NCAPG Regulates Myogenesis in Sheep, and SNPs Located in Its Putative Promoter Region Are Associated with Growth and Development Traits
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Sample and Phenotypic Data Collection
2.3. Isolation of Sheep Embryonic Myoblasts
2.4. Plasmid Construction and Small Interfering RNA Synthesis
2.5. Cell Transfection and Induction of Differentiation
2.6. Total RNA Extraction, Reverse Transcription, and RT-PCR
2.7. Cell Proliferation Detection
2.7.1. CCK-8 Assay
2.7.2. EdU Proliferation Assay
2.7.3. Flow Cytometry for Cell Cycle Analysis
2.8. Cell Differentiation Detection
2.8.1. Cell Immunofluorescence Staining
- (1)
- Discard the cell culture medium; rinse 1–2 times with 1 × PBS on a destaining shaker; add 500 μL cell fixative to each well and incubate at room temperature for 15–30 min;
- (2)
- Discard the fixative and rinse with 1 × PBS destaining shaker 3–4 times, 5 min/time; add 500 μL of 0.5% Triton X-100 to each well, and incubate with destaining shaker for 20–30 min to enhance cell membrane permeability;
- (3)
- Discard the permeabilization solution and rinse with 1 × PBS decolorizing shaker 3–4 times, 5 min/time; add 500 μL of freshly prepared 10% goat serum working solution to each well, and incubate at 37 °C in the dark for 60 min;
- (4)
- Discard the blocking solution, add 300 μL of freshly prepared primary antibody working solution (MyHC, 1:200) to each well, and incubate overnight at 4 °C in the dark;
- (5)
- Discard the primary antibody working solution, and rinse with 1 × PBS decolorizing shaker 3–4 times, 5 min/time; add 300 μL of freshly prepared secondary antibody working solution (Goat Anti-Rabbit IgG (H + L)) to each well, 1:1000), incubate at 37 °C in the dark for 60 min;
- (6)
- Discard the secondary antibody working solution and rinse with 1 × PBS destaining shaker 3–4 times, 5 min/time;
- (7)
- Add 300 μL of DAPI working solution to each well, and incubate at room temperature in the dark for 3–5 min; rinse with 1 × PBS decolorizing shaker 3–4 times, 5 min each time, and immediately place it under an inverted fluorescence microscope to take pictures and observe.
2.8.2. MRF Quantification
2.9. Sequencing and Genotyping of Putative NCAPG Promoter Region
2.10. Statistical Analysis
3. Results
3.1. NCAPG Was Highly Expressed in Embryonic Sheep Muscle
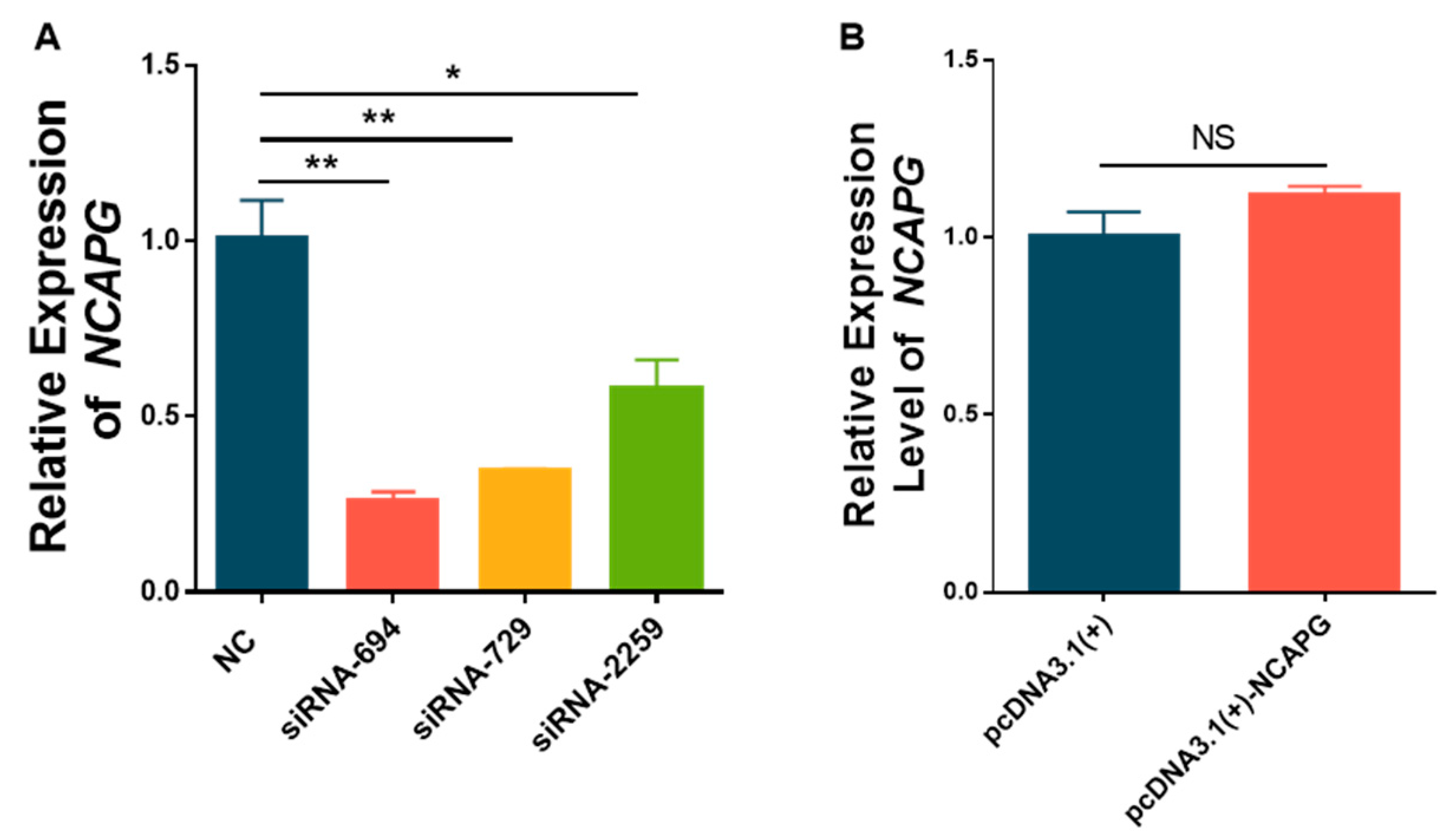
3.2. The Expression of NCAPG Was Reduced by siRNA Knockdown
3.3. Interfering with NCAPG Inhibits the Proliferation of Sheep Fetal Myoblasts
3.4. Interfering with NCAPG Hinders the Differentiation of Sheep Fetal Myoblasts
3.5. Five SNPs in the Putative Promoter Region of NCAPG
3.6. SNPs in NCAPG Promoter Region Associated with Sheep Growth and Developmental Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Meuwissen, T.; Hayes, B.; Goddard, M. Genomic selection: A paradigm shift in animal breeding. Anim. Front. 2016, 6, 6–14. [Google Scholar] [CrossRef]
- Ducrocq, V.; Laloe, D.; Swaminathan, M.; Rognon, X.; Tixier-Boichard, M.; Zerjal, T. Genomics for Ruminants in Developing Countries: From Principles to Practice. Front. Genet. 2018, 9, 251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, J.; Zhao, F.; Ren, H.; Xu, L.; Lu, J.; Zhang, S.; Zhang, X.; Wei, C.; Lu, G.; et al. Genome-Wide Association Studies for Growth and Meat Production Traits in Sheep. PLoS ONE 2013, 8, e66569. [Google Scholar] [CrossRef] [PubMed]
- Al-Mamun, H.A.; Kwan, P.; Clark, S.A.; Ferdosi, M.H.; Tellam, R.; Gondro, C. Genome-wide association study of body weight in Australian Merino sheep reveals an orthologous region on OAR6 to human and bovine genomic regions affecting height and weight. Genet. Sel. Evol. 2015, 47, 66. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, M.; Rahimi-Mianji, G.; Nejati-Javaremi, A. Genomewide association study of body weight traits in Baluchi sheep. J. Genet. 2015, 94, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Bolormaa, S.; Hayes, B.J.; van der Werf, J.H.J.; Pethick, D.; Goddard, M.E.; Daetwyler, H.D. Detailed phenotyping identifies genes with pleiotropic effects on body composition. BMC Genom. 2016, 17, 224. [Google Scholar] [CrossRef] [PubMed]
- Matika, O.; Riggio, V.; Anselme-Moizan, M.; Law, A.S.; Pong-Wong, R.; Archibald, A.L.; Bishop, S.C. Genome-wide association reveals QTL for growth, bone and in vivo carcass traits as assessed by computed tomography in Scottish Blackface lambs. Genet. Sel. Evol. 2016, 48, 11. [Google Scholar] [CrossRef]
- Kominakis, A.; Hager-Theodorides, A.L.; Zoidis, E.; Saridaki, A.; Antonakos, G.; Tsiamis, G. Combined GWAS and ‘guilt by association’-based prioritization analysis identifies functional candidate genes for body size in sheep. Genet. Sel. Evol. 2017, 49, 41. [Google Scholar] [CrossRef]
- Ghasemi, M.; Zamani, P.; Vatankhah, M.; Abdoli, R. Genome-wide association study of birth weight in sheep. Animal 2019, 13, 1797–1803. [Google Scholar] [CrossRef]
- Almasi, M.; Zamani, P.; Mirhoseini, S.Z.; Moradi, M.H. Genome-Wide Association Study of Weaning Traits in Lori-Bakhtiari Sheep. Ann. Anim. Sci. 2020, 20, 811–824. [Google Scholar] [CrossRef]
- Cao, Y.; Song, X.; Shan, H.; Jiang, J.; Xiong, P.; Wu, J.; Shi, F.; Jiang, Y. Genome-Wide Association Study of Body Weights in Hu Sheep and Population Verification of Related Single-Nucleotide Polymorphisms. Front. Genet. 2020, 11, 588. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Yue, Y.; Yuan, C.; Liu, J.; Chen, Z.; Niu, C.; Sun, X.; Zhu, S.; Zhao, H.; Guo, T.; et al. Genome-Wide Association Study of Body Weight Traits in Chinese Fine-Wool Sheep. Animals 2020, 10, 170. [Google Scholar] [CrossRef] [PubMed]
- Pasandideh, M.; Gholizadeh, M.; Rahimi-Mianji, G. Identification of two novel SNPs affecting lambing traits in sheep by using a 50K SNP-Chip. Small Rumin. Res. 2020, 191, 106193. [Google Scholar] [CrossRef]
- Pasandideh, M.; Gholizadeh, M.; Rahimiimianji, G. A genome-wide association study revealed five SNPs affecting 8-month weight in sheep. Anim. Genet. 2020, 51, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; He, X.Y.; Pan, L.X.; Wang, J.W.; Gan, S.Q.; Chu, M.X.; Genomeiimianji, G. Genome-wide association study of body weight and conformation traits in neonatal sheep. Anim. Genet. 2020, 51, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Almasi, M.; Zamani, P.; Mirhoseini, S.Z.; Moradi, M.H. Genome-wide association study for postweaning weight traits in Lori-Bakhtiari sheep. Trop. Anim. Health Prod. 2021, 53, 163. [Google Scholar] [CrossRef]
- Jiang, J.; Cao, Y.; Shan, H.; Wu, J.; Song, X.; Jiang, Y. The GWAS Analysis of Body Size and Population Verification of Related SNPs in Hu Sheep. Front. Genet. 2021, 12, 642552. [Google Scholar] [CrossRef]
- Krivoruchko, A.; Yatsyk, O.; Kanibolockaya, A.; Kulintsev, V. Genome-wide association study (GWAS) of high productivity classes in the Karachaevsky sheep breed. J. Cent. Eur. Agric. 2021, 22, 669–677. [Google Scholar] [CrossRef]
- McLaren, A.; Kaseja, K.; McLean, K.A.; Boon, S.; Lambe, N.R. Genetic analyses of novel traits derived from CT scanning for implementation in terminal sire sheep breeding programmes. Livest. Sci. 2021, 250, 104555. [Google Scholar] [CrossRef]
- Tao, L.; Liu, Y.F.; Zhang, H.; Li, H.Z.; Zhao, F.P.; Wang, F.Y.; Zhang, R.S.; Di, R.; Chu, M.X. Genomeomem CT scanning for implementation in terminal sirn body size traits in Qira black sheep (Ovis aries). Anim. Genet. 2021, 52, 560–564. [Google Scholar] [CrossRef]
- Yilmaz, O.; Kizilaslan, M.; Arzik, Y.; Behrem, S.; Ata, N.; Karaca, O.; Elmaci, C.; Cemal, I. Genomeomemem CT scanning for implementation ingrowth and in vivo carcass composition traits in Esme sheep. J. Anim. Breed. Genet. 2021, 139, 26–39. [Google Scholar] [CrossRef]
- Kizilaslan, M.; Arzik, Y.; White, S.N.; Piel, L.M.W.; Cinar, M.U. Genetic Parameters and Genomic Regions Underlying Growth and Linear Type Traits in Akkaraman Sheep. Genes 2022, 13, 1414. [Google Scholar] [CrossRef] [PubMed]
- Krivoruchko, A.; Likhovid, A.; Kanibolotskaya, A.; Saprikina, T.; Safaryan, E.; Yatsyk, O. Genome-Wide Search for Associations with Meat Production Parameters in Karachaevsky Sheep Breed Using the Illumina BeadChip 600 K. Genes 2023, 14, 1288. [Google Scholar] [CrossRef] [PubMed]
- Tuersuntuoheti, M.; Zhang, J.; Zhou, W.; Zhang, C.-L.; Liu, C.; Chang, Q.; Liu, S. Exploring the growth trait molecular markers in two sheep breeds based on Genome-wide association analysis. PLoS ONE 2023, 18, e0283383. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, J.; Guo, Y.; Zhang, L.; Xu, L.; Gao, X.; Zhu, B.; Gao, H.; Ni, H.; Chen, Y. Multi-strategy genome-wide association studies identify the DCAF16-NCAPG region as a susceptibility locus for average daily gain in cattle. Sci. Rep. 2016, 6, 38073. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, A.C.; Daetwyler, H.D.; Chamberlain, A.J.; Ponce, C.H.; Sargolzaei, M.; Schenkel, F.S.; Sahana, G.; Govignon-Gion, A.; Boitard, S.; Dolezal, M.; et al. Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat. Genet. 2018, 50, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Duan, X.; Chen, S.; He, H.; Liu, X. NCAPG is differentially expressed during longissimus muscle development and is associated with growth traits in Chinese Qinchuan beef cattle. Genet. Mol. Biol. 2015, 38, 450–456. [Google Scholar] [CrossRef]
- Hu, X.; Xing, Y.; Fu, X.; Yang, Q.; Ren, L.; Wang, Y.; Li, Q.; Li, J.; Zhang, L. NCAPG Dynamically Coordinates the Myogenesis of Fetal Bovine Tissue by Adjusting Chromatin Accessibility. Int. J. Mol. Sci. 2020, 21, 1248. [Google Scholar] [CrossRef]
- Rehfeldt, C.; Kuhn, G. Consequences of birth weight for postnatal growth performance and carcass quality in pigs as related to myogenesis1. J. Anim. Sci. 2006, 84, E113–E123. [Google Scholar] [CrossRef]
- Du, M.; Tong, J.; Zhao, J.; Underwood, K.R.; Zhu, M.; Ford, S.P.; Nathanielsz, P.W. Fetal programming of skeletal muscle development in ruminant animals1. J. Anim. Sci. 2010, 88, E51–E60. [Google Scholar] [CrossRef]
- Du, M.; Zhao, J.X.; Yan, X.; Huang, Y.; Nicodemus, L.V.; Yue, W.; McCormick, R.J.; Zhu, M.J. Fetal muscle development, mesenchymal multipotent cell differentiation, and associated signaling pathways. J. Anim. Sci. 2011, 89, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, M.; Bajard, L.; Chang, T.; Daubas, P.; Hadchouel, J.; Meilhac, S.; Montarras, D.; Rocancourt, D.; Relaix, F. The formation of skeletal muscle: From somite to limb. J. Anat. 2003, 202, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Wang, B.; Fu, X.; Yang, Q.; Zhu, M.-J. Fetal programming in meat production. Meat Sci. 2015, 109, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, H.; Hu, J.; Li, S.; Luo, Y.; Hickford, J.G.H. Two single nucleotide polymorphisms in the promoter of the ovine myostatin gene (MSTN) and their effect on growth and carcass muscle traits in New Zealand Romney sheep. J. Anim. Breed. Genet. 2016, 133, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Shishay, G.; Liu, G.; Jiang, X.; Yu, Y.; Teketay, W.; Du, D.; Jing, H.; Liu, C. Variation in the Promoter Region of the MC4R Gene Elucidates the Association of Body Measurement Traits in Hu Sheep. Int. J. Mol. Sci. 2019, 20, 240. [Google Scholar] [CrossRef]
- Maylinda, S.; Busono, W. The accuracy of body weight estimation in Fat Tailed Sheep based on linear body measurements and tail circumference. J. Ilmu-Ilmu Peternak. 2019, 29, 193–199. [Google Scholar] [CrossRef]
- Ge, L.; Su, P.; Wang, S.; Gu, Y.; Cao, X.; Lv, X.; Wang, S.; Getachew, T.; Mwacharo, J.M.; Haile, A.; et al. New Insight into the Role of the Leucine Aminopeptidase 3 (LAP3) in Cell Proliferation and Myogenic Differentiation in Sheep Embryonic Myoblasts. Genes 2022, 13, 1438. [Google Scholar] [CrossRef]
- Han, Y.; Guo, W.; Su, R.; Zhang, Y.; Yang, L.; Borjigin, G.; Duan, Y. Effects of sheep slaughter age on myogenic characteristics in skeletal muscle satellite cells. Anim. Biosci. 2022, 35, 614–623. [Google Scholar] [CrossRef]
- Lalitha, S. Primer Premier 5. Biotech Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Al-Aama, J.Y.; Liu, Y.; Hu, C.; Liu, C.; Liu, D.; Mei, L.; He, C.; Jiang, L.; Wu, H.; Chen, H.; et al. A rapid improved multiplex ligation detection reaction method for the identification of gene mutations in hereditary hearing loss. PLoS ONE 2019, 14, e0215212. [Google Scholar] [CrossRef]
- Granato, I.S.C.; Galli, G.; de Oliveira Couto, E.G.; e Souza, M.B.; Mendonça, L.F.; Fritsche-Neto, R. snpReady: A tool to assist breeders in genomic analysis. Mol. Breed. 2018, 38, 102. [Google Scholar] [CrossRef]
- Yuan, Z.; Sunduimijid, B.; Xiang, R.; Behrendt, R.; Knight, M.I.; Mason, B.A.; Reich, C.M.; Prowse-Wilkins, C.; Vander Jagt, C.J.; Chamberlain, A.J.; et al. Expression quantitative trait loci in sheep liver and muscle contribute to variations in meat traits. Genet. Sel. Evol. 2021, 53, 8. [Google Scholar] [CrossRef]
- Liu, K.; Li, Y.; Yu, B.; Wang, F.; Mi, T.; Zhao, Y. Silencing non-SMC chromosome-associated polypeptide G inhibits proliferation and induces apoptosis in hepatocellular carcinoma cells. Can. J. Physiol. Pharmacol. 2018, 96, 1246–1254. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Ai, J.; Wang, J.; Sun, C.; Lu, H.; He, A.; Li, M.; Liao, Y.; Lei, J.; Zhou, F.; et al. NCAPG promotes the proliferation of hepatocellular carcinoma through the CKII-dependent regulation of PTEN. J. Transl. Med. 2022, 20, 325. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ge, C.; Fang, D.; Wei, W.; Li, L.; Wei, Q.; Yu, H. NCAPG facilitates colorectal cancer cell proliferation, migration, invasion and epithelial–mesenchymal transition by activating the Wnt/β-catenin signaling pathway. Cancer Cell Int. 2022, 22, 119. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Li, Y.; Shi, X.; Liu, P.; Zhang, Y.; Tian, H. NCAPG Promotes Pulmonary Artery Smooth Muscle Cell Proliferation as a Promising Therapeutic Target of Idiopathic Pulmonary Hypertension: Bioinformatics Analysis and Experiment Verification. Int. J. Mol. Sci. 2022, 23, 11762. [Google Scholar] [CrossRef]
- Pang, D.; Yu, S.; Yang, X. A mini-review of the role of condensin in human nervous system diseases. Front. Mol. Neurosci. 2022, 15, 889796. [Google Scholar] [CrossRef]
- Lin, J.; Li, G.; Bai, Y.; Xie, Y. NCAPG as a novel prognostic biomarker in numerous cancers: A meta-analysis and bioinformatics analysis. Aging 2023, 15, 2503–2524. [Google Scholar] [CrossRef]
- Yue, J.; Hou, X.; Liu, X.; Wang, L.; Gao, H.; Zhao, F.; Shi, L.; Shi, L.; Yan, H.; Deng, T.; et al. The landscape of chromatin accessibility in skeletal muscle during embryonic development in pigs. J. Anim. Sci. Biotechnol. 2021, 12, 56. [Google Scholar] [CrossRef]
- Li, Q.; Wang, Y.; Hu, X.; Zhang, Y.; Li, H.; Zhang, Q.; Cai, W.; Wang, Z.; Zhu, B.; Xu, L.; et al. Transcriptional states and chromatin accessibility during bovine myoblasts proliferation and myogenic differentiation. Cell Prolif. 2022, 55, e13219. [Google Scholar] [CrossRef] [PubMed]
- Gan, S.; Du, Z.; Liu, S.; Yang, Y.; Shen, M.; Wang, X.; Yin, J.; Hu, X.; Fei, J.; Fan, J.J.; et al. Association of SNP haplotypes at the myostatin gene with muscular hypertrophy in sheep. Asian-Australas. J. Anim. Sci. 2008, 21, 928–935. [Google Scholar] [CrossRef]
- Kijas, J.W.; McCulloch, R.; Edwards, J.E.H.; Oddy, V.H.; Lee, S.H.; van der Werf, J. Evidence for multiple alleles effecting muscling and fatness at the Ovine GDF8 locus. BMC Genet. 2007, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chi, Z.; Jia, S.; Zhao, S.; Cao, G.; Purev, C.; Cang, M.; Yu, H.; Li, X.; Bao, S.J.T. Effects of novel variants in BMP15 gene on litter size in Mongolia and Ujimqin sheep breeds. Theriogenology 2023, 198, 1–11. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, J.; Qian, Y.; Meng, C.; Wang, H.; Zhong, J.; Cao, S. A T > G Mutation in the NR5A2 Gene Is Associated with Litter Size in Hu Sheep through Upregulation of Promoter Activity by Transcription Factor MTF-1. Front. Genet. 2019, 10, 1011. [Google Scholar] [CrossRef] [PubMed]
- Nkrumah, J.D.; Li, C.; Yu, J.; Hansen, C.; Keisler, D.H.; Moore, S.S. Polymorphisms in the bovine leptin promoter associated with serum leptin concentration, growth, feed intake, feeding behavior, and measures of carcass merit. J. Anim. Sci. 2005, 83, 20–28. [Google Scholar] [CrossRef]
- Wu, S.; Wang, Y.; Ning, Y.; Guo, H.; Wang, X.; Zhang, L.; Khan, R.; Cheng, G.; Wang, H.; Zan, L. Genetic Variants in STAT3 Promoter Regions and Their Application in Molecular Breeding for Body Size Traits in Qinchuan Cattle. Int. J. Mol. Sci. 2018, 19, 1035. [Google Scholar] [CrossRef]
- Guo, H.; Raza, S.H.A.; Schreurs, N.M.; Khan, R.; Wei, D.; Wang, L.; Zhang, S.; Zhang, L.; Wu, S.; Ullah, I.; et al. Genetic variants in the promoter region of the KLF3 gene associated with fat deposition in Qinchuan cattle. Gene 2018, 672, 50–55. [Google Scholar] [CrossRef]
- Li, K.; Liu, Y.; He, X.; Tao, L.; Jiang, Y.; Lan, R.; Hong, Q.; Chu, M. A Novel SNP in the Promoter Region of IGF1 Associated with Yunshang Black Goat Kidding Number via Promoting Transcription Activity by SP1. Front. Cell Dev. Biol. 2022, 10, 873095. [Google Scholar] [CrossRef]
- Deng, N.; Zhou, H.; Fan, H.; Yuan, Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget 2017, 8, 110635–110649. [Google Scholar] [CrossRef]





| Primer Name | Sequence (5′-3′) | Product Length |
|---|---|---|
| NCAPG-F | Gggagaccc1aagct2ggctagcATGGGGAAGGAGAAGAGACTGC | 3054 bp |
| NCAPG-R | agtggatccgagctcggtaccCTAACTTGTATCTTCATTGAGAAATTGTG |
| Name | Primer Name | Sequence (5′-3′) |
|---|---|---|
| NCAPG | siRNA-694 | F: GCGCCAUCAGCAAAGACUUTT |
| R: AAGUCUUUGCUGAUGGCGCTT | ||
| siRNA-729 | F:GCGCACCAAGGAUGUGAAATT | |
| R: UUUCACAUCCUUGGUGCGCTT | ||
| siRNA-2259 | F:GGUCAGCAGCAGGAUUCUUTT | |
| R:AAGAAUCCUGCUGCUGACCTT | ||
| NC | F:UUCUCCGAACGUGUCACGUTT | |
| R:ACGUGACACGUUCGGAGAATT |
| Name | Primer Name | Sequence (5′-3′) | GenBank Accession |
|---|---|---|---|
| NCAPG | NCAPG-F | AGAGACTGCTGCCGATTAAGG | XM_027970895.2 |
| NCAPG-R | ACCTGTTTTGTCATCCACCGAG | ||
| GAPDH | GAPDH-F | TCTCAAGGGCATTCTAGGCTAC | NM_001190390.1 |
| GAPDH-R | GCCGAATTCATTGTCGTACCAG |
| Primer Name | Sequence (5′-3′) | Amplified Region | Product Length |
|---|---|---|---|
| NCAPG-F | CAAGCATATTTCATGTACTCTTAA | R1 | 880 bp |
| NCAPG-R | TATTGTTTTCATGTTCTAAGGTTAT | ||
| NCAPG-F | AGTTATGGGAAAAAGCAGCTTCC | R2 | 360 bp |
| NCAPG-R | TGTTTTCAACAAAGGAGAACACAGA | ||
| NCAPG-F | GGCTGTAATTATGGAGATGTTGATT | R3 | 480 bp |
| NCAPG-R | TCTAATTACTTAAACCTCTCCCCAC | ||
| NCAPG-F | CAGACTCTTCACCAGCAATTTCACT | R4 | 370 bp |
| NCAPG-R | AGTGAGTACCCTGTGGAGATTCG | ||
| NCAPG-F | AGTCCCTGTTCTCACCCATGG | R5 | 435 bp |
| NCAPG-R | ACGAAGCCTGTTCACGTTCG |
| SNP | MAF 1 | He 2 | Ho 3 | PIC 4 | Chisq Value | p 5 |
|---|---|---|---|---|---|---|
| SNP1 | 0.39 | 0.48 | 0.59 | 0.36 | 29.357 | 6.02 × 108 |
| SNP2 | 0.38 | 0.47 | 0.57 | 0.36 | 23.915 | 1.01 × 106 |
| SNP3 | 0.39 | 0.47 | 0.58 | 0.36 | 24.562 | 7.20 × 107 |
| SNP4 | 0.38 | 0.47 | 0.57 | 0.36 | 23.915 | 1.01 × 106 |
| SNP5 | 0.29 | 0.42 | 0.48 | 0.33 | 12.256 | 4.64 × 104 |
| Age | Traits | SNP1 | SNP2 | SNP3 | SNP4 | SNP5 |
|---|---|---|---|---|---|---|
| Birth | Body weight | 0.329 | 0.350 | 0.371 | 0.350 | 0.365 |
| Body height | 0.005 | 0.005 | 0.005 | 0.005 | 0.646 | |
| Body length | 0.038 | 0.038 | 0.039 | 0.038 | 0.498 | |
| Shin circumference | 0.018 | 0.019 | 0.020 | 0.019 | 0.540 | |
| One month | Body weight | 0.306 | 0.310 | 0.308 | 0.310 | 0.296 |
| Body height | 0.004 | 0.004 | 0.005 | 0.004 | 0.598 | |
| Body length | 0.011 | 0.011 | 0.012 | 0.011 | 0.547 | |
| Shin circumference | 0.008 | 0.009 | 0.010 | 0.009 | 0.533 | |
| Two months | Body weight | 0.020 | 0.025 | 0.025 | 0.025 | 0.947 |
| Body height | 0.004 | 0.004 | 0.004 | 0.004 | 0.985 | |
| Body length | 0.004 | 0.005 | 0.005 | 0.005 | 0.984 | |
| Shin circumference | 0.006 | 0.006 | 0.007 | 0.006 | 0.898 | |
| Three months | Body weight | 0.010 | 0.013 | 0.014 | 0.013 | 0.853 |
| Body height | 0.002 | 0.002 | 0.002 | 0.002 | 0.802 | |
| Body length | 0.001 | 0.002 | 0.002 | 0.002 | 0.895 | |
| Shin circumference | 0.007 | 0.009 | 0.009 | 0.009 | 0.637 | |
| Six months | Body weight | 0.007 | 0.015 | 0.016 | 0.015 | 0.041 |
| Body height | 0.001 | 0.002 | 0.002 | 0.002 | 0.014 | |
| Body length | 0.001 | 0.003 | 0.003 | 0.003 | 0.032 | |
| Shin circumference | 0.001 | 0.003 | 0.003 | 0.003 | 0.056 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Z.; Ge, L.; Su, P.; Gu, Y.; Chen, W.; Cao, X.; Wang, S.; Lv, X.; Getachew, T.; Mwacharo, J.M.; et al. NCAPG Regulates Myogenesis in Sheep, and SNPs Located in Its Putative Promoter Region Are Associated with Growth and Development Traits. Animals 2023, 13, 3173. https://doi.org/10.3390/ani13203173
Yuan Z, Ge L, Su P, Gu Y, Chen W, Cao X, Wang S, Lv X, Getachew T, Mwacharo JM, et al. NCAPG Regulates Myogenesis in Sheep, and SNPs Located in Its Putative Promoter Region Are Associated with Growth and Development Traits. Animals. 2023; 13(20):3173. https://doi.org/10.3390/ani13203173
Chicago/Turabian StyleYuan, Zehu, Ling Ge, Pengwei Su, Yifei Gu, Weihao Chen, Xiukai Cao, Shanhe Wang, Xiaoyang Lv, Tesfaye Getachew, Joram M. Mwacharo, and et al. 2023. "NCAPG Regulates Myogenesis in Sheep, and SNPs Located in Its Putative Promoter Region Are Associated with Growth and Development Traits" Animals 13, no. 20: 3173. https://doi.org/10.3390/ani13203173
APA StyleYuan, Z., Ge, L., Su, P., Gu, Y., Chen, W., Cao, X., Wang, S., Lv, X., Getachew, T., Mwacharo, J. M., Haile, A., & Sun, W. (2023). NCAPG Regulates Myogenesis in Sheep, and SNPs Located in Its Putative Promoter Region Are Associated with Growth and Development Traits. Animals, 13(20), 3173. https://doi.org/10.3390/ani13203173






