Genome-Wide Association Study of Fiber Diameter in Alpacas
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. DNA Samples and Genotyping
2.3. Data Quality Control
2.4. Genome-Wide Association Study
2.4.1. Method 1. Linear Model
2.4.2. Method 2. Haplotype and Marker Analysis
2.4.3. Method 3. Selection Signatures
2.5. Candidate Regions
3. Results
3.1. Data Quality Control
3.2. Genome-Wide Association Study
3.2.1. Method 1. Linear Model
3.2.2. Method 2. Haplotype and Marker Analysis
3.2.3. Method 3. Selection Signatures
3.3. Candidate Regions
4. Discussion
4.1. Sample
4.2. Genome-Wide Association Study
4.3. Discussion by Method
4.3.1. Method 1. Linear Model
4.3.2. Method 2. Haplotype and Marker Analysis
4.3.3. Method 3. Selection Signatures
4.4. Gene Annotation
4.5. Candidate Regions
4.6. Genome Annotation
4.7. Future Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, H.; Guang, X.; Al-Fageeh, M.B.; Cao, J.; Pan, S.; Zhou, H.; Zhang, L.; Abutarboush, M.H.; Xing, Y.; Xie, Z.; et al. Camelid genomes reveal evolution and adaptation to desert environments. Nat. Commun. 2014, 5, 5188. [Google Scholar] [CrossRef] [PubMed]
- Richardson, M.F.; Munyard, K.; Croft, L.J.; Allnutt, T.R.; Jackling, F.; Alshanbari, F.; Jevit, M.; Wright, G.A.; Cransberg, R.; Tibary, A.; et al. Chromosome-Level Alpaca Reference Genome VicPac3.1 Improves Genomic Insight into the Biology of New World Camelids. Front. Genet. 2019, 10, 586. [Google Scholar] [CrossRef] [PubMed]
- Balmus, G.; Trifonov, V.A.; Biltueva, L.S.; O’Brien, P.C.M.; Alkalaeva, E.S.; Fu, B.; Skidmore, J.A.; Allen, T.; Graphodatsky, A.S.; Yang, F.; et al. Cross-species chromosome painting among camel, cattle, pig and human: Further insights into the putative Cetartiodactyla ancestral karyotype. Chromosome Res. 2007, 15, 499–514. [Google Scholar] [CrossRef] [PubMed]
- Avila, F.; Baily, M.P.; Perelman, P.; Das, P.J.; Pontius, J.; Chowdhary, R.; Owens, E.; Johnson, W.E.; Merriwether, D.A.; Raudsepp, T. A Comprehensive Whole-Genome Integrated Cytogenetic Map for the Alpaca (Lama pacos). Cytogenet. Genome Res. 2014, 144, 196–207. [Google Scholar] [CrossRef]
- Mendoza, M.N.; Raudsepp, T.; Alshanbari, F.; Gutiérrez, G.; Ponce de León, F.A. Chromosomal Localization of Candidate Genes for Fiber Growth and Color in Alpaca (Vicugna pacos). Front. Genet. 2019, 10, 583. [Google Scholar] [CrossRef]
- Mendoza, M.N.; Raudsepp, T.; More, M.J.; Gutiérrez, G.A.; Ponce de León, F.A. Cytogenetic Mapping of 35 New Markers in the Alpaca (Vicugna pacos). Genes 2020, 11, 522. [Google Scholar] [CrossRef]
- Pauciullo, A.; Shuiep, E.T.; Ogah, M.D.; Cosenza, G.; Di Stasio, L.; Erhardt, G. Casein Gene Cluster in Camelids: Comparative Genome Analysis and New Findings on Haplotype Variability and Physical Mapping. Front. Genet. 2019, 10, 748. [Google Scholar] [CrossRef]
- More, M.; Gutiérrez, G.; Rothschild, M.; Bertolini, F.; Ponce de León, F.A. Evaluation of SNP Genotyping in Alpacas Using the Bovine HD Genotyping Beadchip. Front. Genet. 2019, 10, 361. [Google Scholar] [CrossRef]
- Guridi, M.; Soret, B.; Alfonso, L.; Arana, A. Single nucleotide polymorphisms in the Melanocortin 1 Receptor gene are linked with lightness of fibre colour in Peruvian Alpaca (Vicugna pacos). Anim. Genet. 2011, 42, 679–682. [Google Scholar] [CrossRef]
- Chandramohan, B.; Renieri, C.; La Manna, V.; La Terza, A. The alpaca agouti gene: Genomic locus, transcripts and causative mutations of eumelanic and pheomelanic coat color. Gene 2013, 521, 303–310. [Google Scholar] [CrossRef]
- Chandramohan, B.; Renieri, C.; La Manna, V.; La Terza, A. The Alpaca Melanocortin 1 Receptor: Gene Mutations, Transcripts, and Relative Levels of Expression in Ventral Skin Biopsies. Sci. World J. 2015, 2015, e265751. [Google Scholar] [CrossRef] [PubMed]
- Feeley, N.L.; Bottomley, S.; Munyard, K.A. Novel mutations in Vicugna pacos (alpaca) Tyrp1 are not correlated with brown fibre colour phenotypes. Small Rumin. Res. 2016, 143, 29–34. [Google Scholar] [CrossRef]
- Fernández, A.G.; Gutiérrez, G.A.; Ponce de León, F.A. Identificación bioinformática de Polimorfismos de Nucleótido Simple (PNSs) en genes candidatos para las características de la fibra en alpacas (Vicugna pacos). Rev. Peru. Biol. 2019, 26, 087–094. [Google Scholar] [CrossRef]
- Salas, W.; Delgado, I.; Rodríguez, J.; Paucar-Chanca, R. Genetic variation in HOXC13 and KRT31 genes of alpaca (Vicugna pacos). Manglar 2020, 17, 159–162. [Google Scholar] [CrossRef]
- Calderon, M.; More, M.J.; Gutierrez, G.A.; Ponce de León, F.A. Development of a 76k Alpaca (Vicugna pacos) Single Nucleotide Polymorphisms (SNPs) Microarray. Genes 2021, 12, 291. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, H.; Yang, H.; Wang, S.; Rong, E.; Pei, W.; Li, H.; Wang, N. Genome-Wide Association Study for Wool Production Traits in a Chinese Merino Sheep Population. PLoS ONE 2014, 9, e107101. [Google Scholar] [CrossRef]
- Zhao, H.; Guo, T.; Lu, Z.; Liu, J.; Zhu, S.; Qiao, G.; Han, M.; Yuan, C.; Wang, T.; Li, F.; et al. Genome-wide association studies detects candidate genes for wool traits by re-sequencing in Chinese fine-wool sheep. BMC Genom. 2021, 22, 127. [Google Scholar] [CrossRef]
- Zhao, H.; Zhu, S.; Guo, T.; Han, M.; Chen, B.; Qiao, G.; Wu, Y.; Yuan, C.; Liu, J.; Lu, Z.; et al. Whole-genome re-sequencing association study on yearling wool traits in Chinese fine-wool sheep. J. Anim. Sci. 2021, 99, skab210. [Google Scholar] [CrossRef]
- Bolormaa, S.; Swan, A.A.; Stothard, P.; Khansefid, M.; Moghaddar, N.; Duijvesteijn, N.; van der Werf, J.H.J.; Daetwyler, H.D.; MacLeod, I.M. A conditional multi-trait sequence GWAS discovers pleiotropic candidate genes and variants for sheep wool, skin wrinkle and breech cover traits. Genet. Sel. Evol. 2021, 53, 58. [Google Scholar] [CrossRef]
- Qiao, X.; Su, R.; Wang, Y.; Wang, R.; Yang, T.; Li, X.; Chen, W.; He, S.; Jiang, Y.; Xu, Q.; et al. Genome-wide Target Enrichment-aided Chip Design: A 66 K SNP Chip for Cashmere Goat. Sci. Rep. 2017, 7, 8621. [Google Scholar] [CrossRef]
- Wang, F.H.; Zhang, L.; Gong, G.; Yan, X.C.; Zhang, L.T.; Zhang, F.T.; Liu, H.F.; Lv, Q.; Wang, Z.Y.; Wang, R.J.; et al. Genome-wide association study of fleece traits in Inner Mongolia Cashmere goats. Anim. Genet. 2021, 52, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Su, R.; Wan, W.; Zhang, W.; Jiang, H.; Qiao, X.; Fan, Y.; Zhang, Y.; Wang, R.; Liu, Z.; et al. Identification of selection signals by large-scale whole-genome resequencing of cashmere goats. Sci. Rep. 2017, 7, 15142. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Lu, J.; Fei, X.; Lu, Z.; Quan, K.; Liu, Y.; Chu, M.; Di, R.; Wang, H.; Wei, C. Genetic Signatures of Selection for Cashmere Traits in Chinese Goats. Animals 2020, 10, 1905. [Google Scholar] [CrossRef] [PubMed]
- Paredes, M.M.; Membrillo, A.; Gutiérrez, J.P.; Cervantes, I.; Azor, P.J.; Morante, R.; Alonso-Moraga, A.; Molina, A.; Muñoz-Serrano, A. Association of microsatellite markers with fiber diameter trait in Peruvian alpacas (Vicugna pacos). Livest. Sci. 2014, 161, 6–16. [Google Scholar] [CrossRef]
- IWTO-12-2012; Measurement of the Mean and Distribution of Fibre Diameter Using the Sirolan-Laserscan Fibre Diameter Analyser. International Wool Textile Organization: Brussels, Belgium, 2017.
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef]
- Jiang, L.; Zheng, Z.; Qi, T.; Kemper, K.E.; Wray, N.R.; Visscher, P.M.; Yang, J. A resource-efficient tool for mixed model association analysis of large-scale data. Nat. Genet. 2019, 51, 1749–1755. [Google Scholar] [CrossRef]
- Prakapenka, D.; Wang, C.; Liang, Z.; Bian, C.; Tan, C.; Da, Y. GVCHAP: A Computing Pipeline for Genomic Prediction and Variance Component Estimation Using Haplotypes and SNP Markers. Front. Genet. 2020, 11, 282. [Google Scholar] [CrossRef]
- Sabeti, P.C.; Varilly, P.; Fry, B.; Lohmueller, J.; Hostetter, E.; Cotsapas, C.; Xie, X.; Byrne, E.H.; McCarroll, S.A.; Gaudet, R.; et al. Genome-wide detection and characterization of positive selection in human populations. Nature 2007, 449, 913–918. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Sul, J.H.; Martin, L.S.; Eskin, E. Population structure in genetic studies: Confounding factors and mixed models. PLoS Genet. 2018, 14, e1007309. [Google Scholar] [CrossRef]
- Browning, S.R.; Browning, B.L. Rapid and Accurate Haplotype Phasing and Missing-Data Inference for Whole-Genome Association Studies by Use of Localized Haplotype Clustering. Am. J. Hum. Genet. 2007, 81, 1084–1097. [Google Scholar] [CrossRef] [PubMed]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, S.B.; Schaffner, S.F.; Nguyen, H.; Moore, J.M.; Roy, J.; Blumenstiel, B.; Higgins, J.; DeFelice, M.; Lochner, A.; Faggart, M.; et al. The Structure of Haplotype Blocks in the Human Genome. Science 2002, 296, 2225–2229. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Fan, H.; Wang, Y.; Zhang, L.; Gao, X.; Chen, Y.; Li, J.; Ren, H.; Gao, H. Genome-Wide Association Studies Using Haplotypes and Individual SNPs in Simmental Cattle. PLoS ONE 2014, 9, e109330. [Google Scholar] [CrossRef] [PubMed]
- Szpiech, Z.A.; Hernandez, R.D. selscan: An Efficient Multithreaded Program to Perform EHH-Based Scans for Positive Selection. Mol. Biol. Evol. 2014, 31, 2824–2827. [Google Scholar] [CrossRef]
- Binns, D.; Dimmer, E.; Huntley, R.; Barrell, D.; O’Donovan, C.; Apweiler, R. QuickGO: A web-based tool for Gene Ontology searching. Bioinformatics 2009, 25, 3045–3046. [Google Scholar] [CrossRef]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, gkn923. [Google Scholar] [CrossRef]
- Wang, Y.; Niu, Z.; Zeng, Z.; Jiang, Y.; Jiang, Y.; Ding, Y.; Tang, S.; Shi, H.; Ding, X. Using High-Density SNP Array to Reveal Selection Signatures Related to Prolificacy in Chinese and Kazakhstan Sheep Breeds. Animals 2020, 10, 1633. [Google Scholar] [CrossRef]
- Cardon, L.R.; Palmer, L.J. Population stratification and spurious allelic association. Lancet 2003, 361, 598–604. [Google Scholar] [CrossRef] [PubMed]
- Freedman, M.L.; Reich, D.; Penney, K.L.; McDonald, G.J.; Mignault, A.A.; Patterson, N.; Gabriel, S.B.; Topol, E.J.; Smoller, J.W.; Pato, C.N.; et al. Assessing the impact of population stratification on genetic association studies. Nat. Genet. 2004, 36, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Balding, D.J. A tutorial on statistical methods for population association studies. Nat. Rev. Genet. 2006, 7, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Voight, B.F.; Pritchard, J.K. Confounding from Cryptic Relatedness in Case-Control Association Studies. PLoS Genet. 2005, 1, e32. [Google Scholar] [CrossRef]
- Astle, W.; Balding, D.J. Population Structure and Cryptic Relatedness in Genetic Association Studies. Stat. Sci. 2009, 24, 451–471. [Google Scholar] [CrossRef]
- Marees, A.T.; de Kluiver, H.; Stringer, S.; Vorspan, F.; Curis, E.; Marie-Claire, C.; Derks, E.M. A tutorial on conducting genome-wide association studies: Quality control and statistical analysis. Int. J. Methods Psychiatr. Res. 2018, 27, e1608. [Google Scholar] [CrossRef]
- Öner, Y.; Serrano, M.; Sarto, P.; Iguácel, L.P.; Piquer-Sabanza, M.; Estrada, O.; Juan, T.; Calvo, J.H. Genome-Wide Association Studies of Somatic Cell Count in the Assaf Breed. Animals 2021, 11, 1531. [Google Scholar] [CrossRef]
- Li, Y.; Levran, O.; Kim, J.; Zhang, T.; Chen, X.; Suo, C. Extreme sampling design in genetic association mapping of quantitative trait loci using balanced and unbalanced case-control samples. Sci. Rep. 2019, 9, 15504. [Google Scholar] [CrossRef]
- Sahana, G.; Guldbrandtsen, B.; Bendixen, C.; Lund, M.S. Genome-wide association mapping for female fertility traits in Danish and Swedish Holstein cattle. Anim. Genet. 2010, 41, 579–588. [Google Scholar] [CrossRef]
- Chen, Z.; Yao, Y.; Ma, P.; Wang, Q.; Pan, Y. Haplotype-based genome-wide association study identifies loci and candidate genes for milk yield in Holsteins. PLoS ONE 2018, 13, e0192695. [Google Scholar] [CrossRef]
- Nascimento, A.V.d.; Romero, Â.R.d.S.; Utsunomiya, Y.T.; Utsunomiya, A.T.H.; Cardoso, D.F.; Neves, H.H.R.; Carvalheiro, R.; Garcia, J.F.; Grisolia, A.B. Genome-wide association study using haplotype alleles for the evaluation of reproductive traits in Nelore cattle. PLoS ONE 2018, 13, e0201876. [Google Scholar] [CrossRef] [PubMed]
- Cuyabano, B.C.D.; Su, G.; Lund, M.S. Genomic prediction of genetic merit using LD-based haplotypes in the Nordic Holstein population. BMC Genom. 2014, 15, 1171. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Sun, W.; Guo, T.; Li, J.; Zhu, S.; Lu, Z.; Qiao, G.; Han, M.; Zhao, H.; Yang, B.; et al. Genome-Wide Selective Signatures Reveal Candidate Genes Associated with Hair Follicle Development and Wool Shedding in Sheep. Genes 2021, 12, 1924. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xue, X.; Liu, Y.; Abied, A.; Ding, Y.; Zhao, S.; Wang, W.; Ma, L.; Guo, J.; Guan, W.; et al. Genome-wide comparative analyses reveal selection signatures underlying adaptation and production in Tibetan and Poll Dorset sheep. Sci. Rep. 2021, 11, 2466. [Google Scholar] [CrossRef]
- Abied, A.; Bagadi, A.; Bordbar, F.; Pu, Y.; Augustino, S.M.A.; Xue, X.; Xing, F.; Gebreselassie, G.; Han, J.-L.; Mwacharo, J.M.; et al. Genomic Diversity, Population Structure, and Signature of Selection in Five Chinese Native Sheep Breeds Adapted to Extreme Environments. Genes 2020, 11, 494. [Google Scholar] [CrossRef]
- Song, Y.; Luo, Y.; Zhou, H.; Liu, X.; Li, S.; Hao, Z.; Shen, J.; Zhen, H.; Li, L.; Wang, J.; et al. Variation in caprine KRTAP1-3 and its association with cashmere fibre diameter. Gene 2022, 823, 146341. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, N.; Liu, K.; He, J.; Yu, J.; Bu, R.; Cheng, M.; De, W.; Liu, J.; Li, H. Identification of genes and proteins associated with anagen wool growth. Anim. Genet. 2017, 48, 67–79. [Google Scholar] [CrossRef]
- Lv, X.; Chen, W.; Sun, W.; Hussain, Z.; Wang, S.; Wang, J. Analysis of lncRNAs Expression Profiles in Hair Follicle of Hu Sheep Lambskin. Animals 2020, 10, 1035. [Google Scholar] [CrossRef]
- Rishikaysh, P.; Dev, K.; Diaz, D.; Qureshi, W.; Filip, S.; Mokry, J. Signaling Involved in Hair Follicle Morphogenesis and Development. Int. J. Mol. Sci. 2014, 15, 1647–1670. [Google Scholar] [CrossRef]
- Chen, Y.; Fan, Z.; Wang, X.; Mo, M.; Zeng, S.B.; Xu, R.-H.; Wang, X.; Wu, Y. PI3K/Akt signaling pathway is essential for de novo hair follicle regeneration. Stem Cell Res. Ther. 2020, 11, 144. [Google Scholar] [CrossRef]
- Zhao, B.; Luo, H.; He, J.; Huang, X.; Chen, S.; Fu, X.; Zeng, W.; Tian, Y.; Liu, S.; Li, C.-J.; et al. Comprehensive transcriptome and methylome analysis delineates the biological basis of hair follicle development and wool-related traits in Merino sheep. BMC Biol. 2021, 19, 197. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Han, J.; Yuan, C.; Liu, J.; Niu, C.; Lu, Z.; Yue, Y.; Yang, B. Comparative proteomics reveals genetic mechanisms underlying secondary hair follicle development in fine wool sheep during the fetal stage. J. Proteom. 2020, 223, 103827. [Google Scholar] [CrossRef] [PubMed]
- Di Meo, G.P.; Perucatti, A.; Incarnato, D.; Ferretti, L.; Di Berardino, D.; Caputi Jambrenghi, A.; Vonghia, G.; Iannuzzi, L. Comparative mapping of twenty-eight bovine loci in sheep (Ovis aries, 2n = 54) and river buffalo (Bubalus bubalis, 2n = 50) by FISH. Cytogenet. Genome Res. 2002, 98, 262–264. [Google Scholar] [CrossRef]
- Pauciullo, A.; Perucatti, A.; Cosenza, G.; Iannuzzi, A.; Incarnato, D.; Genualdo, V.; Di Berardino, D.; Iannuzzi, L. Sequential Cross-Species Chromosome Painting among River Buffalo, Cattle, Sheep and Goat: A Useful Tool for Chromosome Abnormalities Diagnosis within the Family Bovidae. PLoS ONE 2014, 9, e110297. [Google Scholar] [CrossRef] [PubMed]
- Schibler, L.; Di Meo, G.P.; Cribiu, E.P.; Iannuzzi, L. Molecular Cytogenetics and Comparative Mapping in Goats (Capra hircus, 2n = 60). Cytogenet. Genome Res. 2009, 126, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Brown, G.R.; Hiatt, S.M.; Thibaud-Nissen, F.; Astashyn, A.; Ermolaeva, O.; Farrell, C.M.; Hart, J.; Landrum, M.J.; McGarvey, K.M.; et al. RefSeq: An update on mammalian reference sequences. Nucleic Acids Res. 2014, 42, D756–D763. [Google Scholar] [CrossRef] [PubMed]
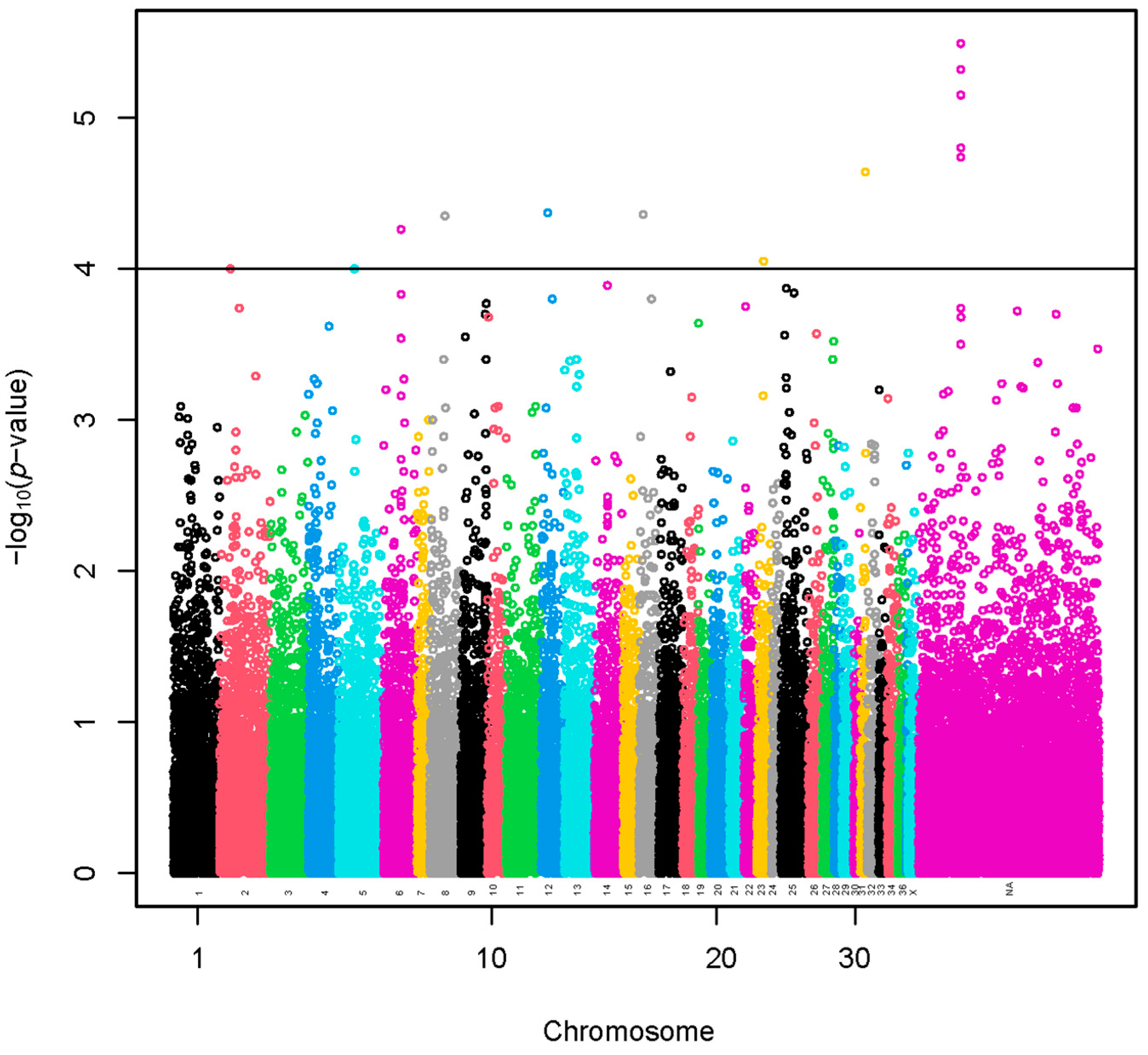
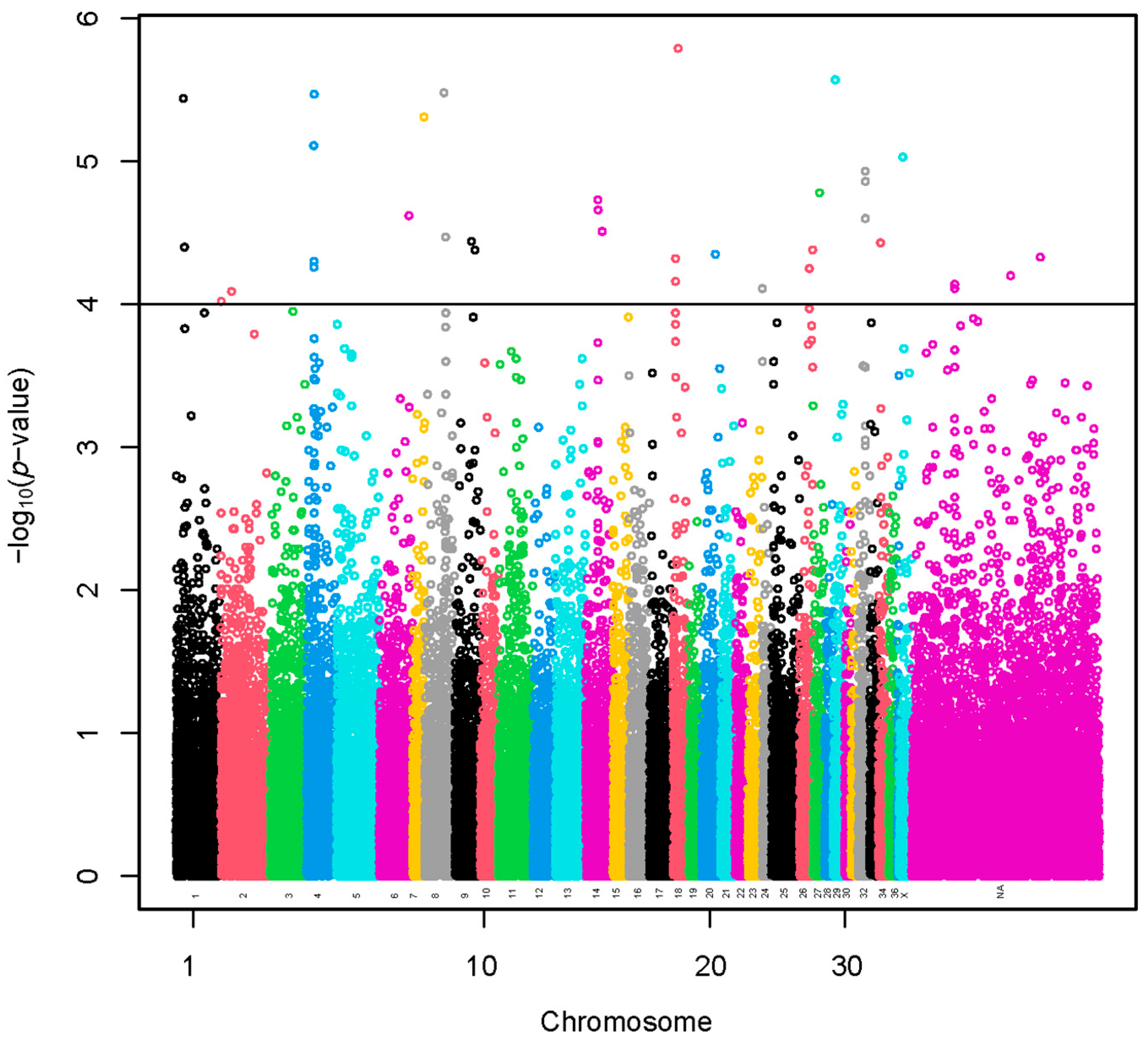


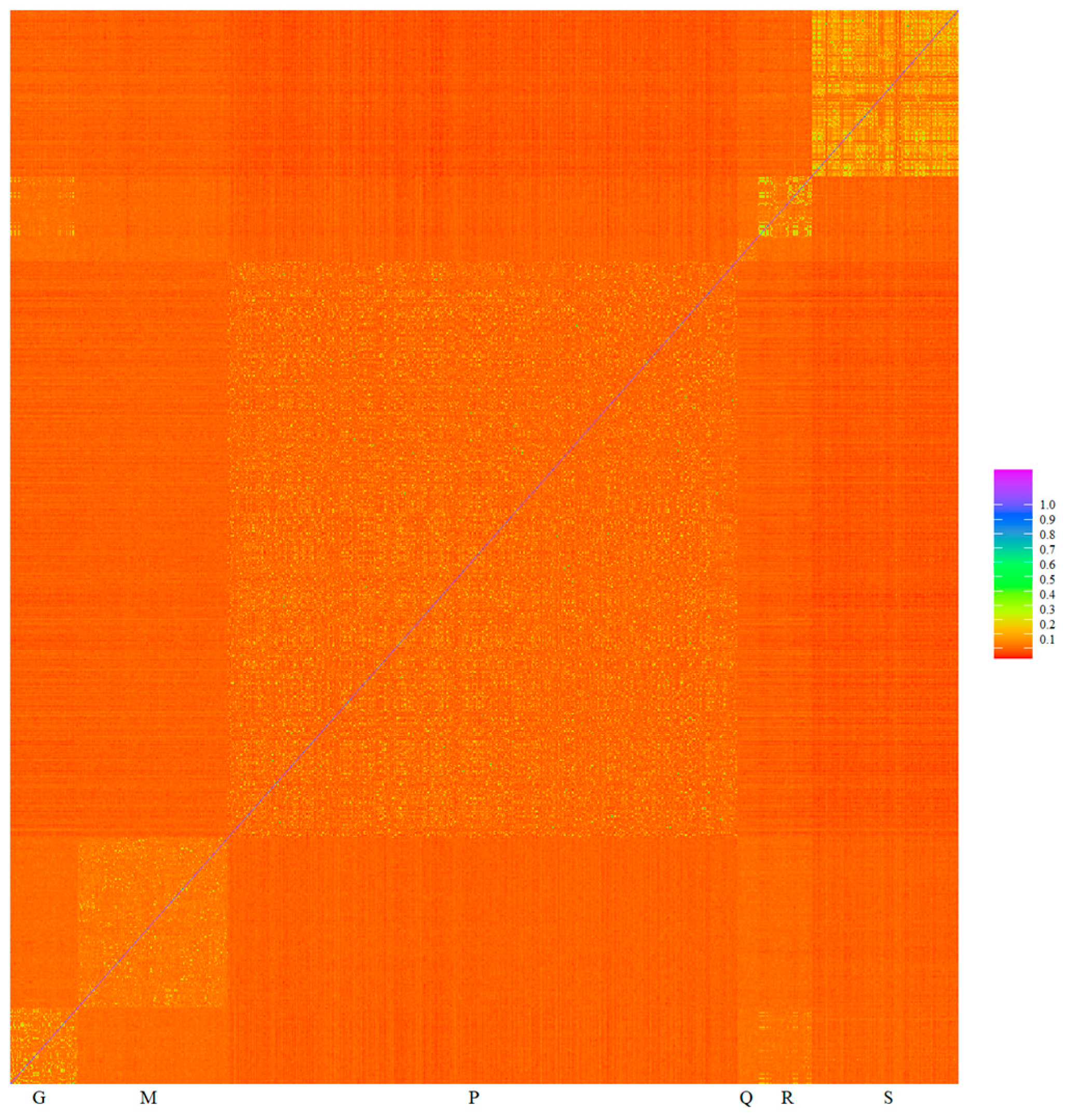
| VPA | Scaffolds | Start | End | Methods | SNPs |
|---|---|---|---|---|---|
| 6 | ABRR03077257.1 | 41319229 | 41714500 | Linear model—All the alpacas and selection signatures | AX-468770712 AX-417339934 AX-468704215 (*) AX-468766588 AX-417339050 AX-468763899 AX-468788057 AX-468767891 AX-417339948 AX-417339057 AX-468770136 AX-417339951 AX-468709166 |
| 9 | ABRR03004037.1 | 19580657 | 19719247 | Linear model—Extreme alpacas and selection signatures | AX-417305708 AX-468712525 |
| 29 | ABRR03000003.1 | 4613849 | 4659732 | Linear model—Extreme alpacas and selection signatures | AX-468751446 AX-417267477 AX-417266118 (*) |
| na | ABRR03000033.1 | 13561792 | 14451734 | Linear model—All the alpacas and extreme alpacas | AX-417286284 AX-432730912 AX-417286293 AX-417284656 AX-417280615 AX-417286315 AX-417284681 |
| VPA | Gene Name | Alpaca Scaffold | SNP | Distance (bp) Gene Marker | Method |
|---|---|---|---|---|---|
| 2 | TACC3 | ABRR03000026.1 | AX-417277871 | 289,181 | Linear model—Extreme alpacas |
| 7 | PSMA2 | ABRR03008474.1 | AX-468769383 | 279,078 | Linear model—Extreme alpacas |
| 7 | POLD2 | ABRR03008474.1 | AX-468769383 | 446,076 | Linear model—Extreme alpacas |
| 27 | NTRK3 | ABRR03000011.1 | AX-468719480 | 127,219 | Linear model—Extreme alpacas |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
More, M.; Veli, E.; Cruz, A.; Gutiérrez, J.P.; Gutiérrez, G.; Ponce de León, F.A. Genome-Wide Association Study of Fiber Diameter in Alpacas. Animals 2023, 13, 3316. https://doi.org/10.3390/ani13213316
More M, Veli E, Cruz A, Gutiérrez JP, Gutiérrez G, Ponce de León FA. Genome-Wide Association Study of Fiber Diameter in Alpacas. Animals. 2023; 13(21):3316. https://doi.org/10.3390/ani13213316
Chicago/Turabian StyleMore, Manuel, Eudosio Veli, Alan Cruz, Juan Pablo Gutiérrez, Gustavo Gutiérrez, and F. Abel Ponce de León. 2023. "Genome-Wide Association Study of Fiber Diameter in Alpacas" Animals 13, no. 21: 3316. https://doi.org/10.3390/ani13213316
APA StyleMore, M., Veli, E., Cruz, A., Gutiérrez, J. P., Gutiérrez, G., & Ponce de León, F. A. (2023). Genome-Wide Association Study of Fiber Diameter in Alpacas. Animals, 13(21), 3316. https://doi.org/10.3390/ani13213316








