Characterization of the Ruminal Microbiome of Water Buffaloes (Bubalus bubalis) Kept in Different Ecosystems in the Eastern Amazon
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Aspects
2.2. Ecosystems
2.2.1. Ilha do Marajó (IM)
2.2.2. Baixo Amazonas (BA)
2.2.3. Continente do Pará (CP)
2.2.4. Tomé-Açu (TA)
2.3. Pasture Sampling and Analysis
2.4. Animals
2.5. Sampling of Rumen Contents and DNA Extraction
2.6. Sequencing
2.7. Bioinformatics Processing
2.8. Statistical Analysis
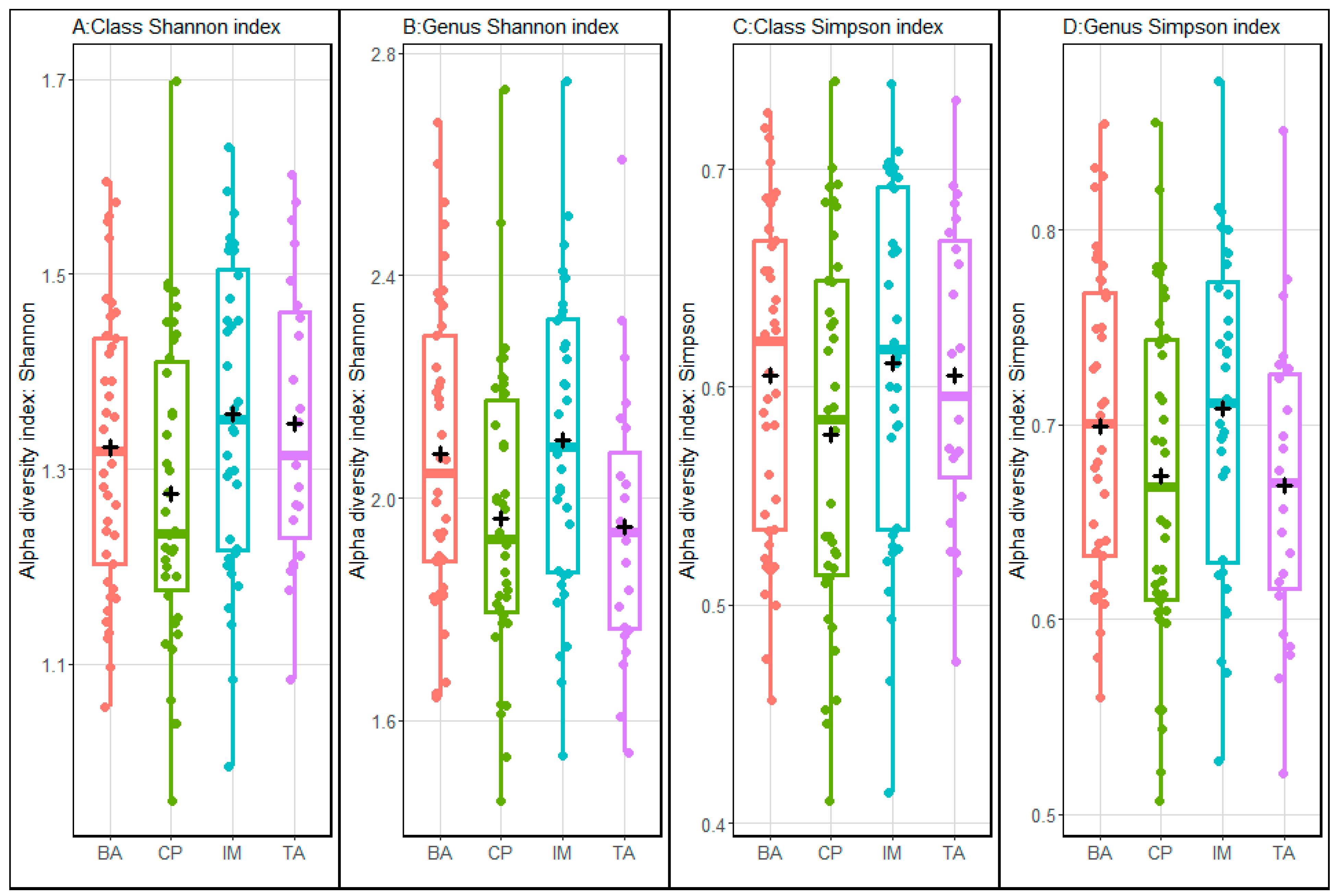
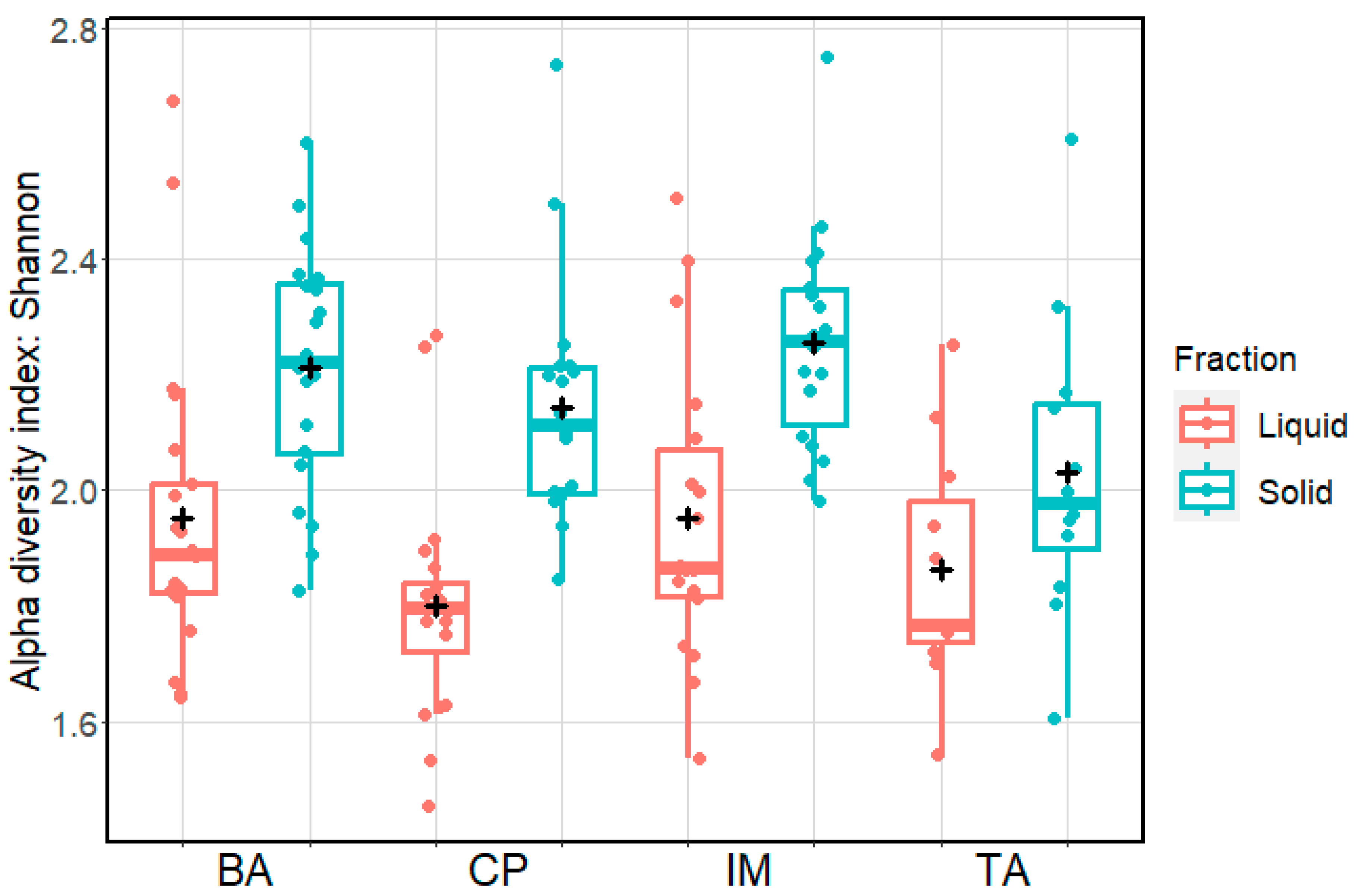
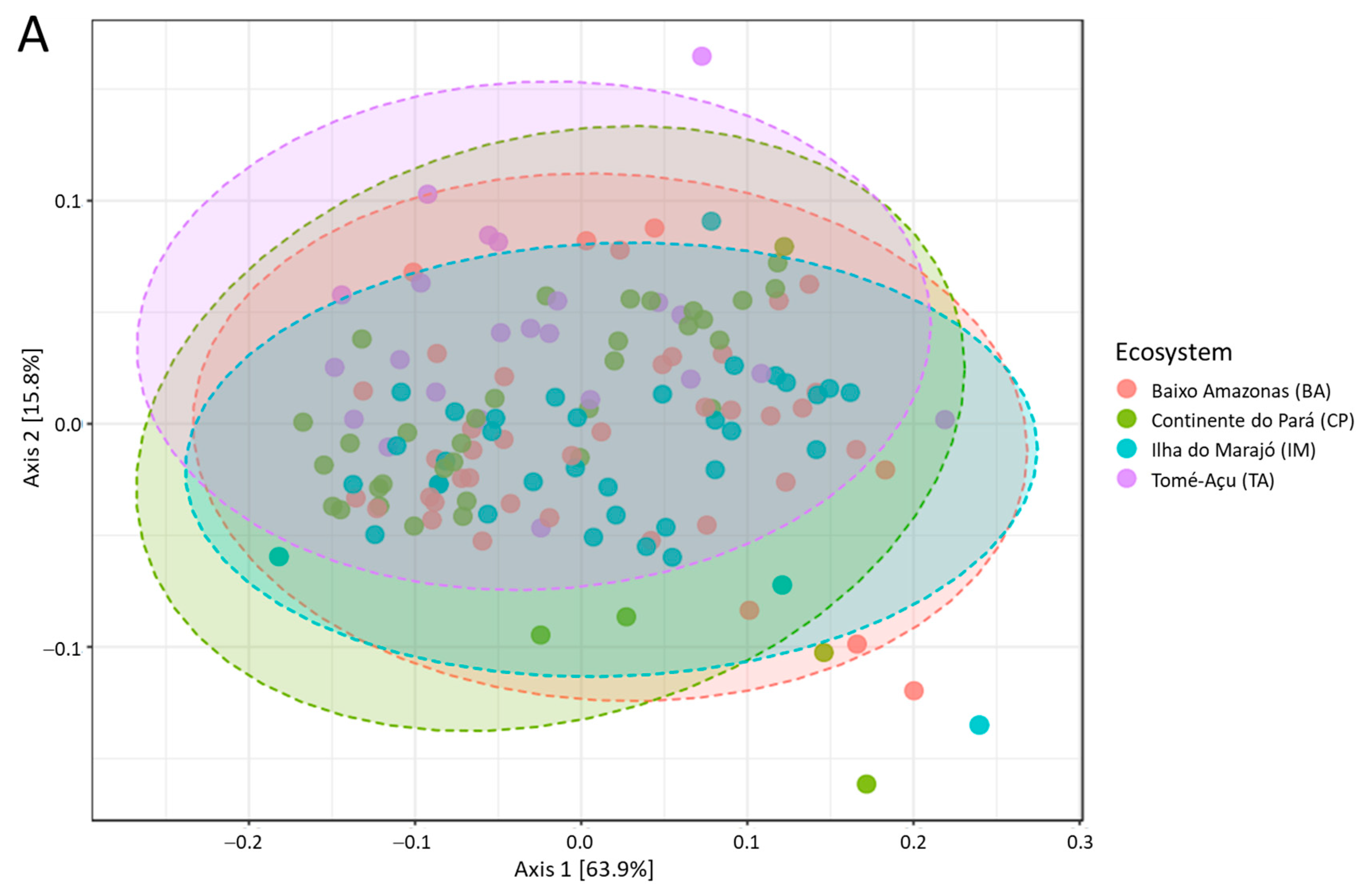
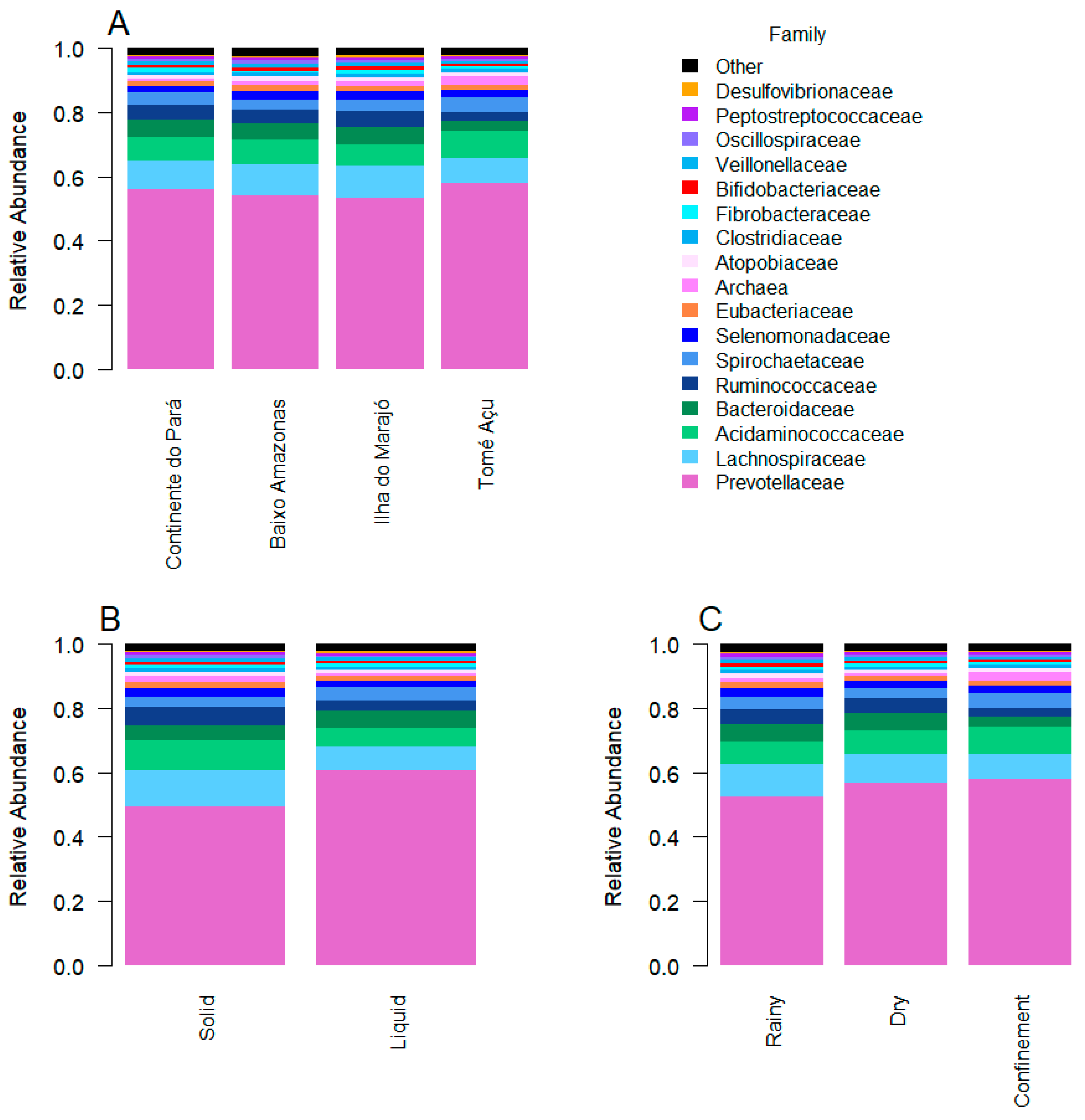
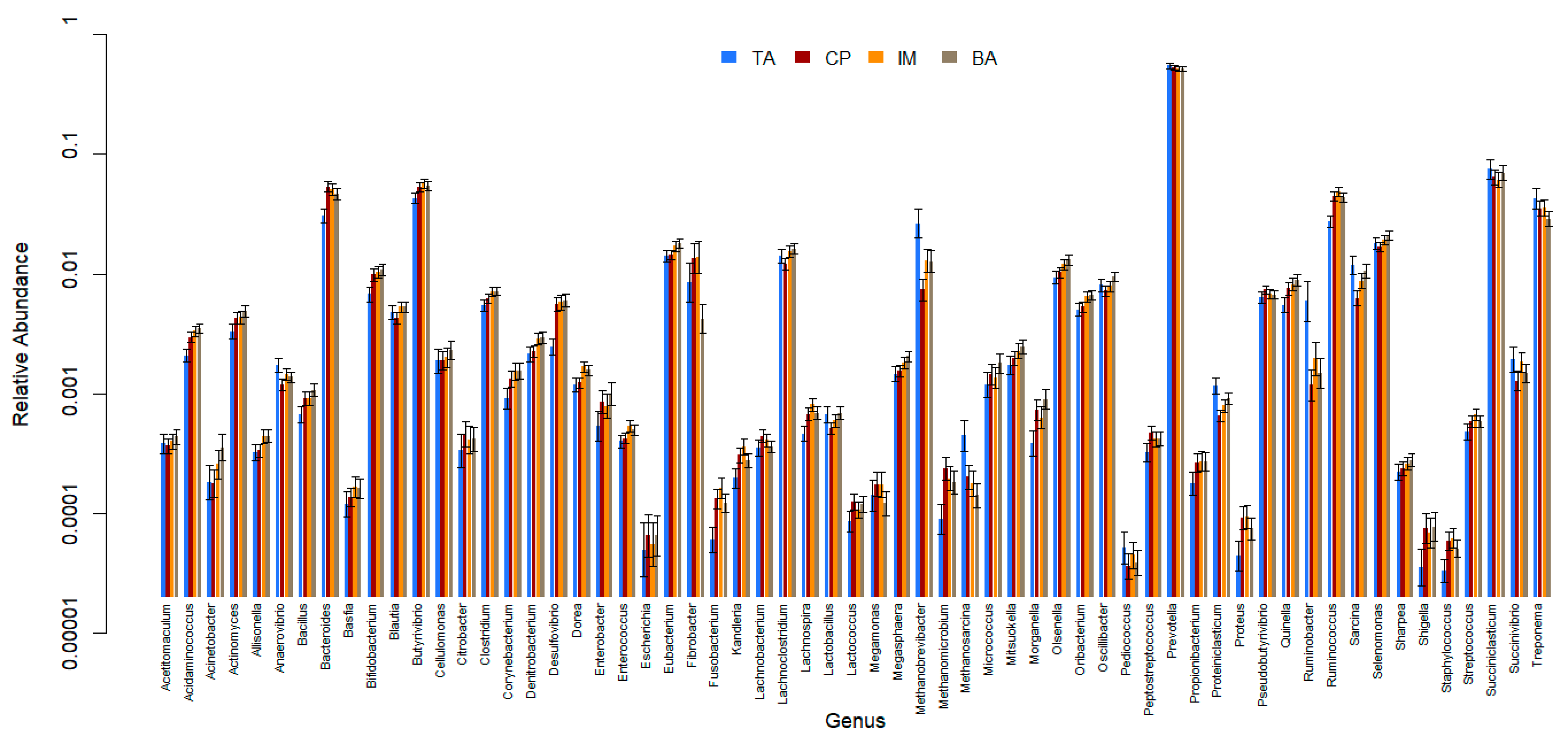
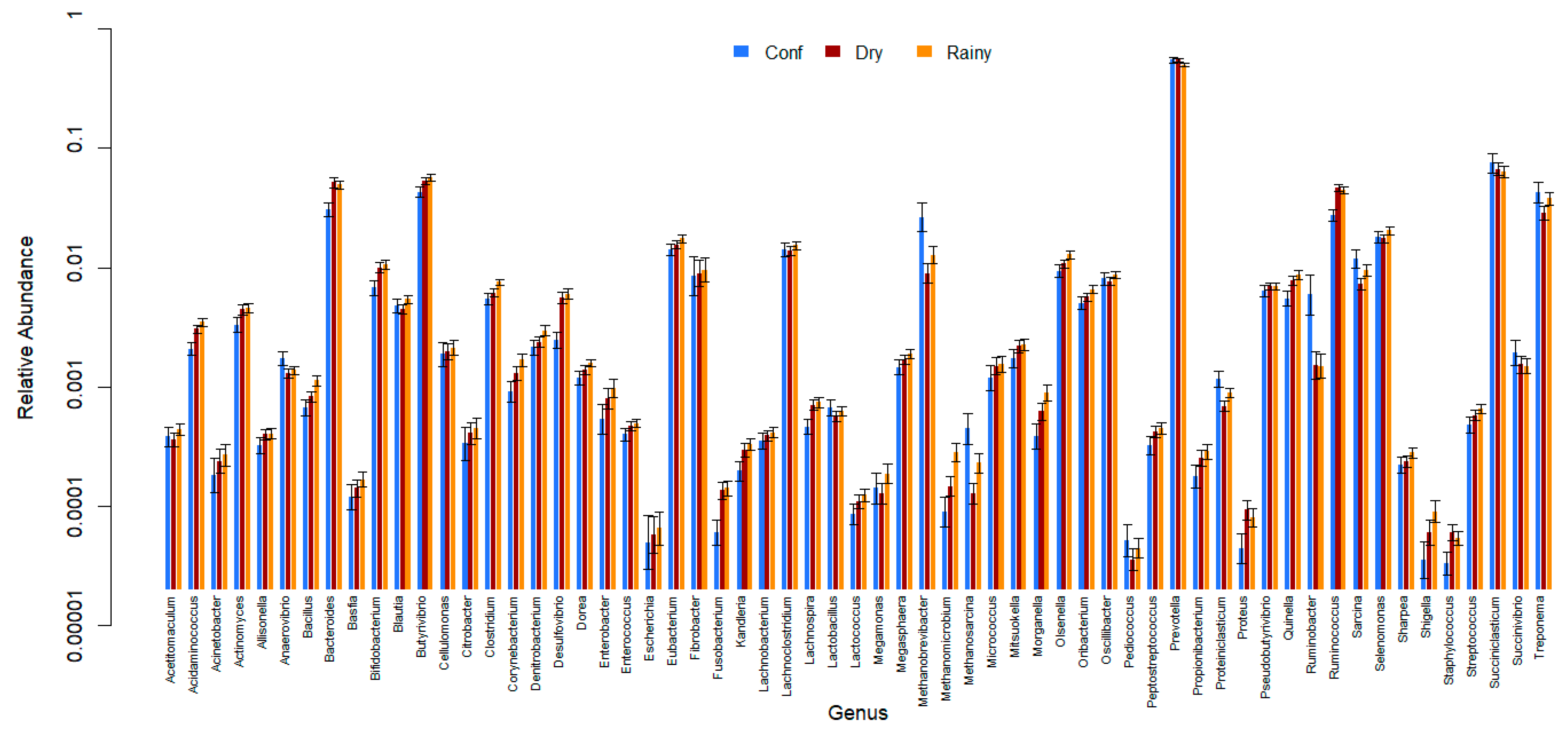
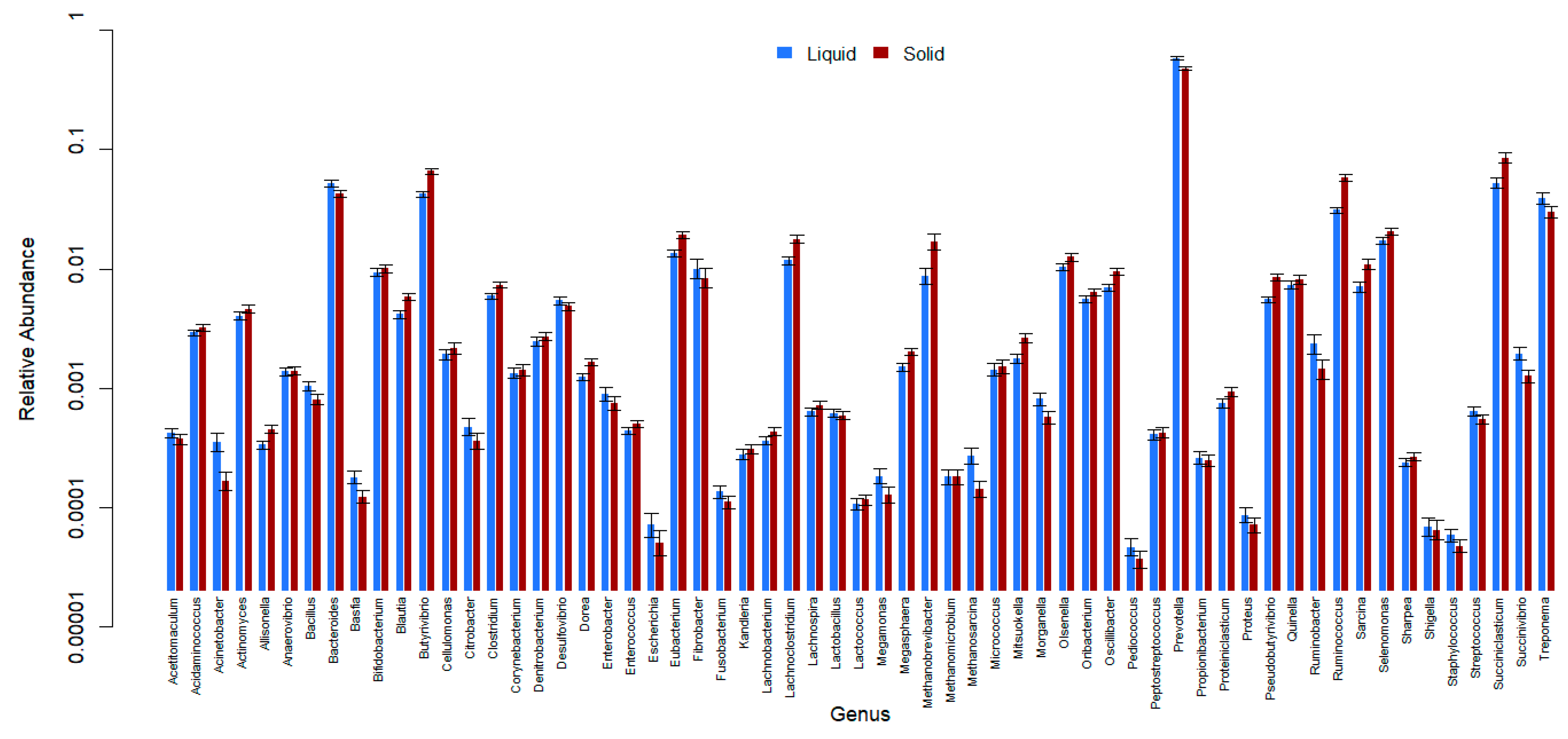
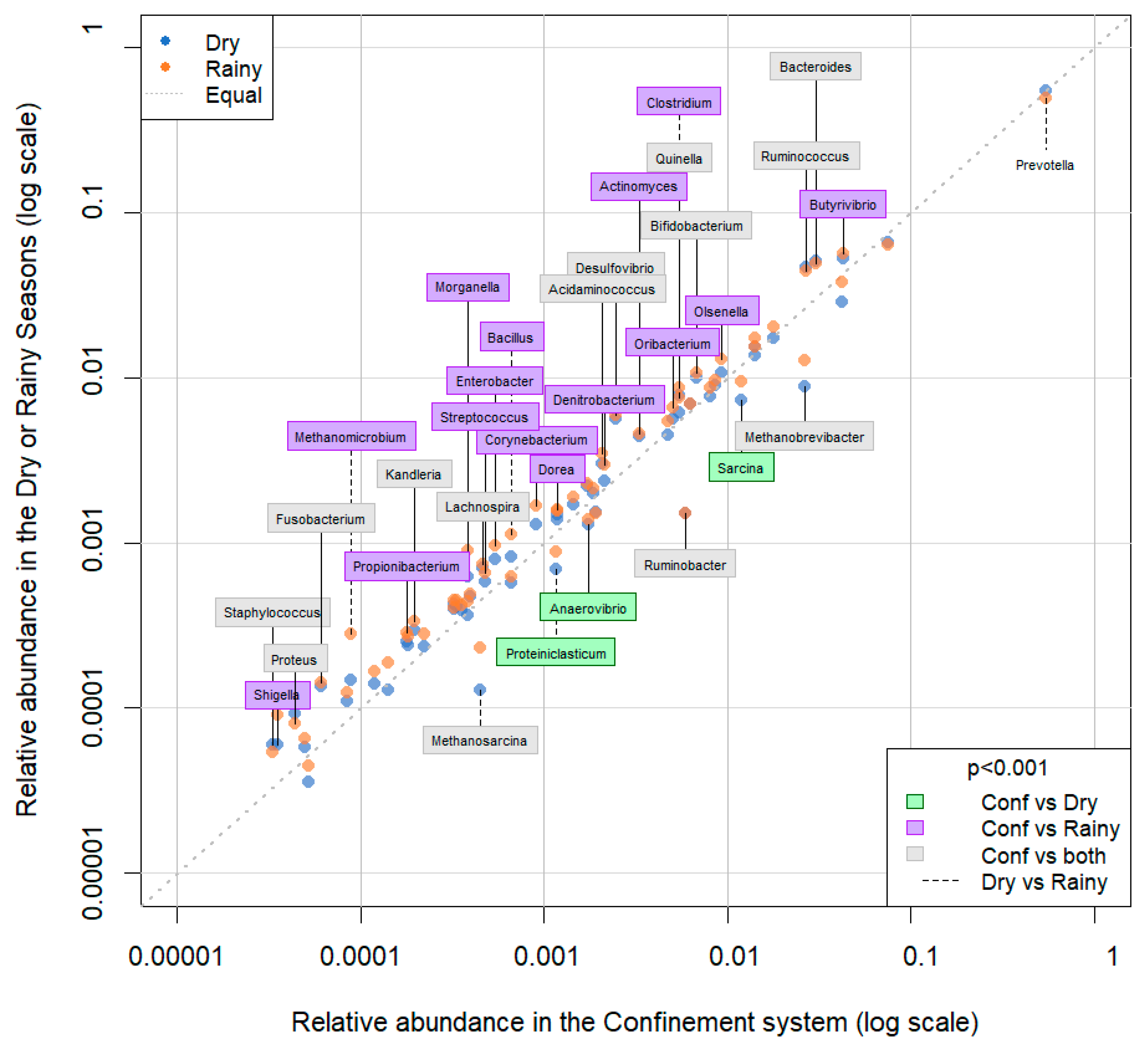
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Institute Brasileiro de Geografia e Estatística. Rebanho de Búfalos no Brasil. 2023. Available online: https://www.ibge.gov.br/explica/producao-agropecuaria/bubalinos/br (accessed on 12 December 2023).
- Vieira, J.N.; Teixeira, C.S.; Kuabara, M.Y.; de Oliveira, D.A.A. Bubalinocultura no Brasil. Pubvet 2011, 5, 1003. [Google Scholar] [CrossRef]
- National Reasearch Council. The Water Buffalo: New Prospects for an Underutilized Animal, 2nd ed.; National Academy Press: Washington, DC, USA, 1984.
- Schader, C.; Muller, A.; El-Hage Scialabba, N.; Hecht, J.; Isensee, A.; Erb, K.H.; Smith, P.; Makkar, H.P.S.; Klocke, P.; Leiber, F.; et al. Impacts of feeding less food-competing feedstuffs to livestock on global food system sustainability. J. R. Soc. Interface 2015, 12, 20150891. [Google Scholar] [CrossRef] [PubMed]
- Bergman, E.N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990, 70, 567–590. [Google Scholar] [CrossRef] [PubMed]
- Bach, A.; Calsamiglia, S.; Stern, M.D. Nitrogen metabolism in the rumen. J. Dairy Sci. 2005, 88, E9–E21. [Google Scholar] [CrossRef] [PubMed]
- Lan, W.; Yang, C. Ruminal methane production: Associated microorganisms and the potential of applying hydrogen-utilizing bacteria for mitigation. Sci. Total Environ. 2019, 654, 1270–1283. [Google Scholar] [CrossRef] [PubMed]
- Delseny, M.; Han, B.; Hsing, Y.I. High throughput DNA sequencing: The new sequencing revolution. Plant Sci. 2010, 179, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Ansorge, W.J. Next-generation DNA sequencing techniques. Nat. Biotechnol. 2009, 25, 195–203. [Google Scholar] [CrossRef]
- Hess, M.K.; Rowe, S.J.; Van Stijn, T.C.; Henry, H.M.; Hickey, S.M.; Brauning, R.; McCulloch, A.F.; Hess, A.S.; Kirk, M.R.; Kumar, S.; et al. A restriction enzyme reduced representation sequencing approach for low-cost, high-throughput metagenome profiling. PLoS ONE 2020, 15, e0219882. [Google Scholar] [CrossRef]
- Hess, M.K.; Hodgkinson, H.E.; Hess, A.S.; Zetouni, L.; Budel, J.C.; Henry, H.; Donaldson, A.; Bilton, T.P.; van Stijn, T.C.; Kirk, M.R.; et al. Large-scale analysis of sheep rumen metagenome profiles captured by reduced representation sequencing reveals individual profiles are influenced by the environment and genetics of the host. BMC Genom. 2023, 24, 551. [Google Scholar] [CrossRef]
- Alvares, C.A.; Stape, J.L.; Sentelhas, P.C.; Moraes, G.J.L.; Sparovek, G. Köppen’s climate classification map for Brazil. Meteorol. Z. 2013, 22, 711–728. [Google Scholar] [CrossRef]
- Francez, D.C.; Rosa, L.S. Viabilidade econômica de sistemas agroflorestais em áreas de agricultores familiares no Pará, Brasil. Rev. Ciências Agrárias 2011, 54, 178–187. [Google Scholar] [CrossRef]
- Pacheco, N.A.; Bastos, T.X. Caracterização Climática do Município de Tomé-Açu, PA; Embrapa Amazônia Oriental: Belém of Pará, Brazil, 2001; Available online: https://ainfo.cnptia.embrapa.br/digital/bitstream/item/63489/1/Oriental-Doc87.pdf (accessed on 12 December 2023).
- Salman, A.K.D.; Soares, J.P.G.; Canesin, R.C. Métodos de Amostragem para Avaliação Quantitativa de Pastagens; EMBRAPA Embrapa: Porto Velho, Rondônia, Brazil, 2006; Available online: https://ainfo.cnptia.embrapa.br/digital/bitstream/item/24669/1/ct84-pastagem.pdf (accessed on 12 December 2023).
- Detmann, E.; Souza, M.A.; Valadares Filho, S.C.; Queiroz, A.C.; Berchielli, T.T.; Saliba, E.O.S.; Azevedo, J.A.G. Métodos para Análise de Alimentos; Instituto Nacional de Ciência e Tecnologia de Ciência Animal: Visconde do Rio Branco, Minas Gerais, Brazil, 2012. [Google Scholar]
- Stevenson, D.M.; Weimer, P.J. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 2007, 75, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A Robust, Simple Genotyping-by-Sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Seshadri, R.; Leahy, S.C.; Attwood, G.T.; Teh, K.H.; Lambie, S.C.; Cookson, A.L.; Eloe-Fadrosh, E.A.; Pavlopoulos, G.A.; Hadjithomas, M.; Varghese, N.J.; et al. Cultivation and sequencing of rumen microbiome members from the Hungate1000 Collection. Nat. Biotechnol. 2018, 36, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Altermann, E.; Leahy, S.C.; Jauregui, R.; Jonker, A.; Henderson, G.; Kittelmann, S.; Attwood, G.T.; Kamke, J.; Waters, S.M.; et al. Genomic insights into the physiology of Quinella, an iconic uncultured rumen bacterium. Nat. Commun. 2022, 13, 6240. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 22, e61217. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar]
- RStudio Team. RStudio: Integrated Sevelopment for R; RStudio: Boston, MA, USA, 2020. [Google Scholar]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 48. [Google Scholar] [CrossRef]
- Luo, D.; Ganesh, S.; Koolaard, J. Predictmeans: Calculate Predicted Means for Linear Models. 2018. Available online: https://cran.r-hub.io/web/packages/predictmeans/index.html (accessed on 12 December 2023).
- Kuznetsova, A.; Brockhoff, P.B.; Christensen, R.H. lmerTest package: Tests in linear mixed effects models. J. Stat. Softw. 2017, 82, 1–26. [Google Scholar] [CrossRef]
- Iqbal, M.W.; Zhang, Q.; Yang, Y.; Li, L.; Zou, C.; Huang, C.; Lin, B. Comparative study of rumen fermentation and microbial community differences between water buffalo and Jersey cows under similar feeding conditions. J. Appl. Anim. Res. 2018, 46, 740–748. [Google Scholar] [CrossRef]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Global Rumen Census Collaborators; Janssen, P.H. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef] [PubMed]
- AlZahal, O.; Li, F.; Guan, L.L.; Walker, N.D.; McBride, B.W. Factors influencing ruminal bacterial community diversity and composition and microbial fibrolytic enzyme abundance in lactating dairy cows with a focus on the role of active dry yeast. J. Dairy Sci. 2017, 100, 4377–4393. [Google Scholar] [CrossRef] [PubMed]
- Nathani, N.M.; Patel, A.K.; Mootapally, C.S.; Reddy, B.; Shah, S.V.; Lunagaria, P.M.; Kothari, R.K.; Joshi, C.G. Effect of roughage on rumen microbiota composition in the efficient feed converter and sturdy Indian Jaffrabadi buffalo (Bubalus bubalis). BMC Genom. 2015, 16, 1116. [Google Scholar] [CrossRef] [PubMed]
- Betancur-Murillo, C.L.; Aguilar-Marín, S.B.; Jovel, J. Prevotella: A key player in ruminal metabolism. Microorganisms 2023, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Wallace, R.J.; McKain, N.; Broderick, G.A. Breakdown of different peptides by Prevotella (Bacteroides) ruminicola and mixed microorganisms from the sheep rumen. Curr. Microbiol. 1993, 26, 333–336. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.B. Fermentation of Peptides by Bacteroides ruminicola B14. Appl. Environ. Microbiol. 1983, 45, 1566–1574. [Google Scholar] [CrossRef]
- Bi, Y.; Zeng, S.; Zhang, R.; Diao, Q.; Tu, Y. Effects of dietary energy levels on rumen bacterial community composition in Holstein heifers under the same forage to concentrate ratio condition. BMC Microbiol. 2018, 18, 69. [Google Scholar] [CrossRef]
- Fernando, S.C.; Purvis, H.T.; Najar, F.Z.; Sukharnikov, L.O.; Krehbiel, C.R.; Nagaraja, T.G.; Roe, B.A.; DeSilva, U. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 2010, 76, 7482–7490. [Google Scholar] [CrossRef]
- Franzolin, R.; St-Pierre, B.; Northwood, K.; Wright, A.-D.G. Analysis of rumen methanogen diversity in water buffaloes (Bubalus bubalis) under three different diets. Microb. Ecol. 2012, 64, 131–139. [Google Scholar] [CrossRef]
- Kumar, S.; Dagar, S.S.; Agrawal, R.K.; Puniya, A.K. Comparative diversity analysis of ruminal methanogens in Murrah buffaloes (Bubalus bubalis) in four states of North India. Anaerobe 2018, 52, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.; Patel, A.K.; Parmar, N.R.; Patel, A.B.; Reddy, B.; Joshi, C.G. Characterization of the rumen microbiome of Indian Kankrej cattle (Bos indicus) adapted to different forage diet. Appl. Microbiol. Biotechnol. 2014, 98, 9749–9761. [Google Scholar] [CrossRef] [PubMed]
- Rooke, J.A.; Wallace, R.J.; Duthie, C.-A.; McKain, N.; de Souza, S.M.; Hyslop, J.J.; Ross, D.W.; Waterhouse, T.; Roehe, R. Hydrogen and methane emissions from beef cattle and their rumen microbial community vary with diet, time after feeding and genotype. Br. J. Nutr. 2014, 112, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Pandit, R.J.; Hinsu, A.T.; Patel, S.H.; Jakhesara, S.J.; Koringa, P.G.; Bruno, F.; Psifidi, A.; Shah, S.V.; Joshi, C.G. Microbiota composition, gene pool and its expression in Gir cattle (Bos indicus) rumen under different forage diets using metagenomic and metatranscriptomic approaches. Syst. Appl. Microbiol. 2018, 41, 374–385. [Google Scholar] [CrossRef]
- Bowen, J.M.; McCabe, M.S.; Lister, S.J.; Cormican, P.; Dewhurst, R.J. Evaluation of microbial communities associated with the liquid and solid phases of the rumen of cattle offered a diet of perennial ryegrass or white clover. Front. Microbiol. 2018, 9, 1–8. [Google Scholar] [CrossRef]
- De Mulder, T.; Goossens, K.; Peiren, N.; Vandaele, L.; Haegeman, A.; De Tender, C.; Ruttink, T.; Van de Wiele, T.; De Campeneere, S. Exploring the methanogen and bacterial communities of rumen environments: Solid adherent, fluid and epimural. FEMS Microbiol. Ecol. 2017, 93, 1–12. [Google Scholar] [CrossRef]




| Ecosystem | Lattitude | Longitude | Altitude (m) | Köppen Classification | Rainfall (mm) | Av. Temp. (°C) | Av Humidity (%) |
|---|---|---|---|---|---|---|---|
| IM | 0°39′27.89″ S | 48°42′35.01″ W | 7 | Am | 2500 | 27 | 85 |
| BA | 02°41′48.83″ S | 54°38′35.43″ W | 108 | Am | 2000 | 26 | 86 |
| CP | 01°12′52.63″ S | 47°24′30.94″ W | 53 | Am | 2467 | 26 | 86 |
| TA | 02°17′28.32″ S | 48°05′56.11″ W | Am |
| Ecosystem | Weight (kg) | Age (Months) | ||
|---|---|---|---|---|
| Rainy | Dry | Rainy | Dry | |
| Ilha do Marajó (IM) | 396 | 418 | 24 | 36 |
| Baixo Amazonas (BA) | 445 | 454 | 24 | 36 |
| Continente do Pará (CP) | 212 | 432 | 24 | 36 |
| Tomé-Açu (TA) | 433 | 18 | ||
| Items | BA 1 | IM 2 | CP 3 | TA 4 | ||||
|---|---|---|---|---|---|---|---|---|
| DS | RS | DS | RS | DS | RS | Forage | BR | |
| Proximate composition (% dry matter) | ||||||||
| Dry matter | 23.97 | 24.1 | 23.87 | 18.31 | 23.32 | 26.12 | 24.71 | 39 |
| Organic matter | 91.53 | 90.98 | 89.45 | 84.86 | 91.79 | 96.88 | 95.35 | 94.77 |
| Crude protein | 7.86 | 8.72 | 7.56 | 8.86 | 7.73 | 28.03 | 9.38 | 8.27 |
| NDF | 73.23 | 79.07 | 68.72 | 70.91 | 75.73 | 55.69 | 68.96 | 54.36 |
| NFC | 9.07 | 1.79 | 11.79 | 3.09 | 6.27 | 5.16 | 13.85 | 29.58 |
| ADF | 44.99 | 54.9 | 40.05 | 43.9 | 55.8 | 22.55 | 44.51 | 38.08 |
| Ether extract | 1.36 | 1.4 | 1.38 | 1.99 | 1.64 | 8 | 2.06 | 6.52 |
| TDN | 51.48 | 42.2 | 56.09 | 52.5 | 41.35 | 72.48 | 51.93 | 57.94 |
| Ash | 8.47 | 9.02 | 10.55 | 15.14 | 8.21 | 3.12 | 6.17 | 5.23 |
| Mean rumen pH | ||||||||
| 7.18 | 7.53 | 7.23 | 7.35 | 7.36 | 7.13 | 6.86 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noronha, G.N.; Hess, M.K.; Dodds, K.G.; Silva, A.G.M.e.; de Souza, S.M.; da Silva, J.A.R.; Graças, D.A.d.; de Carvalho Rodrigues, T.C.G.; da Silva, W.C.; da Silva, É.B.R.; et al. Characterization of the Ruminal Microbiome of Water Buffaloes (Bubalus bubalis) Kept in Different Ecosystems in the Eastern Amazon. Animals 2023, 13, 3858. https://doi.org/10.3390/ani13243858
Noronha GN, Hess MK, Dodds KG, Silva AGMe, de Souza SM, da Silva JAR, Graças DAd, de Carvalho Rodrigues TCG, da Silva WC, da Silva ÉBR, et al. Characterization of the Ruminal Microbiome of Water Buffaloes (Bubalus bubalis) Kept in Different Ecosystems in the Eastern Amazon. Animals. 2023; 13(24):3858. https://doi.org/10.3390/ani13243858
Chicago/Turabian StyleNoronha, Gerlane Nunes, Melanie K. Hess, Ken G. Dodds, André Guimarães Maciel e Silva, Shirley Motta de Souza, Jamile Andréa Rodrigues da Silva, Diego Assis das Graças, Thomaz Cyro Guimarães de Carvalho Rodrigues, Welligton Conceição da Silva, Éder Bruno Rebelo da Silva, and et al. 2023. "Characterization of the Ruminal Microbiome of Water Buffaloes (Bubalus bubalis) Kept in Different Ecosystems in the Eastern Amazon" Animals 13, no. 24: 3858. https://doi.org/10.3390/ani13243858
APA StyleNoronha, G. N., Hess, M. K., Dodds, K. G., Silva, A. G. M. e., de Souza, S. M., da Silva, J. A. R., Graças, D. A. d., de Carvalho Rodrigues, T. C. G., da Silva, W. C., da Silva, É. B. R., Janssen, P. H., Henry, H. M., Rowe, S. J., de Castro, V. C. G., & Lourenço-Júnior, J. d. B. (2023). Characterization of the Ruminal Microbiome of Water Buffaloes (Bubalus bubalis) Kept in Different Ecosystems in the Eastern Amazon. Animals, 13(24), 3858. https://doi.org/10.3390/ani13243858









