Local Expression Dynamics of Various Adipokines during Induced Luteal Regression (Luteolysis) in the Bovine Corpus Luteum
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Bovine Corpus Luteum during Induced Luteolysis
2.2. RNA Extraction and RT-qPCR
2.3. Determination of Relative mRNA Expression
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schams, D.; Berisha, B. Regulation of Corpus Luteum Function in Cattle—An Overview. Reprod. Domest. Anim. 2004, 39, 241–251. [Google Scholar] [CrossRef]
- Skarzynski, D.; Ferreira-Dias, G.; Okuda, K. Regulation of Luteal Function and Corpus Luteum Regression in Cows: Hormonal Control, Immune Mechanisms and Intercellular Communication. Reprod. Domest. Anim. 2008, 43, 57–65. [Google Scholar] [CrossRef]
- Meidan, R.; Girsh, E.; Mamluk, R.; Levy, N.; Farberov, S. Luteolysis in Ruminants: Past Concepts, New Insights, and Persisting Challenges. In The Life Cycle of the Corpus Luteum; Meidan, R., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 159–182. [Google Scholar]
- Chouhan, V.S.; Panda, R.P.; Yadav, V.P.; Babitha, V.; Khan, F.A.; Das, G.K.; Gupta, M.; Dangi, S.S.; Singh, G.; Bag, S.; et al. Expression and localization of vascular endothelial growth factor and its receptors in the corpus luteum during oestrous cycle in water buffaloes (Bubalus bubalis). Reprod. Domest. Anim. 2013, 48, 810–818. [Google Scholar] [CrossRef]
- Schams, D.; Steinberg, V.; Steffl, M.; Meyer, H.H.; Berisha, B. Expression and possible role of fibroblast growth factor family members in porcine antral follicles during final maturation. Reproduction 2009, 138, 141–149. [Google Scholar] [CrossRef]
- Niswender, G.D.; Juengel, J.L.; Silva, P.J.; Rollyson, M.K.; McIntush, E.W. Mechanisms Controlling the Function and Life Span of the Corpus Luteum. Physiol. Rev. 2000, 80, 1–29. [Google Scholar] [CrossRef]
- Okuda, K.; Uenoyama, Y.; Berisha, B.; Lange, I.G.; Taniguchi, H.; Kobayashi, S.; Kobayashi, S.-I.; Miyamoto, A.; Schams, D. Estradiol-17β Is Produced in Bovine Corpus Luteum. Biol. Reprod. 2001, 65, 1634–1639. [Google Scholar] [CrossRef]
- Watanabe, S.; Shirasuna, K.; Matsui, M.; Yamamoto, D.; Berisha, B.; Schams, D.; Miyamoto, A. Effect of Intraluteal Injection of Endothelin Type A Receptor Antagonist on PGF2alpha-induced Luteolysis in the Cow. J. Reprod. Dev. 2006, 52, 551–559. [Google Scholar] [CrossRef]
- Kobayashi, S.; Miyamoto, A.; Berisha, B.; Schams, D. Growth hormone, but not luteinizing hormone, acts with luteal peptides on prostaglandin F2α and progesterone secretion by bovine corpora lutea in vitro. Prostaglandins Other Lipid Mediat. 2001, 63, 79–92. [Google Scholar] [CrossRef]
- Berisha, B.; Schams, D. Ovarian function in ruminants. Domest. Anim. Endocrinol. 2005, 29, 305–317. [Google Scholar] [CrossRef]
- Berisha, B.; Schams, D.; Rodler, D.; Sinowatz, F.; Pfaffl, M. Expression pattern of HIF1alpha and vasohibins during follicle maturation and corpus luteum function in the bovine ovary. Reprod. Domest. Anim. 2017, 52, 130–139. [Google Scholar] [CrossRef]
- Kobayashi, S.; Acosta, T.J.; Ozawa, T.; Hayashi, K.; Berisha, B.; Ohtani, M.; Schams, D.; Miyamoto, A. Intraluteal Release of Angiotensin II and Progesterone In Vivo During Corpora Lutea Development in the Cow: Effect of Vasoactive Peptides. Biol. Reprod. 2002, 66, 174–179. [Google Scholar] [CrossRef]
- Berisha, B.; Schams, D.; Miyamoto, A. The Expression of Angiotensin and Endothelin System Members in Bovine Corpus Luteum During Estrous Cycle and Pregnancy. Endocrine 2002, 19, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Kurowska, P.; Mlyczyńska, E.; Dupont, J.; Rak, A. Novel Insights on the Corpus Luteum Function: Role of Vaspin on Porcine Luteal Cell Angiogenesis, Proliferation and Apoptosis by Activation of GRP78 Receptor and MAP3/1 Kinase Pathways. Int. J. Mol. Sci. 2020, 21, 6823. [Google Scholar] [CrossRef] [PubMed]
- Berisha, B.; Schams, D.; Rodler, D.; Sinowatz, F.; Pfaffl, M.W. Changes in the expression of prostaglandin family members in bovine corpus luteum during the estrous cycle and pregnancy. Mol. Reprod. Dev. 2018, 85, 622–634. [Google Scholar] [CrossRef]
- Rueda, B.R.; Hendry, I.R.; Hendry, W.J.; Stormshak, F.; Slayden, O.; Davis, J.S. Decreased Progesterone Levels and Progesterone Receptor Antagonists Promote Apoptotic Cell Death in Bovine Luteal Cells. Biol. Reprod. 2000, 62, 269–276. [Google Scholar] [CrossRef]
- Jonńczyk, A.W.; Piotrowska-Tomala, K.K.; Skarzynski, D.J. Effects of prostaglandin F2α (PGF2α) on cell-death pathways in the bovine corpus luteum (CL). BMC Vet. Res. 2019, 15, 416. [Google Scholar] [CrossRef] [PubMed]
- Neuvians, T.P.; Pfaffl, M.W.; Berisha, B.; Schams, D. The mRNA expression of the members of the IGF-system in bovine corpus luteum during induced luteolysis. Domest. Anim. Endocrinol. 2003, 25, 359–372. [Google Scholar] [CrossRef]
- Berisha, B.; Thaqi, G.; Rodler, D.; Schams, D.; Sinowatz, F.; Pfaffl, M.W. Regulatory changes of local produced prostaglandins in corpus luteum after experimentally induced luteolysis in the cow. Anat. Histol. Embryol. 2022, 51, 289–299. [Google Scholar] [CrossRef]
- Zalman, Y.; Klipper, E.; Farberov, S.; Mondal, M.; Wee, G.; Folger, J.K.; Smith, G.W.; Meidan, R. Regulation of Angiogenesis-Related Prostaglandin F2alpha-Induced Genes in the Bovine Corpus Luteum. Biol. Reprod. 2012, 86, 92. [Google Scholar] [CrossRef]
- Miyamoto, A.; Shirasuna, K.; Sasahara, K. Local regulation of corpus luteum development and regression in the cow: Impact of angiogenic and vasoactive factors. Domest. Anim. Endocrinol. 2009, 37, 159–169. [Google Scholar] [CrossRef]
- Ochoa, J.C.; Peñagaricano, F.; Baez, G.M.; Melo, L.F.; Motta, J.C.L.; Garcia-Guerra, A.; Meidan, R.; Pinheiro Ferreira, J.C.; Sartori, R.; Wiltbank, M.C. Mechanisms for rescue of corpus luteum during pregnancy: Gene expression in bovine corpus luteum following intrauterine pulses of prostaglandins E1 and F2α†. Biol. Reprod. 2017, 98, 465–479. [Google Scholar] [CrossRef]
- Mlyczyńska, E.; Kieżun, M.; Kurowska, P.; Dawid, M.; Pich, K.; Respekta, N.; Daudon, M.; Rytelewska, E.; Dobrzyń, K.; Kamińska, B.; et al. New Aspects of Corpus Luteum Regulation in Physiological and Pathological Conditions: Involvement of Adipokines and Neuropeptides. Cells 2022, 11, 957. [Google Scholar] [CrossRef]
- Galvão, A.M.; Skarzynski, D.; Ferreira-Dias, G. Luteolysis and the Auto-, Paracrine Role of Cytokines from Tumor Necrosis Factor α and Transforming Growth Factor β Superfamilies. Vitam. Horm. 2018, 107, 287–315. [Google Scholar] [CrossRef]
- Galvão, A.M.; Ferreira-Dias, G.; Skarzynski, D.J. Cytokines and Angiogenesis in the Corpus Luteum. Mediat. Inflamm. 2013, 2013, 420186. [Google Scholar] [CrossRef]
- Tanaka, J.; Acosta, T.J.; Berisha, B.; Tetsuka, M.; Matsui, M.; Kobayashi, S.; Schams, D.; Miyamoto, A. Relative Changes in mRNA Expression of Angiopoietins and Receptors Tie in Bovine Corpus Luteum during Estrous Cycle and Prostaglandin F2alpha-induced Luteolysis: A Possible Mechanism for the Initiation of Luteal Regression. J. Reprod. Dev. 2004, 50, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Friedman, A.; Weiss, S.; Levy, N.; Meidan, R. Role of Tumor Necrosis Factor α and Its Type I Receptor in Luteal Regression: Induction of Programmed Cell Death in Bovine Corpus Luteum-Derived Endothelial Cells. Biol. Reprod. 2000, 63, 1905–1912. [Google Scholar] [CrossRef] [PubMed]
- Kurowska, P.; Mlyczyńska, E.; Barbe, A.; Mellouk, N.; Dupont, J.; Rak, A. The Adipokines in Domestic Animal Reproduction: Expression and Role in the Regulation of Ovarian Function. In New Insights into Theriogenology; IntechOpen: London, UK, 2018. [Google Scholar]
- Kurowska, P.; Mlyczyńska, E.; Dawid, M.; Sierpowski, M.; Estienne, A.; Dupont, J.; Rak, A. Adipokines change the balance of proliferation/apoptosis in the ovarian cells of human and domestic animals: A comparative review. Anim. Reprod. Sci. 2021, 228, 106737. [Google Scholar] [CrossRef] [PubMed]
- Kurowska, P.; Mlyczyńska, E.; Dawid, M.; Grzesiak, M.; Dupont, J.; Rak, A. The role of vaspin in porcine corpus luteum. J. Endocrinol. 2020, 247, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Michalakis, K.G.; Segars, J.H. The role of adiponectin in reproduction: From polycystic ovary syndrome to assisted reproduction. Fertil. Steril. 2010, 94, 1949–1957. [Google Scholar] [CrossRef]
- Ouchi, N.; Parker, J.L.; Lugus, J.J.; Walsh, K. Adipokines in inflammation and metabolic disease. Nat. Rev. Immunol. 2011, 11, 85–97. [Google Scholar] [CrossRef]
- Chang, M.-L.; Yang, Z.; Yang, S.-S. Roles of Adipokines in Digestive Diseases: Markers of Inflammation, Metabolic Alteration and Disease Progression. Int. J. Mol. Sci. 2020, 21, 8308. [Google Scholar] [CrossRef]
- Reverchon, M.; Ramé, C.; Bertoldo, M.; Dupont, J. Adipokines and the Female Reproductive Tract. Int. J. Endocrinol. 2014, 2014, 232454. [Google Scholar] [CrossRef]
- Thaqi, G.; Berisha, B.; Pfaffl, M.W. Expression of Locally Produced Adipokines and Their Receptors during Different Physiological and Reproductive Stages in the Bovine Corpus Luteum. Animals 2023, 13, 1782. [Google Scholar] [CrossRef]
- Rak, A.; Mellouk, N.; Froment, P.; Dupont, J. Adiponectin and resistin: Potential metabolic signals affecting hypothalamo-pituitary gonadal axis in females and males of different species. Reproduction 2017, 153, R215–R226. [Google Scholar] [CrossRef] [PubMed]
- Kurowska, P.; Sroka, M.; Dawid, M.; Mlyczyńska, E.; Respekta, N.; Jurek, M.; Klimczyk, D.; Grzesiak, M.; Dupont, J.; Rak, A. Expression and role of resistin on steroid secretion in the porcine corpus luteum. Reproduction 2021, 162, 237–248. [Google Scholar] [CrossRef]
- Estienne, A.; Bongrani, A.; Reverchon, M.; Ramé, C.; Ducluzeau, P.-H.; Froment, P.; Dupont, J. Involvement of Novel Adipokines, Chemerin, Visfatin, Resistin and Apelin in Reproductive Functions in Normal and Pathological Conditions in Humans and Animal Models. Int. J. Mol. Sci. 2019, 20, 4431. [Google Scholar] [CrossRef] [PubMed]
- Yart, L.; Dessauge, F.; Finot, L.; Barbey, S.; Marnet, P.; Lollivier, V. Ovariectomy improves lactation persistency in dairy cows. J. Dairy Sci. 2012, 95, 3794–3802. [Google Scholar] [CrossRef] [PubMed]
- Chappat, P. La castration de la vache. Bulletin GTV 1993, 1, 53–63. [Google Scholar]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.0031. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.J.; Fairclough, R.J.; Payne, E.; Smith, J.F. Hormonal changes around bovine luteolysis. Prostaglandins 1975, 10, 675–684. [Google Scholar] [CrossRef]
- Shirasuna, K.; Akabane, Y.; Beindorff, N.; Nagai, K.; Sasaki, M.; Shimizu, T.; Bollwein, H.; Meidan, R.; Miyamoto, A. Expression of prostaglandin F2α (PGF2α) receptor and its isoforms in the bovine corpus luteum during the estrous cycle and PGF2α-induced luteolysis. Domest. Anim. Endocrinol. 2012, 43, 227–238. [Google Scholar] [CrossRef]
- Webb, R.; Woad, K.J.; Armstrong, D.G. Corpus luteum (CL) function: Local control mechanisms. Domest. Anim. Endocrinol. 2002, 23, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Penny, L.A.; Armstrong, D.; Bramley, T.A.; Webb, R.; Collins, R.A.; Watson, E.D. Immune cells and cytokine production in the bovine corpus luteum throughout the oestrous cycle and after induced luteolysis. Reproduction 1999, 115, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Weems, Y.S.; Ma, Y.; Ford, S.P.; Nett, T.M.; Vann, R.C.; Lewis, A.W.; Neuendorff, D.A.; Welsh, T.H.; Randel, R.D.; Weems, C.W. Effects of intraluteal implants of prostaglandin E1 or E2 on angiogenic growth factors in luteal tissue of Angus and Brahman cows. Theriogenology 2014, 82, 1224–1230. [Google Scholar] [CrossRef]
- Liu, K.; Olofsson, J.I.; Wahlberg, P.; Ny, T. Distinct Expression of Gelatinase A [Matrix Metalloproteinase (MMP)-2], Collagenase-3 (MMP-13), Membrane Type MMP 1 (MMP-14), and Tissue Inhibitor of MMPs Type 1 Mediated by Physiological Signals During Formation and Regression of the Rat Corpus Luteum1. Endocrinology 1999, 140, 5330–5338. [Google Scholar] [CrossRef]
- Socha, B.M.; Łada, P.; Jończyk, A.W.; Korzekwa, A.J.; Skarżyński, D.J. The Role of Peroxisome Proliferator-Activated Receptors in PGF2α-Induced Luteolysis in the Bovine Corpus Luteum. Animals 2022, 12, 1542. [Google Scholar] [CrossRef]
- Irving-Rodgers, H.F.; Friden, B.E.; Morris, S.E.; Mason, H.D.; Brannstrom, M.; Sekiguchi, K.; Sanzen, N.; Sorokin, L.M.; Sado, Y.; Ninomiya, Y.; et al. Extracellular matrix of the human cyclic corpus luteum. Mol. Hum. Reprod. 2006, 12, 525–534. [Google Scholar] [CrossRef]
- Różycka, M.; Kurowska, P.; Grzesiak, M.; Kotula-Balak, M.; Tworzydło, W.; Rame, C.; Gregoraszczuk, E.; Dupont, J.; Rak, A. Apelin and apelin receptor at different stages of corpus luteum development and effect of apelin on progesterone secretion and 3β-hydroxysteroid dehydrogenase (3β-HSD) in pigs. Anim. Reprod. Sci. 2018, 192, 251–260. [Google Scholar] [CrossRef]
- Abe, H.; Al-Zi’abi, M.O.; Sekizawa, F.; Acosta, T.J.; Skarzynski, D.J.; Okuda, K. Lymphatic Involvement in the Disappearance of Steroidogenic Cells from the Corpus Luteum during Luteolysis. PLoS ONE 2014, 9, e88953. [Google Scholar] [CrossRef]
- Maranesi, M.; Zerani, M.; Lilli, L.; Dall’Aglio, C.; Brecchia, G.; Gobbetti, A.; Boiti, C. Expression of luteal estrogen receptor, interleukin-1, and apoptosis-associated genes after PGF2α administration in rabbits at different stages of pseudopregnancy. Domest. Anim. Endocrinol. 2010, 39, 116–130. [Google Scholar] [CrossRef] [PubMed]
- Clemente-Suárez, V.J.; Redondo-Flórez, L.; Beltrán-Velasco, A.I.; Martín-Rodríguez, A.; Martínez-Guardado, I.; Navarro-Jiménez, E.; Laborde-Cárdenas, C.C.; Tornero-Aguilera, J.F. The Role of Adipokines in Health and Disease. Biomedicines 2023, 11, 1290. [Google Scholar] [CrossRef]
- Booth, A.; Magnuson, A.; Fouts, J.; Foster, M. Adipose tissue, obesity and adipokines: Role in cancer promotion. Horm. Mol. Biol. Clin. Investig. 2015, 21, 57–74. [Google Scholar] [CrossRef]
- Kim, J.-E.; Kim, J.-S.; Jo, M.-J.; Cho, E.; Ahn, S.-Y.; Kwon, Y.-J.; Ko, G.-J. The Roles and Associated Mechanisms of Adipokines in Development of Metabolic Syndrome. Molecules 2022, 27, 334. [Google Scholar] [CrossRef] [PubMed]
- Heiker, J.T. Vaspin (serpinA12) in obesity, insulin resistance, and inflammation. J. Pept. Sci. 2014, 20, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Ramirez, M.; Ayala, L.; Benavides, E.A.; Xie, F.; Arellano, A.A.; Stanko, R.L.; Garcia, M.R. Adiponectin Influences FGF2 in the Developing Porcine Corpus Luteum. Vet. Sci. 2022, 9, 77. [Google Scholar] [CrossRef] [PubMed]
- Kurowska, P.; Gazdzik, K.; Jasinska, A.; Mlyczynska, E.; Wachowska, D.; Rak, A. Resistin as a new player in the regulation of porcine corpus luteum luteolysis: In vitro effect on proliferation/viability, apoptosis and autophagy. J. Physiol. Pharmacol. 2023, 1, 21–30. [Google Scholar] [CrossRef]
- Heo, Y.J.; Choi, S.-E.; Jeon, J.Y.; Han, S.J.; Kim, D.J.; Kang, Y.; Lee, K.W.; Kim, H.J. Visfatin Induces Inflammation and Insulin Resistance via the NF-κB and STAT3 Signaling Pathways in Hepatocytes. J. Diabetes Res. 2019, 2019, 4021623. [Google Scholar] [CrossRef]
- Fantuzzi, G. Adipose tissue, adipokines, and inflammation. J. Allergy Clin. Immunol. 2005, 115, 911–919. [Google Scholar] [CrossRef]
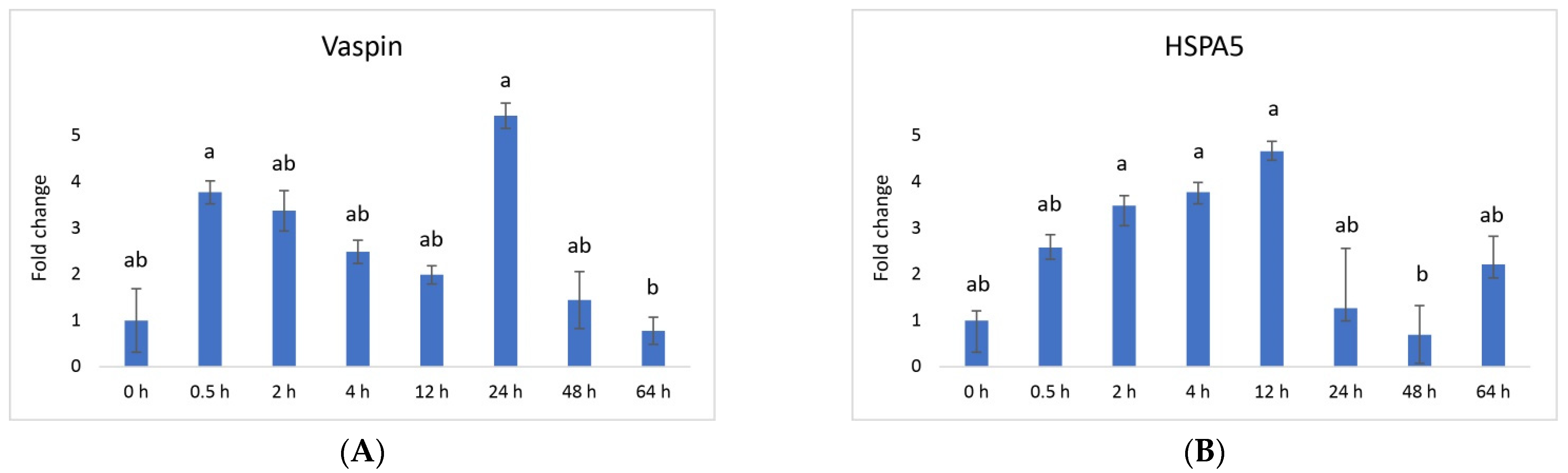
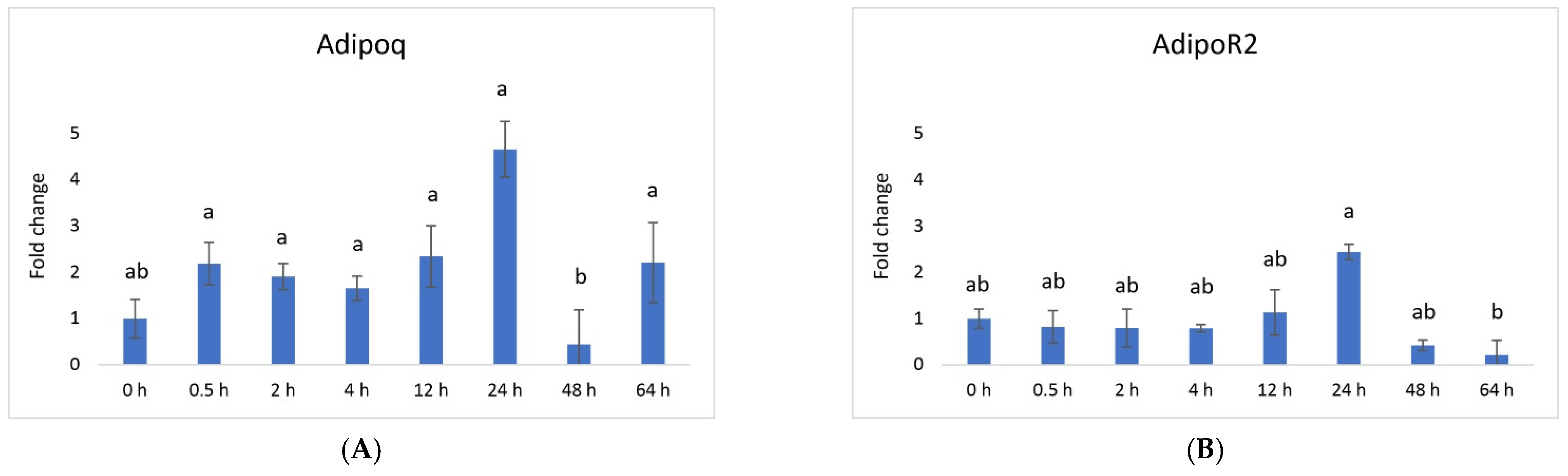
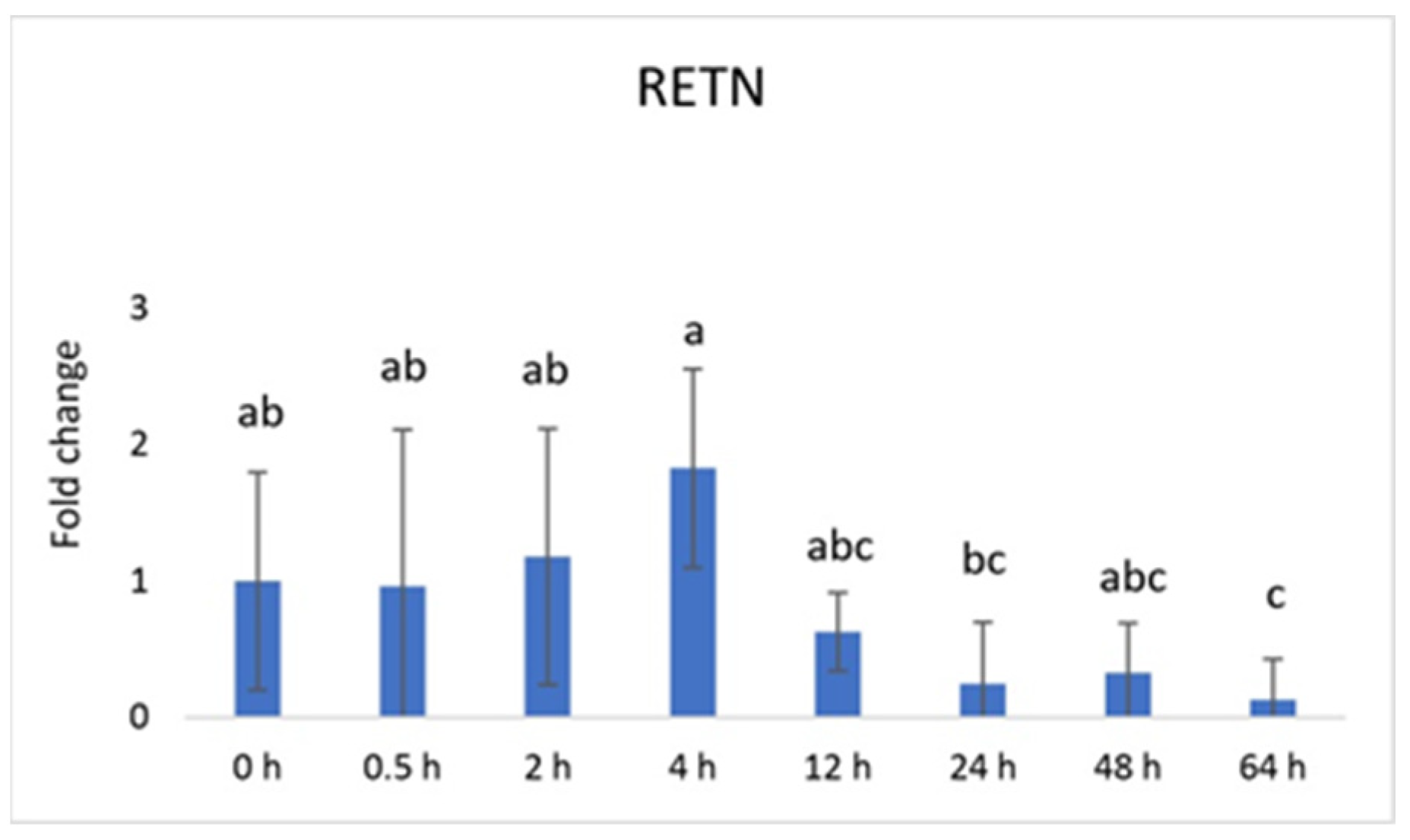
| Genes | Sequence of Nucleotides * | Amplicon Size [bp] | Ensembl Transcript ID |
|---|---|---|---|
| PPIA | For-5′-CTGAGCACTGGAGAGAAAGGA-3′ | 116 | ENSBTAT00000015924.5 |
| Rev-5′-GACTTGCCACCAGTACCATT-3′ | |||
| Hyb-5′-TCCGGGATTTATGTGCCAGGGTGGT-3′ | |||
| UBA52 | For-5′-GGCTGATCTTCGCTGGCA-3′ | 124 | ENSBTAT00000010176.3 |
| Rev-5′-CGGAGGGAAGGCTCGATG-3′ | |||
| Hyb-5′-TGGAGGATGGCCGCACTCTGTCAGA-3′ | |||
| UBC | For-5′-GACCGGGAGTTCAGTCTTCG-3′ | 133 | ENSBTAT00000046011.4 |
| Rev-5′-TTCTCGATGGTGTCACTGGG-3′ | |||
| Hyb-5′-TGTGTTCGCTGCTGACACCACCACT-3′ | |||
| Vaspin | For- 5′-TACCAGAGCAACTTCACGGC-3′ | 146 | ENSBTAT00000045204.4 |
| Rev-5′-GTCAACCTGGGCACAAACAC-3′ | |||
| Hyb-5′-TGAAGCAAGTGGAGCAAGCCCTGGG-3′ | |||
| HSPA5 | For-5′-TTTCTGCCATGGTTCTCACT-3′ | 125 | ENSBTAT00000057533.3 |
| Rev-5′-ATCTTTGGTTGCCTGGCGTT-3′ | |||
| Hyb-5′-AGGAAACTGCTGAGGCTTATTTGGGA-3′ | |||
| Adiponectin | For-5′-GTGAGAAGGGTGAGAAAGGAGA-3′ | 136 | ENSBTAT00000077795.1 |
| Rev-5′-GCACTTTCTCCAGGTTCTCCC-3′ | |||
| Hyb-5′-GAGGCTTTCCAGGAACCCCAGGCAG-3′ | |||
| AdipoR1 | For-5′-CCACACTGTCTACTGTCATTCA-3′ | 150 | ENSBTAT00000040283.5 |
| Rev-5′-GAGAGGTAGATGAGCCGAGG-3′ | |||
| Hyb-5′-TGCTGATCATGGGGAGCTTCGTGCC-3′ | |||
| AdipoR2 | For-5′-GGCGTCTGTCCTTTCTTCCT-3′ | 110 | ENSBTAT00000011923.6 |
| Rev-5′-CTTGCAGGAGAGGGGACATG-3′ | |||
| Hyb-5′-GGAGCGTGAGTGCGATGATGGAGCA-3′ | |||
| Resistin | For-5′-CAGTCGCTGTGCCCCATAG-3′ | 91 | ENSBTAT00000006189.3 |
| Rev-5′-GGCCAATGATCCTTACTGCC-3′ | |||
| Hyb-5′-TGAGAAGATCCAGGAGGTCACCACC-3′ | |||
| Visfatin | For-5′-TCGAAGGGCTACAAGTTGCT-3′ | 143 | ENSBTAT00000020608.4 |
| Rev-5′-GCTCCACCAGAACCAAAGGA-3′ | |||
| Hyb-5′-ACAAGAGATTGTGGAAGGCATGAAGCA-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thaqi, G.; Berisha, B.; Pfaffl, M.W. Local Expression Dynamics of Various Adipokines during Induced Luteal Regression (Luteolysis) in the Bovine Corpus Luteum. Animals 2023, 13, 3221. https://doi.org/10.3390/ani13203221
Thaqi G, Berisha B, Pfaffl MW. Local Expression Dynamics of Various Adipokines during Induced Luteal Regression (Luteolysis) in the Bovine Corpus Luteum. Animals. 2023; 13(20):3221. https://doi.org/10.3390/ani13203221
Chicago/Turabian StyleThaqi, Granit, Bajram Berisha, and Michael W. Pfaffl. 2023. "Local Expression Dynamics of Various Adipokines during Induced Luteal Regression (Luteolysis) in the Bovine Corpus Luteum" Animals 13, no. 20: 3221. https://doi.org/10.3390/ani13203221
APA StyleThaqi, G., Berisha, B., & Pfaffl, M. W. (2023). Local Expression Dynamics of Various Adipokines during Induced Luteal Regression (Luteolysis) in the Bovine Corpus Luteum. Animals, 13(20), 3221. https://doi.org/10.3390/ani13203221






