The Assembly Process of Free-Living and Particle-Attached Bacterial Communities in Shrimp-Rearing Waters: The Overwhelming Influence of Nutrient Factors Relative to Microalgal Inoculation
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Microalgae Culture
2.2. Experimental Design
2.3. Environmental and Bacterial Sample Collection
2.4. Bacterial Illumina HiSeq Sequencing and Data Bioinformatic Analyses
2.5. Data Statistical Analyses
2.6. Co-Occurrence Network Analysis
3. Results
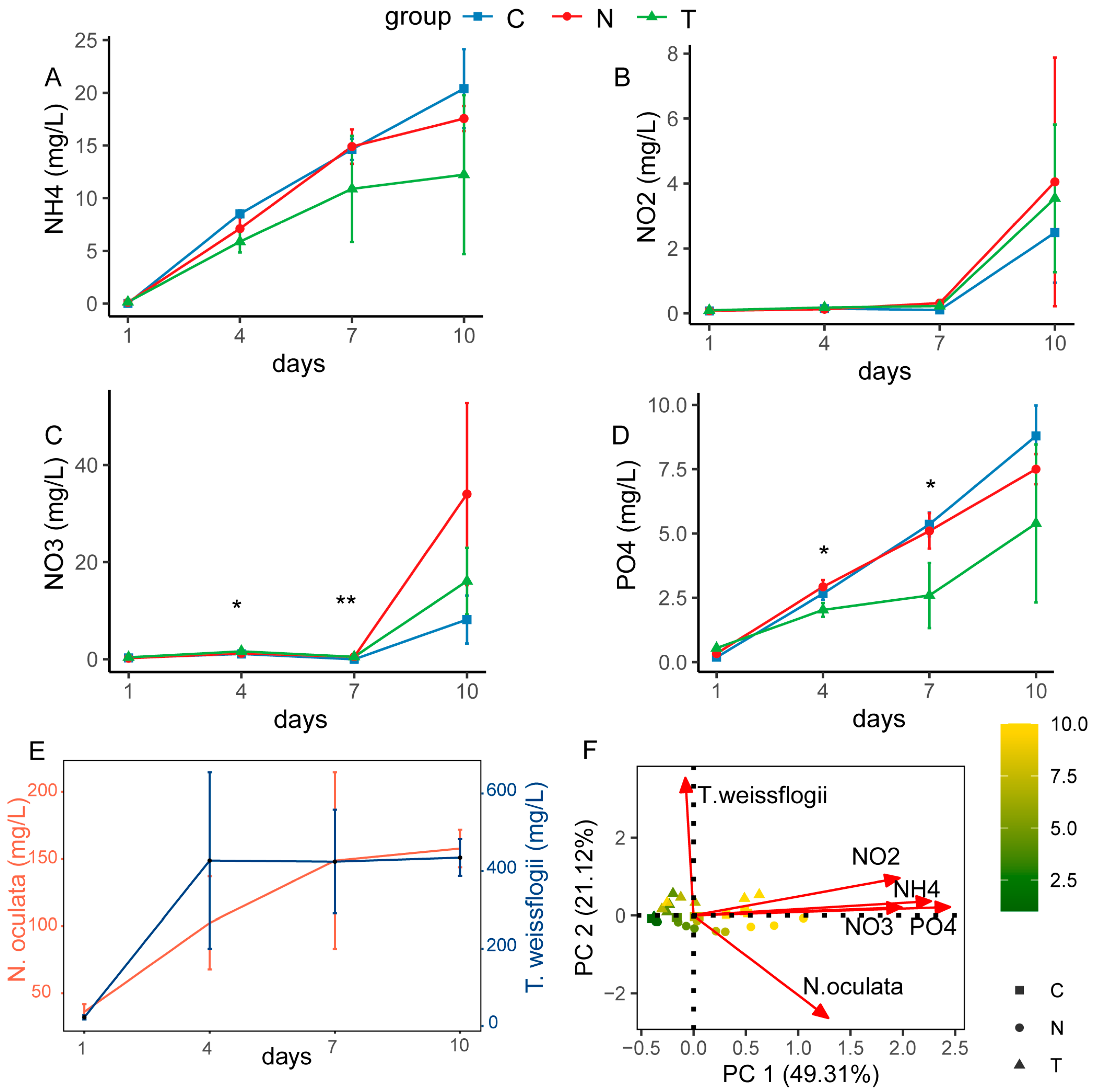
3.1. Variations in Microalgal and Nutrient Factors
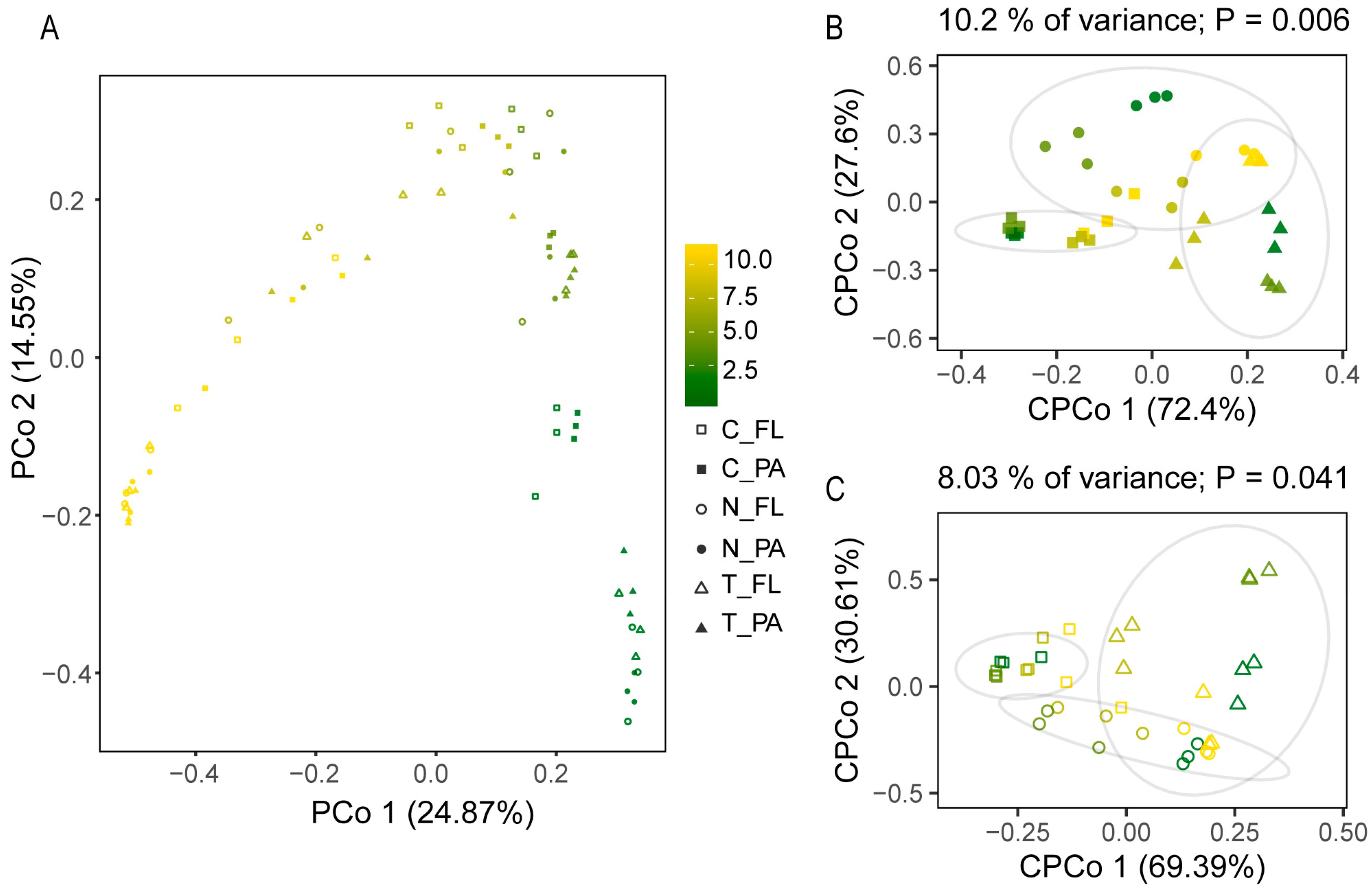
3.2. Dynamics of Bacterial Community Composition and Diversity
3.3. The Representative Bacterial Genera of Different Microalgal Treatments and Their Relationships with the Environment
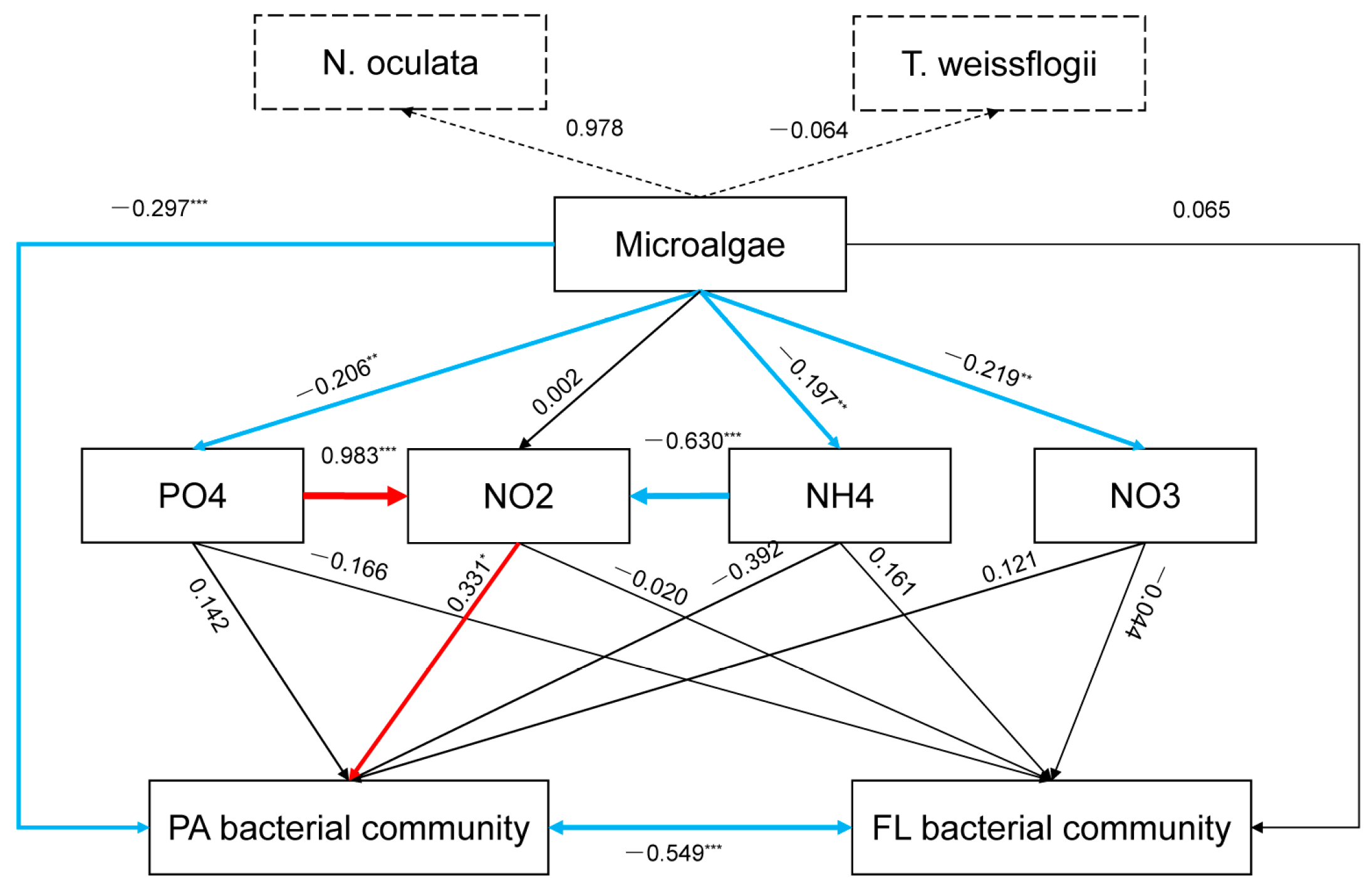
3.4. The Influence of Environmental Factors on Bacterial Communities
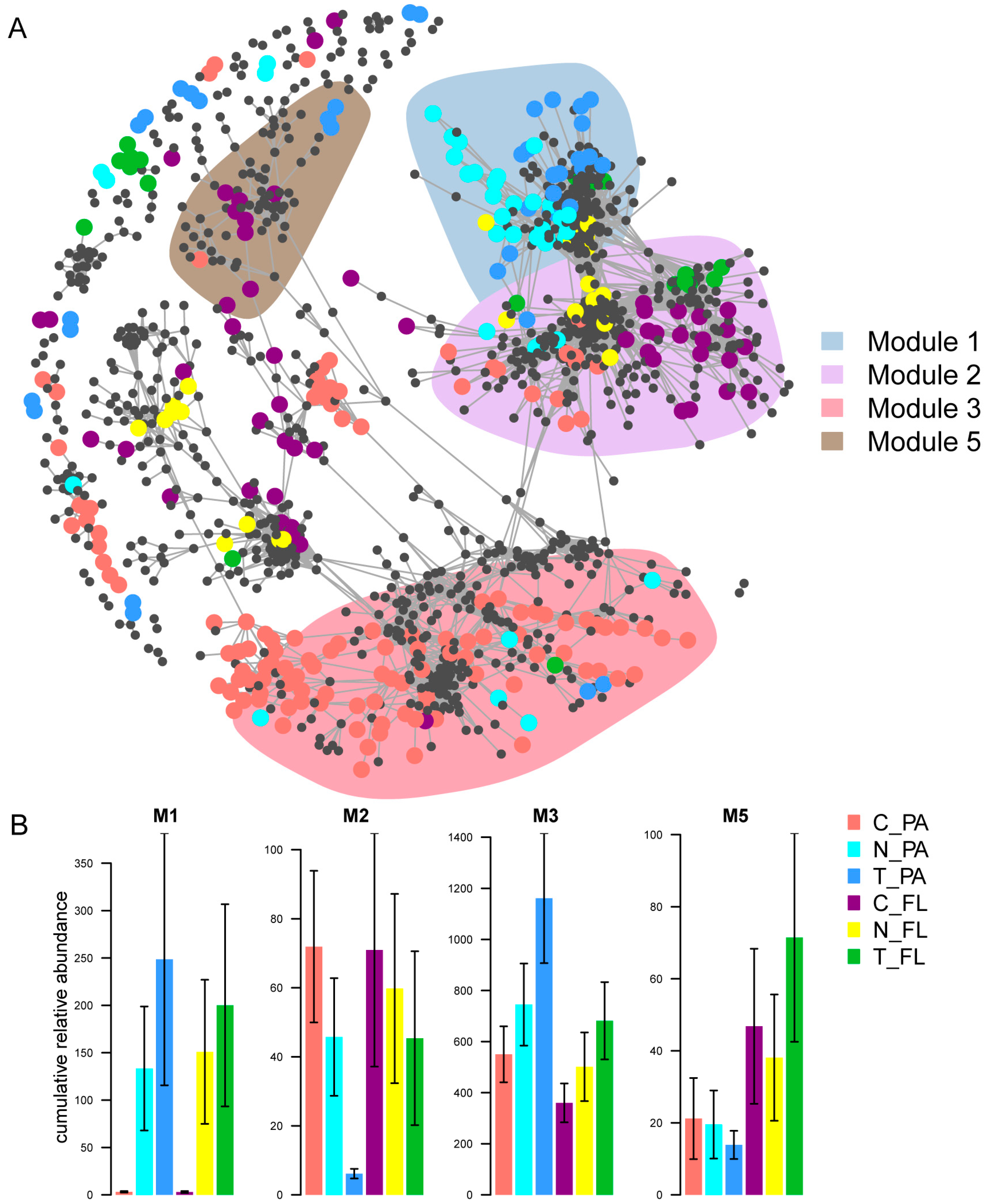
3.5. The Co-Occurrence Patterns of Bacterial Communities and Their Relationships with the Environment
4. Discussion
4.1. The Distinct Patterns of PA and FL Bacterial Communities in Shrimp-Rearing Waters
4.2. The Unequal Effects of Microalgae and Nutrient Factors on Bacterial Community Assembly
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brailo, M.; Schreier, H.J.; McDonald, R.; Marsic-Lucic, J.; Gavrilovic, A.; Pecarevic, M.; Jug-Dujakovic, J. Bacterial community analysis of marine recirculating aquaculture system bioreactors for complete nitrogen removal established from a commercial inoculum. Aquaculture 2019, 503, 198–206. [Google Scholar] [CrossRef]
- Xiong, J.; Zhu, J.; Zhang, D. The application of bacterial indicator phylotypes to predict shrimp health status. Appl. Microbiol. Biotechnol. 2014, 98, 8291–8299. [Google Scholar] [CrossRef]
- Xiong, J.; Xuan, L.; Yu, W.; Zhu, J.; Qiu, Q.; Chen, J. Spatiotemporal successions of shrimp gut microbial colonization: High consistency despite distinct species pool. Environ. Microbiol. 2019, 21, 1383–1394. [Google Scholar] [CrossRef]
- Kumar, V.; Roy, S.; Behera, B.K.; Swain, H.S.; Das, B.K. Biofloc Microbiome With Bioremediation and Health Benefits. Front. Microbiol. 2021, 12, 741164–741180. [Google Scholar] [CrossRef]
- Zheng, Y.; Yu, M.; Liu, Y.; Su, Y.; Xu, T.; Yu, M.; Zhang, X.-H. Comparison of cultivable bacterial communities associated with Pacific white shrimp (Litopenaeus vannamei) larvae at different health statuses and growth stages. Aquaculture 2016, 451, 163–169. [Google Scholar] [CrossRef]
- Palacios, O.A.; López, B.R.; de-Bashan, L.E. Microalga Growth-Promoting Bacteria (MGPB): A formal term proposed for beneficial bacteria involved in microalgal–bacterial interactions. Algal Res. 2022, 61, 102585. [Google Scholar] [CrossRef]
- Nimrat, S.; Suksawat, S.; Boonthai, T.; Vuthiphandchai, V. Potential Bacillus probiotics enhance bacterial numbers, water quality and growth during early development of white shrimp (Litopenaeus vannamei). Vet. Microbiol. 2012, 159, 443–450. [Google Scholar] [CrossRef]
- Godoy, L.C.; Odebrecht, C.; Ballester, E.; Martins, T.G.; Wasielesky, W., Jr. Effect of diatom supplementation during the nursery rearing of Litopenaeus vannamei (Boone, 1931) in a heterotrophic culture system. Aquac. Int. 2012, 20, 559–569. [Google Scholar] [CrossRef]
- Jensen, L.B.M.; Wasielesky, W.; Ballester, E.L.C.; Cavalli, R.O.; Santos, M.H.S. Role of microalgae Thalassiosira fluviatilis in weight gain and survival of the shrimp Farfantepenaeus paulensis reared in indoor nursery tanks. Naulius 2006, 14, 37–42. [Google Scholar]
- Silva, V.F.; Pereira, P.K.M.; Martins, M.A.; Lorenzo, M.A.D.; Cella, H.; Lopes, R.G.; Derner, R.B.; Magallon-Servin, P.; Vieira, F.D.N. Effects of Microalgae Addition and Fish Feed Supplementation in the Integrated Rearing of Pacific White Shrimp and Nile Tilapia Using Biofloc Technology. Animals 2022, 12, 1527. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, D.; Zeng, J.; Xu, H.; Huang, R.; Jiao, C.; Guo, L. Variations of bacterial community during the decomposition of Microcystis under different temperatures and biomass. BMC Microbiol. 2019, 19, 207–216. [Google Scholar] [CrossRef]
- Yang, W.; Zhu, J.; Zheng, C.; Lukwambe, B.; Nicholaus, R.; Lu, K.; Zheng, Z. Succession of phytoplankton community during intensive shrimp (Litopenaeus vannamei) cultivation and its effects on cultivation systems. Aquaculture 2020, 520, 734733. [Google Scholar] [CrossRef]
- Yang, W.; Zheng, C.; Zheng, Z.; Wei, Y.; Lu, K.; Zhu, J. Nutrient enrichment during shrimp cultivation alters bacterioplankton assemblies and destroys community stability. Ecotoxicol. Environ. Saf. 2018, 156, 366–374. [Google Scholar] [CrossRef]
- Wu, W.; Xu, Z.; Dai, M.; Gan, J.; Liu, H.; Richter, C. Homogeneous selection shapes free-living and particle-associated bacterial communities in subtropical coastal waters. Divers. Distrib. 2020, 27, 1904–1917. [Google Scholar] [CrossRef]
- Chun, S.J.; Cui, Y.; Ahn, C.Y.; Oh, H.M. Improving water quality using settleable microalga Ettlia sp. and the bacterial community in freshwater recirculating aquaculture system of Danio rerio. Water Res. 2018, 135, 112–121. [Google Scholar] [CrossRef]
- Wang, C.; Jiang, C.; Gao, T.; Peng, X.; Ma, S.; Sun, Q.; Xia, B.; Xie, X.; Bai, Z.; Xu, S.; et al. Improvement of fish production and water quality in a recirculating aquaculture pond enhanced with bacteria-microalgae association. Aquaculture 2022, 547, 737420. [Google Scholar] [CrossRef]
- Ding, Y.; Chen, N.; Ke, J.; Qi, Z.; Chen, W.; Sun, S.; Zheng, Z.; Xu, J.; Yang, W. Response of the rearing water bacterial community to the beneficial microalga Nannochloropsis oculata cocultured with Pacific white shrimp (Litopenaeus vannamei). Aquaculture 2021, 542, 736895. [Google Scholar] [CrossRef]
- Liu, B.; Eltanahy, E.E.; Liu, H.; Chua, E.T.; Thomas-Hall, S.R.; Wass, T.J.; Pan, K.; Schenk, P.M. Growth-promoting bacteria double eicosapentaenoic acid yield in microalgae. Bioresour. Technol. 2020, 316, 123916. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Edgar, R.C. UNOISE2: Improved error-correction for illumina 16S AND ITS amplicon sequencing. bioRxiv 2016. [Google Scholar] [CrossRef]
- Xu, H.; Zhao, D.; Zeng, J.; Jiao, C.; Yu, Z.; Wu, Q.L. Distinct successional patterns and processes of free-living and particle-attached bacterial communities throughout a phytoplankton bloom. Freshw. Biol. 2020, 65, 1363–1375. [Google Scholar] [CrossRef]
- Xu, H.; Zhao, D.; Huang, R.; Cao, X.; Zeng, J.; Yu, Z.; Hooker, K.V.; Hambright, K.D.; Wu, Q.L. Contrasting Network Features between Free-Living and Particle-Attached Bacterial Communities in Taihu Lake. Microb. Ecol. 2018, 76, 303–313. [Google Scholar] [CrossRef]
- Wang, S.; Hou, W.; Jiang, H.; Dong, H.; Huang, L.; Chen, S.; Wang, B.; Chen, Y.; Lin, B.; Deng, Y. The Lifestyle-Dependent Microbial Interactions Vary Between Upstream and Downstream of the Three Gorges Dam. Front. Ecol. Evol. 2021, 9, 624476. [Google Scholar] [CrossRef]
- Hu, Y.; Xie, G.; Jiang, X.; Shao, K.; Tang, X.; Gao, G. The Relationships Between the Free-Living and Particle-Attached Bacterial Communities in Response to Elevated Eutrophication. Front. Microbiol. 2020, 11, 423–436. [Google Scholar] [CrossRef]
- Liu, M.; Liu, L.; Chen, H.; Yu, Z.; Yang, J.R.; Xue, Y.; Huang, B.; Yang, J. Community dynamics of free-living and particle-attached bacteria following a reservoir Microcystis bloom. Sci. Total Environ. 2019, 660, 501–511. [Google Scholar] [CrossRef]
- Ling, T.; Zhang, Y.-F.; Cao, J.-Y.; Xu, J.-L.; Kong, Z.-Y.; Zhang, L.; Liao, K.; Zhou, C.-X.; Yan, X.-J. Analysis of bacterial community diversity within seven bait-microalgae. Algal Res. 2020, 51, 102033. [Google Scholar] [CrossRef]
- Sun, F.; Wang, C.; Wang, Y.; Tu, K.; Zheng, Z.; Lin, X. Diatom red tide significantly drive the changes of microbiome in mariculture ecosystem. Aquaculture 2020, 520, 734742. [Google Scholar] [CrossRef]
- Li, C.; Liu, J.; Chen, X.; Ren, H.; Su, B.; Ma, K.; Tu, Q. Determinism governs the succession of disturbed bacterioplankton communities in a coastal maricultural ecosystem. Sci. Total Environ. 2022, 828, 154457–154467. [Google Scholar] [CrossRef]
- Paix, B.; Ezzedine, J.A.; Jacquet, S. Diversity, Dynamics, and Distribution of Bdellovibrio and Like Organisms in Perialpine Lakes. Appl. Environ. Microb. 2019, 85, 6. [Google Scholar] [CrossRef]
- Riemann, L.; Winding, A. Community Dynamics of Free-living and Particle-associated Bacterial Assemblages during a Freshwater Phytoplankton Bloom. Microb. Ecol. 2001, 42, 274–285. [Google Scholar] [CrossRef]
- Mestre, M.; Borrull, E.; Sala, M.; Gasol, J.M. Patterns of bacterial diversity in the marine planktonic particulate matter continuum. ISME J. 2017, 11, 999–1010. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, C.; Zhu, Y.; Yuan, C.; Zhao, T. Effect of bacteria-to-algae volume ratio on treatment performance and microbial community of a novel heterotrophic nitrification-aerobic denitrification bacteria-chlorella symbiotic system. Bioresour. Technol. 2021, 342, 126025. [Google Scholar] [CrossRef]
- Cheng, R.; Zhu, H.; Shutes, B.; Yan, B. Treatment of microcystin (MC-LR) and nutrients in eutrophic water by constructed wetlands: Performance and microbial community. Chemosphere 2021, 263, 128139–128149. [Google Scholar] [CrossRef]
- Ramos e Silva, C.A.; Sternberg, L.d.S.L.; Dávalos, P.B.; Souza, F.E.S.d. The impact of organic and intensive farming on the tropical estuary. Ocean. Coast. Manag. 2017, 141, 55–64. [Google Scholar] [CrossRef]
- Thomas, Y.; Courties, C.; El Helwe, Y.; Herbland, A.; Lemonnier, H. Spatial and temporal extension of eutrophication associated with shrimp farm wastewater discharges in the New Caledonia lagoon. Mar. Pollut. Bull. 2010, 61, 387–398. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, L.; Dong, H.; Duan, Y.; Li, Z.; Wen, G.; Chen, J.; Feng, Z.; Xu, W.; Xie, J. Artificial substrates in zero-water-exchange culture system regulate the rearing performance of Pacific white shrimp Litopenaeus vannamei (Boone, 1931) under the winter indoor condition. Aquac. Res. 2016, 47, 91–100. [Google Scholar] [CrossRef]
- Li, J.; Li, J.; Li, W.; Sun, Y.; Liu, X.; Liu, M.; Cheng, Y. JuvenileProcambarus clarkiifarmed using biofloc technology or commercial feed in zero-water exchange indoor tanks: A comparison of growth performance, enzyme activity and proximate composition. Aquac. Res. 2019, 50, 1834–1843. [Google Scholar] [CrossRef]
- Lukwambe, B.; Zhao, L.; Nicholaus, R.; Yang, W.; Zhu, J.; Zheng, Z. Bacterioplankton community in response to biological filters (clam, biofilm, and macrophytes) in an integrated aquaculture wastewater bioremediation system. Environ. Pollut. 2019, 254, 113035. [Google Scholar] [CrossRef]
- Lukwambe, B.; Nicholaus, R.; Yang, W.; Zheng, Z. Blood clams (Tegillarca granosa) bioturbation alter succession of bacterioplankton community and nutrient removal performance in an aquaculture wastewater bioremediation system. Aquaculture 2020, 516, 734520. [Google Scholar] [CrossRef]
- Zhao, L.; Zheng, Y.; Nicholaus, R.; Lukwambe, B.; Zhu, J.; Yang, W.; Zheng, Z. Bioturbation by the razor clam Sinonovacula constricta affects benthic nutrient fluxes in aquaculture wastewater treatment ecosystems. Aquac. Environ. Interact. 2019, 11, 87–96. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, G.; Yu, D.; Qiu, Y.; Li, S.; Guo, E. Novel nitrogen removal process in marine aquaculture wastewater treatment using Enteromorpha ferment liquid as carbon. Bioresour. Technol. 2023, 377, 128913. [Google Scholar] [CrossRef]
- Ren, Z.; Qu, X.; Peng, W.; Yu, Y.; Zhang, M. Nutrients Drive the Structures of Bacterial Communities in Sediments and Surface Waters in the River-Lake System of Poyang Lake. Water 2019, 11, 930. [Google Scholar] [CrossRef]
- Tian, W.; Zhang, H.; Guo, Y.; Wang, Z.; Huang, T. Temporal and Spatial Patterns of Sediment Microbial Communities and Driving Environment Variables in a Shallow Temperate Mountain River. Microorganisms 2022, 10, 816. [Google Scholar] [CrossRef]
- Acinas, S.G.; Sanchez, P.; Salazar, G.; Cornejo-Castillo, F.M.; Sebastian, M.; Logares, R.; Royo-Llonch, M.; Paoli, L.; Sunagawa, S.; Hingamp, P.; et al. Deep ocean metagenomes provide insight into the metabolic architecture of bathypelagic microbial communities. Commun. Biol. 2021, 4, 604–618. [Google Scholar] [CrossRef]
- Ma, S.N.; Wang, H.J.; Wang, H.Z.; Zhang, M.; Li, Y.; Bian, S.J.; Liang, X.M.; Sondergaard, M.; Jeppesen, E. Effects of nitrate on phosphorus release from lake sediments. Water Res. 2021, 194, 116894–116903. [Google Scholar] [CrossRef]
- Ma, S.N.; Wang, H.J.; Wang, H.Z.; Li, Y.; Liu, M.; Liang, X.M.; Yu, Q.; Jeppesen, E.; Sondergaard, M. High ammonium loading can increase alkaline phosphatase activity and promote sediment phosphorus release: A two-month mesocosm experiment. Water Res. 2018, 145, 388–397. [Google Scholar] [CrossRef]
- Natrah, F.M.I.; Bossier, P.; Sorgeloos, P.; Yusoff, F.M.; Defoirdt, T. Significance of microalgal-bacterial interactions for aquaculture. Rev. Aquac. 2014, 6, 48–61. [Google Scholar] [CrossRef]
- Wang, H.; Qi, M.; Bo, Y.; Zhou, C.; Yan, X.; Wang, G.; Cheng, P. Treatment of fishery wastewater by co-culture of Thalassiosira pseudonana with Isochrysis galbana and evaluation of their active components. Algal Res. Biomass Biofuels Bioprod. 2021, 60, 102498. [Google Scholar] [CrossRef]
- Saxena, A.; Singh, P.K.; Bhatnagar, A.; Tiwari, A. Growth of marine diatoms on aquaculture wastewater supplemented with nanosilica. Bioresour. Technol. 2022, 344, 126210. [Google Scholar] [CrossRef]
- Collao, J.; Morales-Amaral, M.D.M.; Acien-Fernandez, F.G.; Bolado-Rodriguez, S.; Fernandez-Gonzalez, N. Effect of operational parameters, environmental conditions, and biotic interactions on bacterial communities present in urban wastewater treatment photobioreactors. Chemosphere 2021, 284, 131271–131282. [Google Scholar] [CrossRef] [PubMed]
- Tisserand, L.; Dadaglio, L.; Intertaglia, L.; Catala, P.; Panagiotopoulos, C.; Obernosterer, I.; Joux, F. Use of organic exudates from two polar diatoms by bacterial isolates from the Arctic Ocean. Philos. Trans. A Math. Phys. Eng. Sci. 2020, 378, 20190356–20190375. [Google Scholar] [CrossRef] [PubMed]
- Tada, Y.; Suzuki, K. Changes in the community structure of free-living heterotrophic bacteria in the open tropical Pacific Ocean in response to microalgal lysate-derived dissolved organic matter. FEMS Microbiol. Ecol. 2016, 92, fiw099. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.; Schimmel, P.; Sanchez-Garcia, S.; Wijffels, R.H.; Smidt, H.; Sipkema, D. Different co-occurring bacteria enhance or decrease the growth of the microalga Nannochloropsis sp. CCAP211/78. Microb. Biotechnol. 2021, 14, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Biondi, N.; Cheloni, G.; Rodolfi, L.; Viti, C.; Giovannetti, L.; Tredici, M.R. Tetraselmis suecica F&M-M33 growth is influenced by its associated bacteria. Microb. Biotechnol. 2018, 11, 211–223. [Google Scholar] [CrossRef]
- Lian, J.; Wijffels, R.H.; Smidt, H.; Sipkema, D. The effect of the algal microbiome on industrial production of microalgae. Microb. Biotechnol. 2018, 11, 806–818. [Google Scholar] [CrossRef]
- Avci, B.; Kruger, K.; Fuchs, B.M.; Teeling, H.; Amann, R.I. Polysaccharide niche partitioning of distinct Polaribacter clades during North Sea spring algal blooms. ISME J. 2020, 14, 1369–1383. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Fraschetti, S.; Alifano, P.; Pizzolante, G.; Stabili, L. The alien species Caulerpa cylindracea and its associated bacteria in the Mediterranean Sea. Mar. Biol. 2016, 163, 4. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, B.; He, C.; Shi, J.; Wu, M.; Guo, J. Sulfur-based Mixotrophic Vanadium (V) Bio-reduction towards Lower Organic Requirement and Sulfate Accumulation. Water Res. 2021, 189, 116655. [Google Scholar] [CrossRef]

| Factors | Whole | PA | FL |
|---|---|---|---|
| Treat | 0.086 *** | 0.120 *** | 0.089 ** |
| Time | 0.221 *** | 0.236 *** | 0.253 *** |
| Lifestyles | 0.083 *** | 0.109 *** | 0.083 ** |
| Whole | PA | FL | |
|---|---|---|---|
| PC | 0.226 *** | 0.221 *** | 0.257 *** |
| PC1 | 0.289 *** | 0.298 *** | 0.360 *** |
| PC2 | 0.031 | 0.012 | 0.009 |
| TM1 | TM2 | TM3 | TM5 | |
|---|---|---|---|---|
| PC1 | −3.827 *** | 5.102 *** | 4.305 *** | 2.163 * |
| PC2 | 1.316 | −1.971 | 3.781 *** | 1.221 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Y.; Wang, X.; Cai, H.; Ke, J.; Zhu, J.; Lu, K.; Zheng, Z.; Yang, W. The Assembly Process of Free-Living and Particle-Attached Bacterial Communities in Shrimp-Rearing Waters: The Overwhelming Influence of Nutrient Factors Relative to Microalgal Inoculation. Animals 2023, 13, 3484. https://doi.org/10.3390/ani13223484
Shi Y, Wang X, Cai H, Ke J, Zhu J, Lu K, Zheng Z, Yang W. The Assembly Process of Free-Living and Particle-Attached Bacterial Communities in Shrimp-Rearing Waters: The Overwhelming Influence of Nutrient Factors Relative to Microalgal Inoculation. Animals. 2023; 13(22):3484. https://doi.org/10.3390/ani13223484
Chicago/Turabian StyleShi, Yikai, Xuruo Wang, Huifeng Cai, Jiangdong Ke, Jinyong Zhu, Kaihong Lu, Zhongming Zheng, and Wen Yang. 2023. "The Assembly Process of Free-Living and Particle-Attached Bacterial Communities in Shrimp-Rearing Waters: The Overwhelming Influence of Nutrient Factors Relative to Microalgal Inoculation" Animals 13, no. 22: 3484. https://doi.org/10.3390/ani13223484
APA StyleShi, Y., Wang, X., Cai, H., Ke, J., Zhu, J., Lu, K., Zheng, Z., & Yang, W. (2023). The Assembly Process of Free-Living and Particle-Attached Bacterial Communities in Shrimp-Rearing Waters: The Overwhelming Influence of Nutrient Factors Relative to Microalgal Inoculation. Animals, 13(22), 3484. https://doi.org/10.3390/ani13223484





