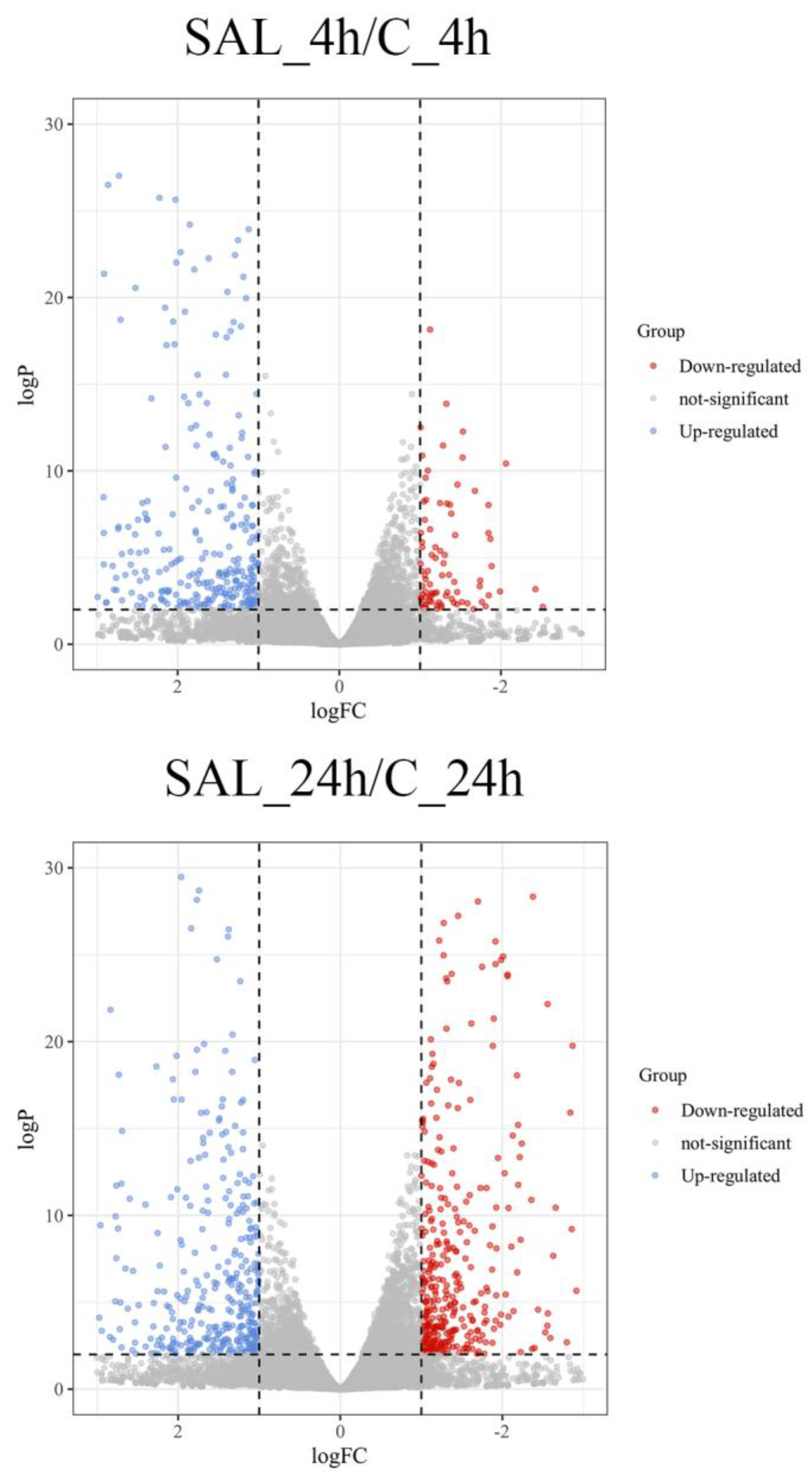



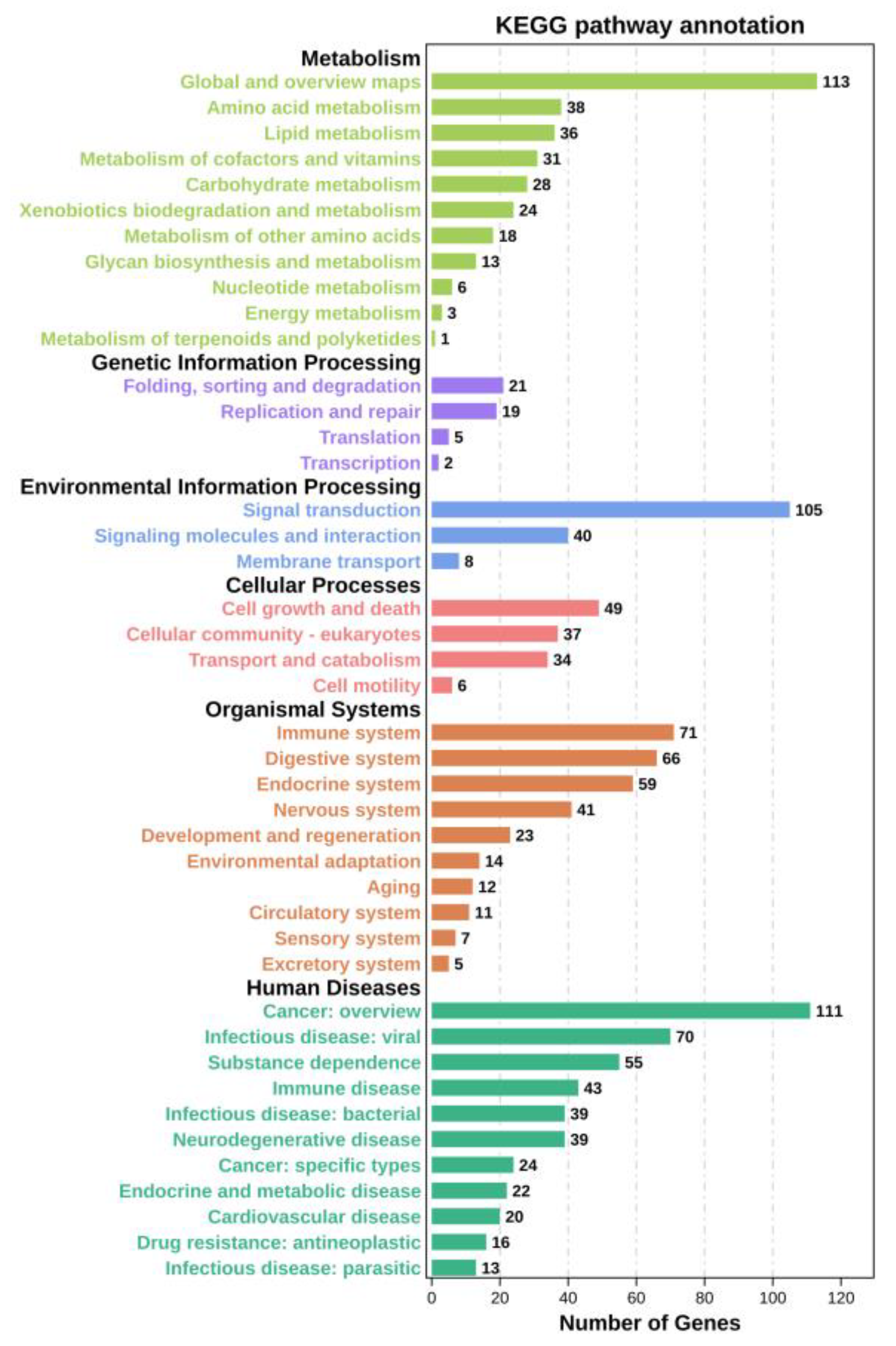
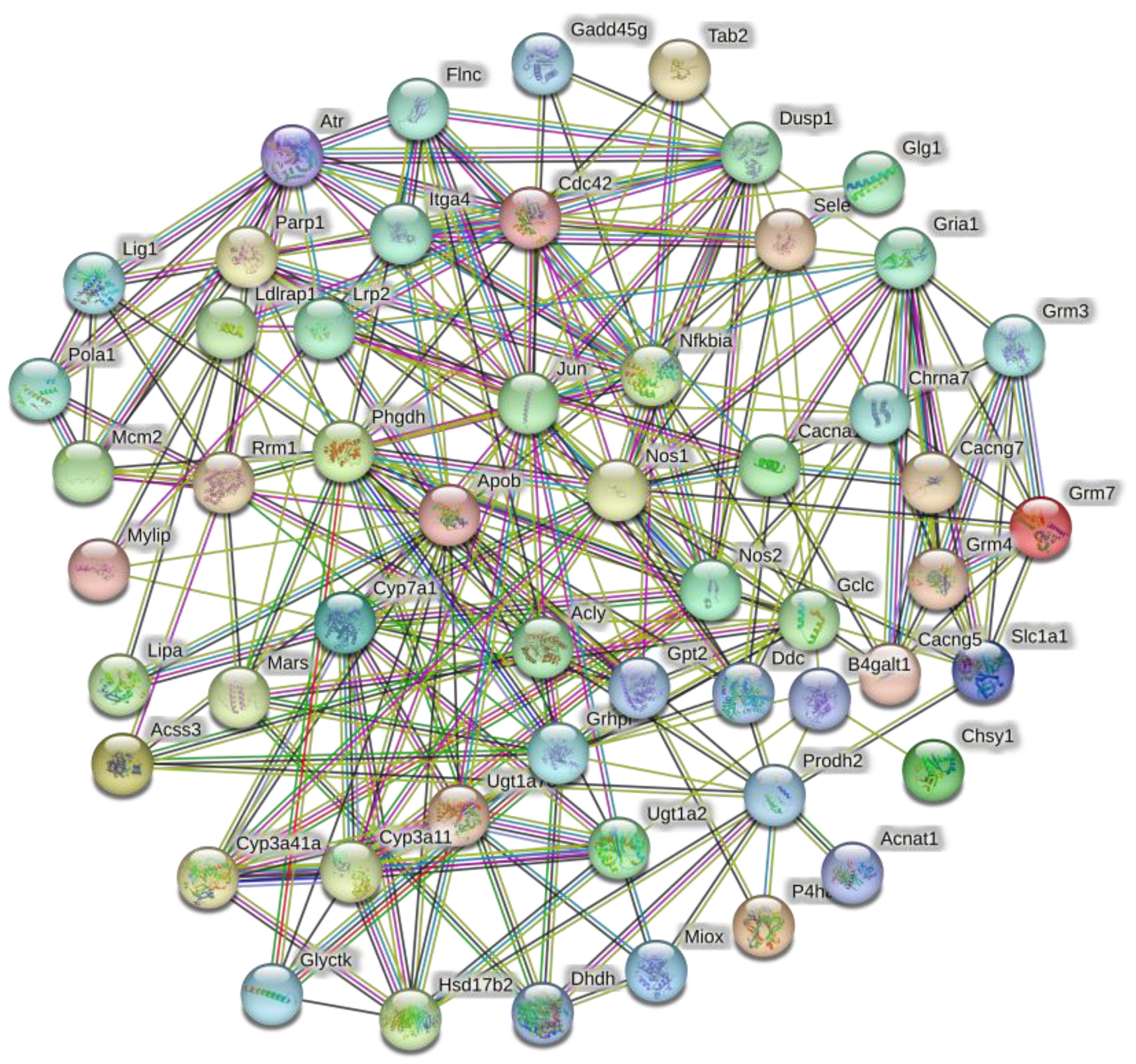

| Pathway | Number of DEGs |
|---|---|
| Apoptosis | 4 |
| Arginine and proline metabolism | 3 |
| Arginine biosynthesis | 2 |
| Ascorbate and aldarate metabolism | 3 |
| Cell adhesion molecules | 3 |
| Cell cycle | 5 |
| Cholesterol metabolism | 6 |
| Cholinergic synapse | 2 |
| DNA replication | 6 |
| Focal adhesion | 2 |
| Glutamatergic synapse | 6 |
| Glycine, serine, and threonine metabolism | 3 |
| MAPK signaling pathway | 8 |
| Metabolic pathways | 27 |
| NOD-like receptor signaling pathway | 3 |
| Toll-like receptor signaling pathway | 2 |
| Stats | |
|---|---|
| Number of nodes | 54 |
| Number of edges | 245 |
| Average nodes | 9.07 |
| Clustering coefficient | 0.558 |
| Number of expected edges | 164 |
| p-value | 1.83 × 10−9 |
| Gene | Gene Name | Number of PPI | Number of KEGG |
|---|---|---|---|
| APOB | apolipoprotein B | 23 | 3 |
| ATR | ATR serine/threonine kinase | 11 | 1 |
| CDC42 | cell division cycle 42 | 17 | 4 |
| CHRNA7 | cholinergic receptor nicotinic alpha 7 subunit | 12 | 1 |
| CYP3A11 | cytochrome P450, family 3, subfamily a, polypeptide 11 | 11 | 5 |
| CYP7A1 | cytochrome P450 family 7 subfamily A member 1 | 15 | 2 |
| DUSP1 | dual specificity phosphatase 1 | 15 | 2 |
| GCLC | glutamate-cysteine ligase catalytic subunit | 13 | 2 |
| GRIA1 | glutamate ionotropic receptor AMPA type subunit 1 | 14 | 2 |
| JUN | Jun proto-oncogene, AP-1 transcription factor subunit | 26 | 6 |
| NFKBIA | NFKB inhibitor alpha | 17 | 5 |
| NOS1 | nitric oxide synthase 1 | 18 | 4 |
| NOS2 | nitric oxide synthase 2 | 14 | 4 |
| PARP1 | poly(ADP-ribose) polymerase 1 | 12 | 1 |
| PHGDH | phosphoglycerate dehydrogenase | 18 | 2 |
| PRODH2 | proline dehydrogenase 2 | 12 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Liu, X.; Wang, W.; Sun, G.; Xu, X.; Feng, Y.; Li, Z.; Yang, J. Investigating the Mechanism of Low-Salinity Environmental Adaptation in Sepia esculenta Larvae through Transcriptome Profiling. Animals 2023, 13, 3139. https://doi.org/10.3390/ani13193139
Wang Y, Liu X, Wang W, Sun G, Xu X, Feng Y, Li Z, Yang J. Investigating the Mechanism of Low-Salinity Environmental Adaptation in Sepia esculenta Larvae through Transcriptome Profiling. Animals. 2023; 13(19):3139. https://doi.org/10.3390/ani13193139
Chicago/Turabian StyleWang, Yongjie, Xiumei Liu, Weijun Wang, Guohua Sun, Xiaohui Xu, Yanwei Feng, Zan Li, and Jianmin Yang. 2023. "Investigating the Mechanism of Low-Salinity Environmental Adaptation in Sepia esculenta Larvae through Transcriptome Profiling" Animals 13, no. 19: 3139. https://doi.org/10.3390/ani13193139




