Population Genetic Structure of Anisakis simplex Infecting the European Hake from North East Atlantic Fishing Grounds
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
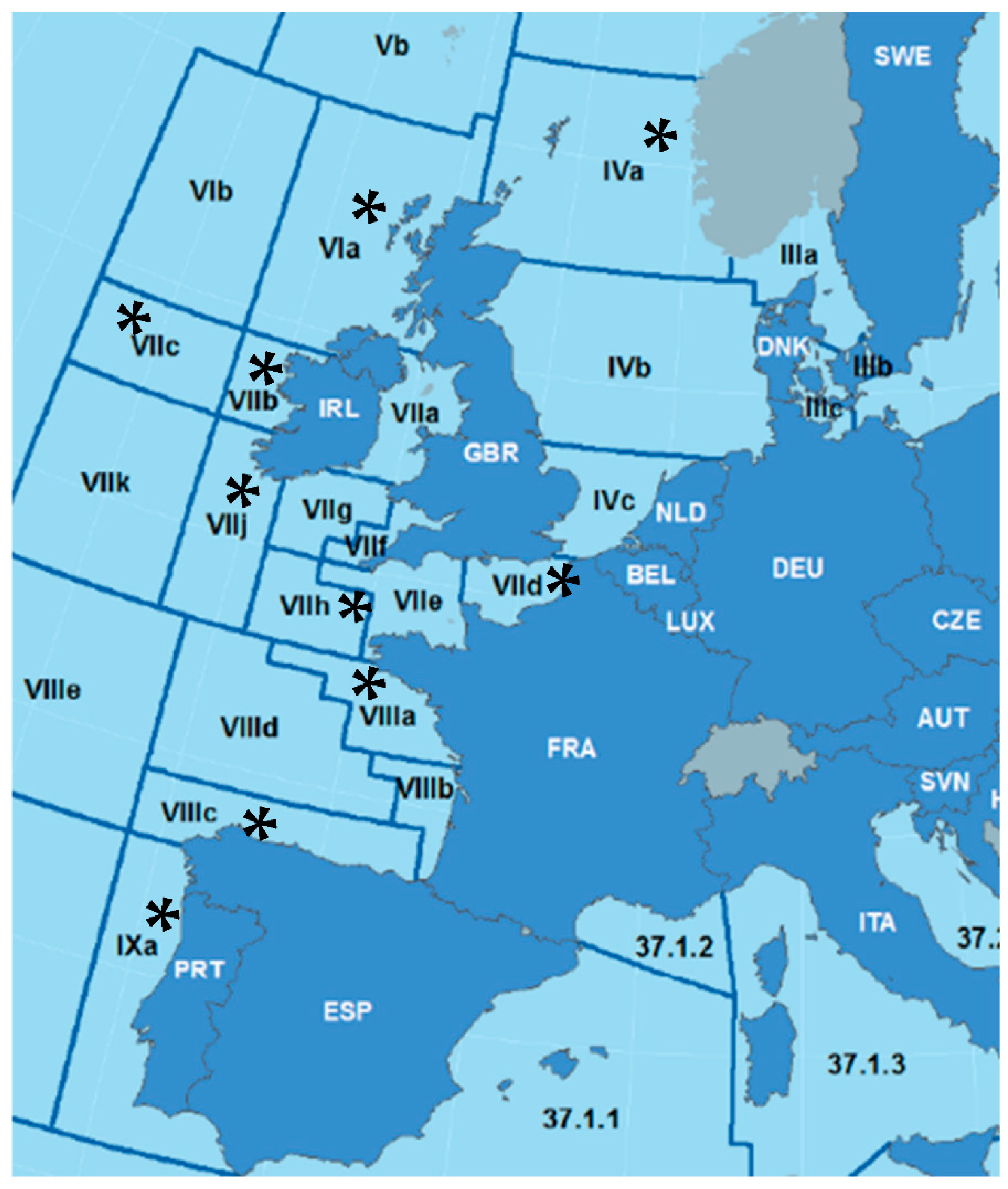
2.1. Fish and Parasite sampling
2.2. Taxonomic Identification of Anisakis spp. L3 Larvae
2.3. Genetic Diversity and Haplotype Analysis
3. Results
3.1. Genetic Identification of Anisakis spp. L3 Larvae
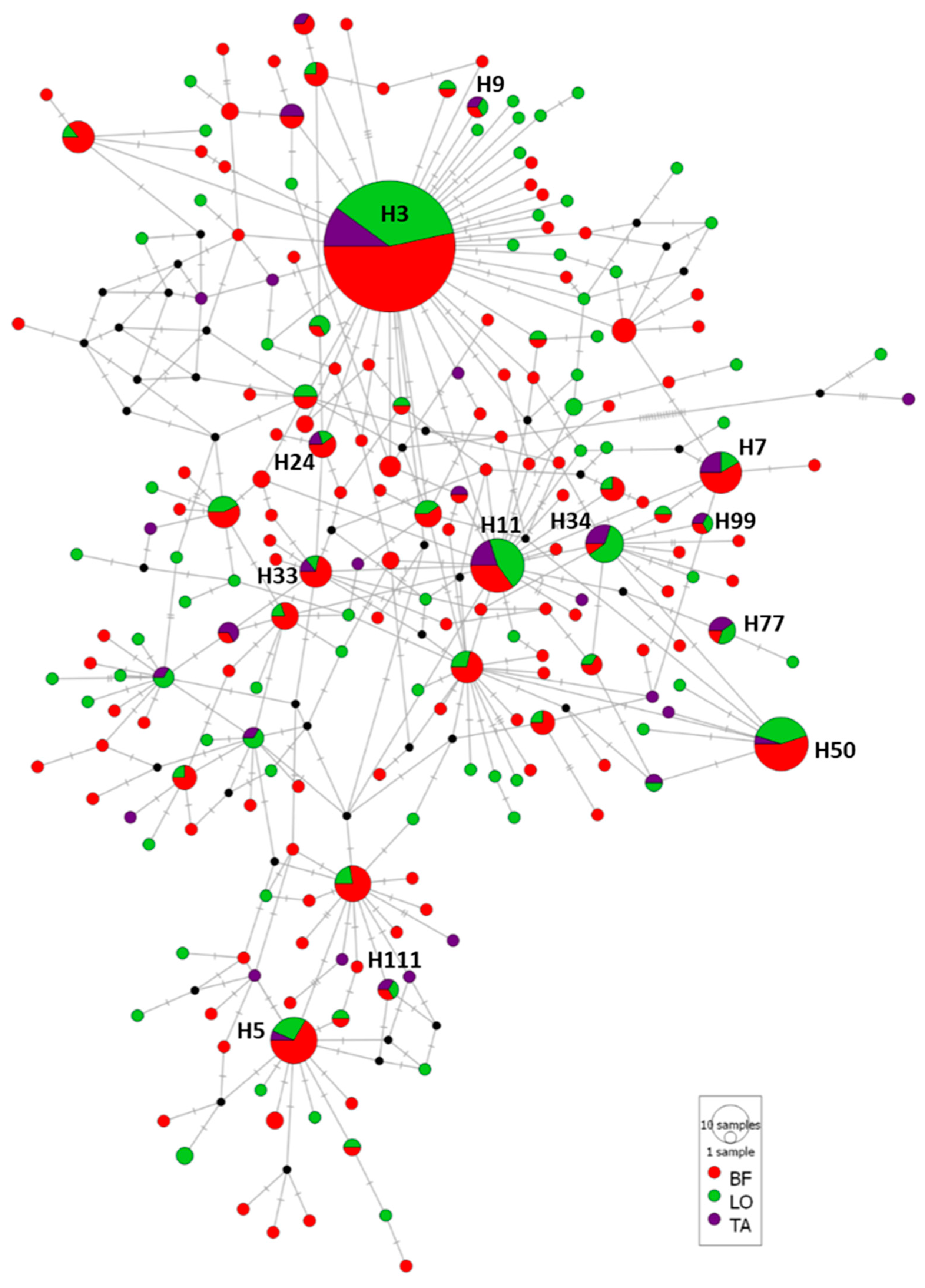
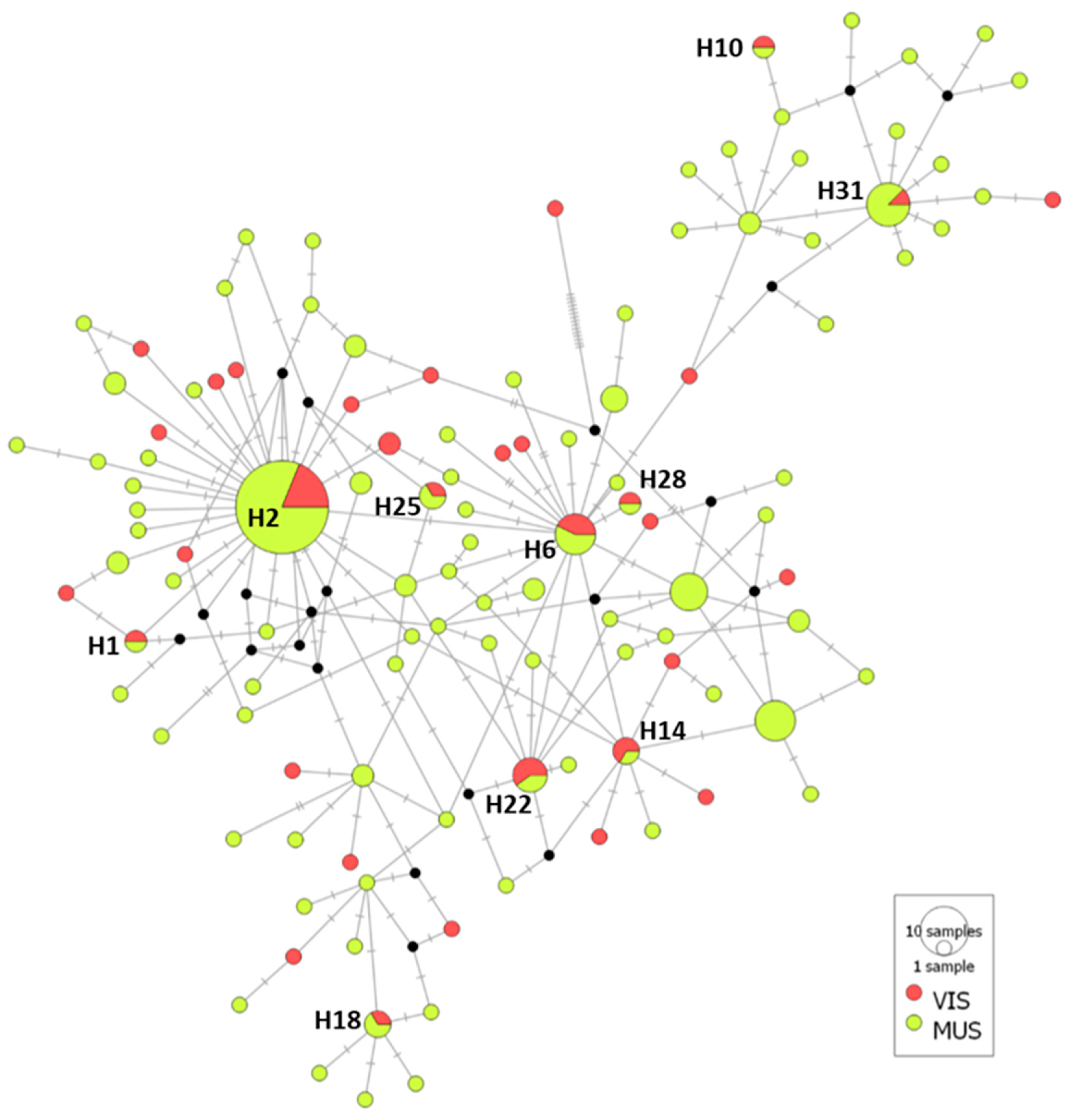
3.2. Genetic Diversity and Population Structure of A. simplex
3.2.1. Case Study 1: A. simplex from Different ICES Divisions
3.2.2. Case Study 2: A. simplex from Different Fish Muscular Sections
3.2.3. Case Study 3: A. simplex from Fish Muscle Versus Viscera
4. Discussion
4.1. Genetic Identification of Anisakis Species of European Hake
4.2. Genetic Diversity and Population Structure of A. simplex Infecting M. merluccius from Different ICES Divisions
4.3. Genetic Diversity and Population Structure of A. simplex from Fish Sections
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ICES. Report of the Working Group on the Assessment of Southern Shelf stocks of Hake, Monk and Megrim (WGHMM). ICES CM 2011/ACOM: 11; ICES: Copenhagen, Denmark, 2011; p. 625. [Google Scholar]
- ICES. Report of the Working Group for the Bay of Biscay and the Iberian Waters Ecoregion (WGBIE). ICES CM 2018/ACOM:12; ICES: Copenhagen, Denmark, 2018; p. 585. [Google Scholar]
- Pascual, S.; Rodríguez, H.; Pierce, G.J.; Hastie, L.C.; González, A.F. The NE Atlantic European hake: A neglected high exposure risk for zoonotic parasites in European fish markets. Fish. Res. 2018, 202, 69–78. [Google Scholar] [CrossRef]
- Rodríguez, H.; Abollo, E.; González, F.; Pascual, S. Scoring the parasite risk in highly-valuable fish species from southern ICES areas. Fish. Res. 2018, 202, 134–139. [Google Scholar] [CrossRef]
- Diez, G.; Chust, G.; Andonegi, E.; Santurtún, M.; Abaroa, C.; Bilbao, E.; Maceira, A.; Mendibil, I. Analysis of potential drivers of spatial and temporal changes in anisakid larvae infection levels in European hake, Merluccius merluccius (L.), from the North-East Atlantic fishing grounds. Fish Parasitol. 2022, 121, 1903–1920. [Google Scholar] [CrossRef] [PubMed]
- Llarena-Reino, M.; Abollo, E.; Regueira, M.; Rodríguez, H.; Pascual, S. Horizon scanning for management of emerging parasitic infections in fishery products. Food Control. 2015, 49, 49–58. [Google Scholar] [CrossRef]
- Abollo, E.; López, A.; Gestal, C.; Benavente, P.; Pascual, S. Macroparasites in cetaceans stranded on the northwestern Spanish Atlantic coast. Dis. Aquat. Organ. 1998, 32, 227–231. [Google Scholar] [CrossRef]
- Abollo, E.; Paggi, L.; Pascual, S.; D’Amelio, S. Occurrence of recombinant genotypes of Anisakis simplex s.s. and Anisakis pegreffii (Nematoda: Anisakidae) in an area of sympatry. Infect. Genet. Evol. 2003, 3, 175–181. [Google Scholar] [CrossRef]
- Mattiucci, S.; Cipriani, P.; Levsen, A.; Paoletti, M.; Nascetti, G. Molecular Epidemiology of Anisakis and Anisakiasis: An Ecological and Evolutionary Road Map. Adv. Parasitol. 2018, 99, 93–263. [Google Scholar] [CrossRef]
- Cipriani, P.; Smaldone, G.; Acerra, V.; D’Angelo, L.; Anastasio, A.; Bellisario, B.; Palma, G.; Nascetti, G.; Mattiucci, S. Genetic identification and distribution of the parasitic larvae of Anisakis pegreffii and Anisakis simplex (s. s.) in European hake Merluccius merluccius from the Tyrrhenian Sea and Spanish Atlantic coast: Implications for food safety. Int. J. Food Microbiol. 2015, 198, 1–8. [Google Scholar] [CrossRef]
- Cipriani, P.; Palomba, M.; Giulietti, L.; Marcer, F.; Mazzariol, S.; Santoro, M.; Alburqueque, R.A.; Covelo, P.; López, A.; Santos, M.B.; et al. Distribution and genetic diversity of Anisakis spp. in cetaceans from the Northeast Atlantic Ocean and the Mediterranean Sea. Sci. Rep. 2022, 12, 13664. [Google Scholar] [CrossRef]
- Gregori, M.; Roura, Á.; Abollo, E.; González, Á.F.; Pascual, S. Anisakis simplex complex (Nematoda: Anisakidae) in zooplankton communities from temperate NE Atlantic waters. J. Nat. Hist. 2015, 49, 755–773. [Google Scholar] [CrossRef]
- Levsen, A.; Svanevik, C.S.; Cipriani, P.; Mattiucci, S.; Gay, M.; Hastie, L.C.; Buselic, I.; Mladineo, I.; Karl, H.G.N.D.; Ostermeyer, U.; et al. A survey of zoonotic nematodes of commercial key fish species from major European fishing grounds—Introducing the FP7 PARASITE exposure assessment study. Fish. Res. 2018, 202, 4–21. [Google Scholar] [CrossRef]
- Abattouy, N.; Valero, A.; Lozano, J.; Barón, S.; Romero, C.; Martín-Sánchez, J. Population genetic analysis of Anisakis simplex s.l. and Anisakis pegreffii (Nematoda, Anisakidae) from parapatric areas and their contact zone. Parasite Epidemiol. Control 2016, 1, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Roca-Geronès, X.; Alcover, M.M.; Godínez-González, C.; Montoliu, I.; Fisa, R. Hybrid Genotype of Anisakis simplex (s.s.) and A. pegreffii Identified in Third- and Fourth-Stage Larvae from Sympatric and Allopatric Spanish Marine Waters. Animals 2021, 11, 2458. [Google Scholar] [CrossRef] [PubMed]
- Mahe, K.; Amara, R.; Bryckaert, T.; Kacher, M.; Brylinski, J.M. Ontogenetic and spatial variation in the diet of hake (Merluccius merluccius) in the Bay of Biscay and the Celtic Sea. ICES J. Mar. Sci. 2007, 64, 1210–1219. [Google Scholar] [CrossRef]
- Perkins, S.L. Phylogeography of Caribbean lizard malaria: Tracing the history of vector-borne parasites. J. Evol. Biol. 2001, 14, 34–45. [Google Scholar] [CrossRef]
- Criscione, C.D.; Poulin, R.; Blouin, M.S. Molecular ecology of parasites: Elucidating ecological and microevolutionary processes. Mol. Ecol. 2005, 14, 2247–2257. [Google Scholar] [CrossRef]
- Gilabert, A.; Wasmuth, J.D. Unravelling parasitic nematode natural history using population genetics. Trends Parasitol. 2013, 29, 438–448. [Google Scholar] [CrossRef]
- MacKenzie, K. Parasites as biological tags in population studies of marine organisms: An update. Parasitology 2002, 124, S153–S163. [Google Scholar] [CrossRef]
- Zhu, X.; D’Amelio, S.; Paggi, L.; Gasser, R.B. Assessing sequence variation in the internal transcribed spacers of ribosomal DNA within and among members of the Contracaecum osculatum complex (Nematoda: Ascaridoidea: Anisakidae). Parasitol. Res. 2000, 86, 677–683. [Google Scholar] [CrossRef]
- Nadler, S.A.; Hudspeth, D.S.S. Phylogeny of the Ascaridoidea (Nematoda: Ascaridida) Based on Three Genes and Morphology: Hypotheses of Structural and Sequence Evolution. J. Parasitol. 2000, 86, 380–393. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 1989, 3, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.-X. Statistical Tests of Neutrality of Mutations Against Population Growth, Hitchhiking and Background Selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Slatkin, M. A measure of population subdivision based on microsatellite allele frequencies. Genetics 1995, 139, 457–462. [Google Scholar] [CrossRef]
- Mattiucci, S.; Acerra, V.; Paoletti, M.; Cipriani, P.; Levsen, A.; Webb, S.C.; Canestrelli, D.; Nascetti, G. No more time to stay ‘single’ in the detection of Anisakis pegreffii, A. simplex (s. s.) and hybridization events between them: A multi-marker nuclear genotyping approach. Parasitology 2016, 143, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Mateos, M.; Merino-Espinosa, G.; Corpas-López, V.; Valero-López, A.; Martín-Sánchez, J. A multi-restriction fragment length polymorphism genotyping approach including the beta-tubulin gene as a new differential nuclear marker for the recognition of the cryptic species Anisakis simplex s.s. and Anisakis pegreffii and their hybridization events. Vet. Parasitol. 2020, 283, 109162. [Google Scholar] [CrossRef] [PubMed]
- Palomba, M.; Paoletti, M.; Webb, S.C.; Nascetti, G.; Mattiucci, S. A novel nuclear marker and development of an ARMS-PCR assay targeting the metallopeptidase 10 (nas 10) locus to identify the species of the Anisakis simplex (s. l.) complex (Nematoda, Anisakidae). Parasite 2020, 27, 39. [Google Scholar] [CrossRef]
- Mattiucci, S.; Cipriani, P.; Webb, S.C.; Paoletti, M.; Marcer, F.; Bellisario, B.; Gibson, D.I.; Nascetti, G. Genetic and Morphological Approaches Distinguish the Three Sibling Species of the Anisakis simplex Species Complex, with a Species Designation as Anisakis berlandi n. sp. For A. simplex sp. C (Nematoda: Anisakidae). J. Parasitol. 2014, 100, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Mladineo, I.; Trumbić, Ž.; Radonić, I.; Vrbatović, A.; Hrabar, J.; Bušelić, I. Anisakis simplex complex: Ecological significance of recombinant genotypes in an allopatric area of the Adriatic Sea inferred by genome-derived simple sequence repeats. Int. J. Parasitol. 2017, 47, 215–223. [Google Scholar] [CrossRef]
- Mattiucci, S.; Bello, E.; Paoletti, M.; Webb, S.C.; Timi, J.T.; Levsen, A.; Cipriani, P.; Nascetti, G. Novel polymorphic microsatellite loci in Anisakis pegreffii and A. simplex (s. s.) (Nematoda: Anisakidae): Implications for species recognition and population genetic analysis. Parasitology 2019, 146, 1387–1403. [Google Scholar] [CrossRef] [PubMed]
- Steinauer, M.L.; Hanelt, B.; Mwangi, I.N.; Maina, G.M.; Agola, L.E.; Kinuthia, J.M.; Mutuku, M.W.; Mungai, B.N.; Wilson, W.D.; Mkoji, G.M.; et al. Introgressive hybridization of human and rodent schistosome parasites in western Kenya. Mol. Ecol. 2008, 17, 5062–5074. [Google Scholar] [CrossRef]
- Baldwin, R.E.; Rew, M.B.; Johansson, M.L.; Banks, M.A.; Jacobson, K.C. Population Structure of Three Species of Anisakis Nematodes Recovered from Pacific Sardines (Sardinops sagax) Distributed Throughout the California Current System. J. Parasitol. 2011, 97, 545–554. [Google Scholar] [CrossRef]
- Klapper, R.; Kochmann, J.; O’Hara, R.B.; Karl, H.; Kuhn, T. Parasites as Biological Tags for Stock Discrimination of Beaked Redfish (Sebastes mentella): Parasite Infra-Communities vs. Limited Resolution of Cytochrome Markers. PLoS ONE 2016, 11, e0153964. [Google Scholar] [CrossRef]
- Mattiucci, S.; Giulietti, L.; Paoletti, M.; Cipriani, P.; Gay, M.; Levsen, A.; Klapper, R.; Karl, H.; Bao, M.; Pierce, G.J.; et al. Population genetic structure of the parasite Anisakis simplex (s. s.) collected in Clupea harengus L. from North East Atlantic fishing grounds. Fish. Res. 2018, 202, 103–111. [Google Scholar] [CrossRef]
- Gu, X.-B.; Wang, B.-J.; Zhao, X.-B.; Li, Y.-F.; Yang, G.-Y.; Lai, W.-M.; Zhong, Z.-J.; Peng, G.-N. Genetic variation in mitochondrial cox2 of Heterakis gallinarum from poultry in Sichuan, China. Mitochondr. DNA Part A 2018, 29, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Huang, H.Q.; Wang, H.Y.; Zhang, D.; Chu, H.J.; Ma, X.P.; Ge, Y.; Ente, M.; Li, K. Genetic diversity of common Gasterophilus spp. from distinct habitats in China. Parasites Vectors 2018, 11, 474. [Google Scholar] [CrossRef] [PubMed]
- Amor, N.; Farjallah, S.; Piras, M.C.; Burreddu, C.; Garippa, G.; Merella, P. New insights into the coexistence of Contracaecum rudolphii A and Contracaecum rudolphii B (Nematoda: Anisakidae) in Phalacrocorax carbo sinensis from Sardinia: Genetic variability and phylogenetic analysis. Parasitology 2020, 147, 1538–1551. [Google Scholar] [CrossRef]
- Azrizal-Wahid, N.; Sofian-Azirun, M.; Low, V.L. New insights into the haplotype diversity of the cosmopolitan cat flea Ctenocephalides felis (Siphonaptera: Pulicidae). Vet. Parasitol. 2020, 281, 109102. [Google Scholar] [CrossRef] [PubMed]
- Mattiucci, S.; Farina, V.; Campbell, N.; MacKenzie, K.; Ramos, P.; Pinto, A.L.; Abaunza, P.; Nascetti, G. Anisakis spp. larvae (Nematoda: Anisakidae) from Atlantic horse mackerel: Their genetic identification and use as biological tags for host stock characterization. Fish. Res. 2008, 89, 146–151. [Google Scholar] [CrossRef]
- Quiazon, K.M.A.; Yoshinaga, T.; Ogawa, K. Experimental challenge of Anisakis simplex sensu stricto and Anisakis pegreffii (Nematoda: Anisakidae) in rainbow trout and olive flounder. Parasitol. Int. 2011, 60, 126–131. [Google Scholar] [CrossRef]
- Arizono, N.; Yamada, M.; Tegoshi, T.; Yoshikawa, M. Anisakis simplex sensu stricto and Anisakis pegreffii: Biological Characteristics and Pathogenetic Potential in Human Anisakiasis. Foodborne Pathog. Dis. 2012, 9, 517–521. [Google Scholar] [CrossRef]
- Romero, M.D.C.; Valero, A.; Navarro-Moll, M.C.; Martín-Sánchez, J. Experimental comparison of pathogenic potential of two sibling species Anisakis simplex s.s. and Anisakis pegreffii in Wistar rat. Trop. Med. Int. Health 2013, 18, 979–984. [Google Scholar] [CrossRef] [PubMed]
- Cipriani, P.; Sbaraglia, G.L.; Paoletti, M.; Giulietti, L.; Bellisario, B.; Palomba, M.; Bušelić, I.; Mladineo, I.; Nascetti, G.; Mattiucci, S. The Mediterranean European hake, Merluccius merluccius: Detecting drivers influencing the Anisakis spp. larvae distribution. Fish. Res. 2018, 202, 79–89. [Google Scholar] [CrossRef]
- Santoro, M.; Palomba, M.; Mattiucci, S.; Osca, D.; Crocetta, F. New Parasite Records for the Sunfish Mola mola in the Mediterranean Sea and Their Potential Use as Biological Tags for Long-Distance Host Migration. Front. Vet. Sci. 2020, 7, 579728. [Google Scholar] [CrossRef] [PubMed]
- Pons-Bordas, C.; Hazenberg, A.; Hernandez-Gonzalez, A.; Pool, R.V.; Covelo, P.; Sánchez-Hermosin, P.; López, A.; Saavedra, C.; Fraija-Fernández, N.; Fernández, M.; et al. Recent increase of ulcerative lesions caused by Anisakis spp. in cetaceans from the north-east Atlantic. J. Helminthol. 2020, 94, e127. [Google Scholar] [CrossRef] [PubMed]
- Blažeković, K.; Pleić, I.L.; Đuras, M.; Gomerčić, T.; Mladineo, I. Three Anisakis spp. isolated from toothed whales stranded along the eastern Adriatic Sea coast. Int. J. Parasitol. 2015, 45, 17–31. [Google Scholar] [CrossRef]
- Cole, R.; Viney, M. The population genetics of parasitic nematodes of wild animals. Parasites Vectors 2018, 11, 590. [Google Scholar] [CrossRef]
- Irigoitia, M.M.; Palomba, M.; Braicovich, P.E.; Lanfranchi, A.L.; Denuncio, P.E.; Gana, J.C.M.; Mattiucci, S.; Timi, J.T. Genetic identification of Anisakis spp. (Nematoda: Anisakidae) from cetaceans of the Southwestern Atlantic Ocean: Ecological and zoogeographical implications. Parasitol. Res. 2021, 120, 1699–1711. [Google Scholar] [CrossRef] [PubMed]
- Tanner, S.E.; Vasconcelos, R.P.; Cabral, H.N.; Thorrold, S.R. Testing an otolith geochemistry approach to determine population structure and movements of European hake in the northeast Atlantic Ocean and Mediterranean Sea. Fish. Res. 2012, 125–126, 198–205. [Google Scholar] [CrossRef]
- Pita, A.; Pérez, M.; Cerviño, S.; Presa, P. What can gene flow and recruitment dynamics tell us about connectivity between European hake stocks in the Eastern North Atlantic? Cont. Shelf Res. 2011, 31, 376–387. [Google Scholar] [CrossRef]
- Pita, A.; Pérez, M.; Balado, M.; Presa, P. Out of the Celtic cradle: The genetic signature of European hake connectivity in South-western Europe. J. Sea Res. 2014, 93, 90–100. [Google Scholar] [CrossRef]
- Leone, A.; Álvarez, P.; García, D.; Saborido-Rey, F.; Rodriguez-Ezpeleta, N. Genome-wide SNP based population structure in European hake reveals the need for harmonizing biological and management units. ICES J. Mar. Sci. 2019, 76, 2260–2266. [Google Scholar] [CrossRef]
- Mäkeläinen, P.; Esteban, R.; Foote, A.D.; Kuningas, S.; Nielsen, J.; Samarra, F.I.; Similä, T.; van Geel, N.C.; Víkingsson, G.A. A comparison of pigmentation features among North Atlantic killer whale (Orcinus orca) populations. J. Mar. Biol. Assoc. UK 2014, 94, 1335–1341. [Google Scholar] [CrossRef]
- Cheney, B.; Thompson, P.M.; Ingram, S.N.; Hammond, P.S.; Stevick, P.T.; Durban, J.W.; Culloch, R.M.; Elwen, S.H.; Mandleberg, L.; Janik, V.M.; et al. Integrating multiple data sources to assess the distribution and abundance of bottlenose dolphins Tursiops truncatus in Scottish waters. Mammal Rev. 2012, 43, 71–88. [Google Scholar] [CrossRef]
- Hodgins, N.K.; Dolman, S.J.; Weir, C.R. Potential hybridism between free-ranging Risso’s dolphins (Grampus griseus) and bottlenose dolphins (Tursiops truncatus) off north-east Lewis (Hebrides, UK). Mar. Biodivers. Rec. 2014, 7, E97. [Google Scholar] [CrossRef]
- Russell, D.J.F.; Morris, C.D.; Duck, C.D.; Thompson, D.; Hiby, L. Monitoring long-term changes in UK grey seal pup production. Aquat. Conserv. Mar. Freshw. Ecosyst. 2019, 29, 24–39. [Google Scholar] [CrossRef]

| ICES Area | As | Ap | Hyb | Total | |
|---|---|---|---|---|---|
| IVa | BF | 62 | 0 | 3 | 65 |
| LO | 8 | 0 | 0 | 8 | |
| TA | 1 | 0 | 0 | 1 | |
| Total | 71 | 0 | 3 | 74 | |
| VIa | BF | 57 | 0 | 0 | 57 |
| LO | 13 | 0 | 1 | 14 | |
| TA | 1 | 0 | 0 | 1 | |
| Total | 71 | 0 | 1 | 72 | |
| VIIb | BF | 59 | 0 | 0 | 59 |
| LO | 11 | 0 | 1 | 12 | |
| TA | 4 | 0 | 0 | 4 | |
| Total | 74 | 0 | 1 | 75 | |
| VIIc | BF | 42 | 0 | 0 | 42 |
| LO | 23 | 0 | 1 | 24 | |
| TA | 9 | 0 | 1 | 10 | |
| Total | 74 | 0 | 2 | 76 | |
| VIIj | BF | 24 | 0 | 0 | 24 |
| LO | 31 | 0 | 2 | 33 | |
| TA | 6 | 0 | 0 | 6 | |
| Total | 61 | 0 | 2 | 63 | |
| VIIh | BF | 37 | 0 | 1 | 38 |
| LO | 35 | 0 | 0 | 35 | |
| TA | 2 | 0 | 0 | 2 | |
| Total | 74 | 0 | 1 | 75 | |
| VIIIa | BF | 2 | 0 | 0 | 2 |
| LO | 49 | 2 | 3 | 54 | |
| TA | 12 | 1 | 2 | 15 | |
| Total | 63 | 3 | 5 | 71 | |
| VIIIc | BF | 37 | 0 | 0 | 37 |
| LO | 28 | 2 | 3 | 33 | |
| TA | 6 | 0 | 0 | 6 | |
| Total | 71 | 2 | 3 | 76 | |
| VIIId | BF | 11 | 0 | 0 | 11 |
| LO | 37 | 0 | 1 | 38 | |
| TA | 25 | 0 | 0 | 25 | |
| Total | 73 | 0 | 1 | 74 | |
| IXa | BF | 61 | 6 | 7 | 74 |
| LO | 1 | 0 | 0 | 1 | |
| TA | 1 | 1 | 0 | 2 | |
| Total | 63 | 7 | 7 | 77 | |
| Overall | 695 | 12 | 26 | 733 |
| ICES Area | As | Ap | Hyb | Total |
|---|---|---|---|---|
| VIIj | 44 | 0 | 0 | 44 |
| VIIIc | 22 | 19 | 10 | 51 |
| IXa | 10 | 24 | 11 | 45 |
| Overall | 76 | 43 | 21 | 140 |
| ICES Areas | N | Nh | Nuh | Pi | Hd ± SD | K | S | Tajima’s D | Fu’s Fs | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| D | P | Fs | P | ||||||||
| IVa | 44 | 27 | 13 | 0.00613 | 0.930 ± 0.030 | 2.96300 | 30 | −1.93422 | 0.00800 * | −24.42734 | 0.00000 * |
| VIa | 64 | 34 | 15 | 0.00496 | 0.848 ± 0.045 | 2.39633 | 30 | −2.00076 | 0.00700 * | −26.84892 | 0.00000 * |
| VIIb | 69 | 43 | 22 | 0.00718 | 0.937 ± 0.024 | 3.46974 | 41 | −1.93982 | 0.00800 * | −26.18191 | 0.00000 * |
| VIIc | 35 | 26 | 14 | 0.00948 | 0.965 ± 0.020 | 4.57815 | 44 | −2.06327 | 0.00200 * | −19.69666 | 0.00000 * |
| VIIj | 50 | 33 | 19 | 0.00740 | 0.953 ± 0.021 | 3.57633 | 34 | −1.77822 | 0.01000 * | −26.07247 | 0.00000 * |
| VIIh | 18 | 13 | 6 | 0.00755 | 0.902 ± 0.066 | 3.64706 | 17 | −1.00685 | 0.12800 | −6.41729 | 0.00100 * |
| VIIIa | 59 | 45 | 25 | 0.00860 | 0.970 ± 0.015 | 4.15546 | 57 | −2.25027 | 0.00100 * | −25.85918 | 0.00000 * |
| VIIIc | 60 | 42 | 19 | 0.00752 | 0.956 ± 0.019 | 3.63277 | 36 | −1.74418 | 0.01700 * | −26.07312 | 0.00000 * |
| VIIId | 65 | 38 | 18 | 0.00675 | 0.938 ± 0.022 | 3.25962 | 32 | −1.66792 | 0.02500 * | −26.27973 | 0.00000 * |
| IXa | 46 | 33 | 17 | 0.00764 | 0.966 ± 0.018 | 3.69082 | 32 | −1.67002 | 0.02700 * | −26.01534 | 0.00000 * |
| Overall | 510 | 215 | - | 0.00723 | 0.938 ± 0.009 | 3.49440 | 115 | −2.31355 | 0.00000 * | −25.42466 | 0.00100 * |
| IVa | VIa | VIIb | VIIc | VIIj | VIIh | VIIIa | VIIIc | VIIId | |
|---|---|---|---|---|---|---|---|---|---|
| IVa | - | ||||||||
| VIa | 0.02373 * | - | |||||||
| VIIb | −0.00631 | 0.02802 * | - | ||||||
| VIIc | 0.00118 | 0.02149 * | −0.00203 | - | |||||
| VIIj | 0.01078 | 0.04431 * | 0.00325 | −0.00585 | - | ||||
| VIIh | 0.00614 | 0.06270 * | 0.00116 | −0.00112 | 0.01541 | - | |||
| VIIIa | −0.00411 | 0.01503 * | −0.00565 | −0.01071 | −0.00473 | 0.00953 | - | ||
| VIIIc | 0.00025 | 0.04427 * | 0.00058 | 0.00612 | −0.00073 | 0.00587 | −0.00036 | - | |
| VIIId | −0.00296 | 0.02871 * | −0.00479 | −0.00732 | −0.00082 | 0.01133 | −0.00553 | 0.00177 | - |
| IXa | −0.00303 | 0.04545 * | −0.00188 | 0.00673 | 0.01137 | −0.01415 | 0.00173 | −0.00629 | 0.00031 |
| N | Nh | Nuh | Pi | Hd ± SD | K | S | Tajima’s D | Fu’s Fs | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| D | P | Fs | P | ||||||||
| BF | 280 | 135 | 100 | 0.00696 | 0.942 ± 0.011 | 3.36193 | 81 | −2.21675 | 0.00000 * | −25.84468 | 0.00000 * |
| LO | 176 | 97 | 63 | 0.00751 | 0.932 ± 0.016 | 3.62675 | 74 | −2.21567 | 0.00100 * | −25.90587 | 0.00000 * |
| TA | 54 | 33 | 14 | 0.00783 | 0.943 ± 0.023 | 3.78337 | 45 | −2.09405 | 0.00400 * | −25.94050 | 0.00000 * |
| Overall | 510 | 215 | - | 0.00723 | 0.938 ± 0.009 | 3.49440 | 115 | −2.31355 | 0.00000 * | −25.42466 | 0.00100 * |
| N | Nh | Nuh | Pi | Hd ± SD | k | S | Tajima’s D | Fu’s Fs | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| D | P | Fs | P | ||||||||
| VIS | 45 | 33 | 23 | 0.00846 | 0.971 ± 0.016 | 4.08687 | 45 | −2.09847 | 0.00500 * | −25.91039 | 0.00000 * |
| MUS | 156 | 93 | 81 | 0.00752 | 0.958 ± 0.012 | 3.63383 | 61 | −2.04939 | 0.00200 * | −25.84799 | 0.00000 * |
| Overall | 201 | 116 | - | 0.00773 | 0.961 ± 0.010 | 3.73567 | 78 | −2.19753 | 0.00000 * | −25.76160 | 0.00000 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramilo, A.; Rodríguez, H.; Pascual, S.; González, Á.F.; Abollo, E. Population Genetic Structure of Anisakis simplex Infecting the European Hake from North East Atlantic Fishing Grounds. Animals 2023, 13, 197. https://doi.org/10.3390/ani13020197
Ramilo A, Rodríguez H, Pascual S, González ÁF, Abollo E. Population Genetic Structure of Anisakis simplex Infecting the European Hake from North East Atlantic Fishing Grounds. Animals. 2023; 13(2):197. https://doi.org/10.3390/ani13020197
Chicago/Turabian StyleRamilo, Andrea, Helena Rodríguez, Santiago Pascual, Ángel F. González, and Elvira Abollo. 2023. "Population Genetic Structure of Anisakis simplex Infecting the European Hake from North East Atlantic Fishing Grounds" Animals 13, no. 2: 197. https://doi.org/10.3390/ani13020197
APA StyleRamilo, A., Rodríguez, H., Pascual, S., González, Á. F., & Abollo, E. (2023). Population Genetic Structure of Anisakis simplex Infecting the European Hake from North East Atlantic Fishing Grounds. Animals, 13(2), 197. https://doi.org/10.3390/ani13020197







