Antibiotic-Resistant Bacteria Dissemination in the Wildlife, Livestock, and Water of Maiella National Park, Italy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
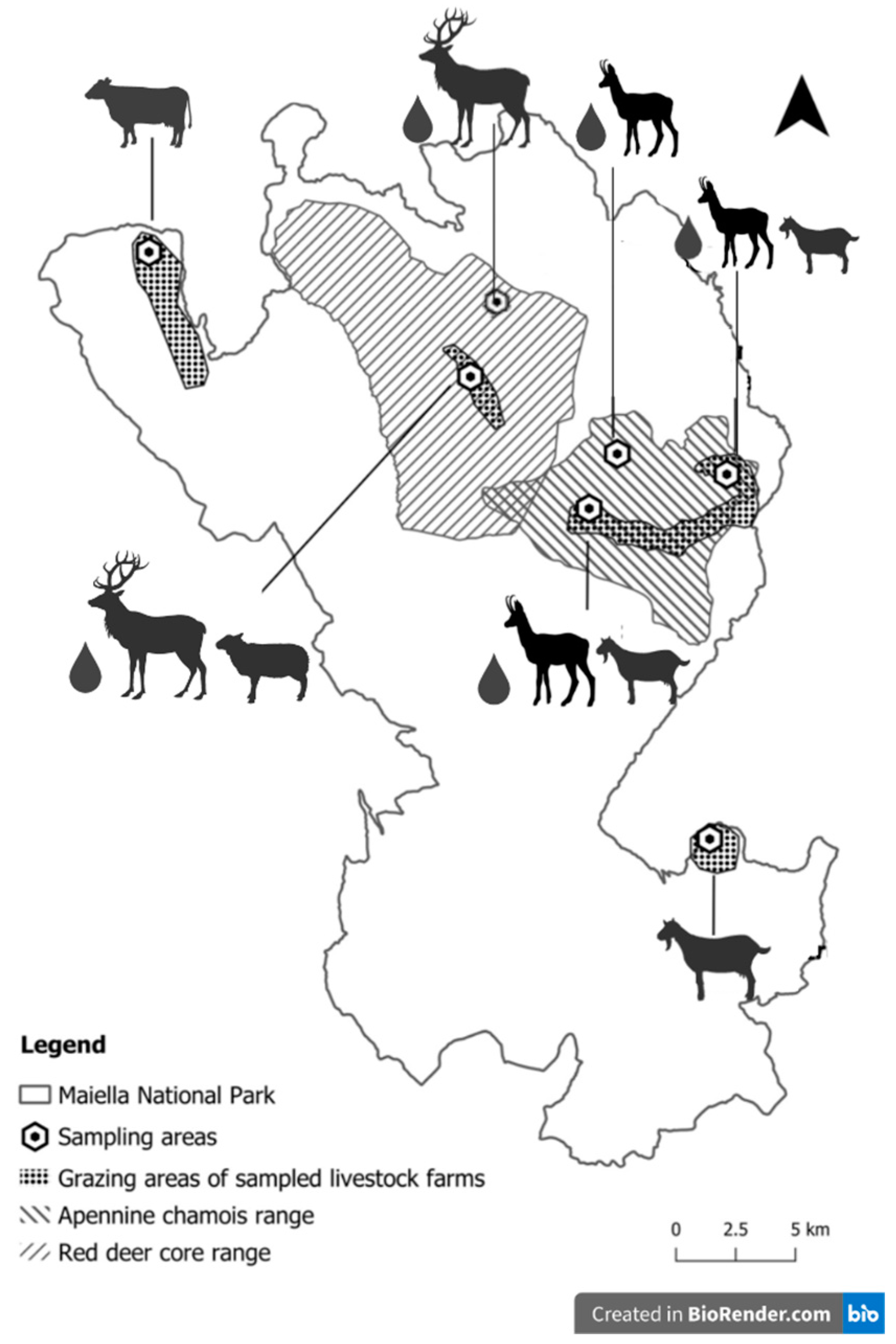
2.1. Study Area
2.2. Sampling Design
2.3. Bacteria Isolation and Antibiotic Susceptibility Test
2.4. Detection of Antibiotic Resistance Genes
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Altamirano, F.L.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I. A brief history of the antibiotic era: Lessons learned and challenges for the future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.; Wertheim, H.F.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef]
- Woolhouse, M.; Ward, M.; van Bunnik, B.; Farrar, J. Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R Soc. Lond. Biol. Sci. 2015, 370, 20140083. [Google Scholar] [CrossRef]
- Walsh, T.R. A one-health approach to antimicrobial resistance. Nat. Microbiol. 2018, 3, 854–855. [Google Scholar] [CrossRef]
- White, A.; Hughes, J.M. Critical Importance of a One Health Approach to Antimicrobial Resistance. EcoHealth 2019, 16, 404–409. [Google Scholar] [CrossRef]
- Vittecoq, M.; Godreuil, S.; Prugnolle, F.; Durand, P.; Brazier, L.; Renaud, N.; Arnal, A.; Aberkane, S.; Jean-Pierre, H.; Gauthier-Clerc, M.; et al. Antimicrobial resistance in wildlife. J. Appl. Ecol. 2016, 53, 519–529. [Google Scholar] [CrossRef]
- Ramey, A.M.; Ahlstrom, C.A. Antibiotic resistant bacteria in wildlife: Perspectives on trends, acquisition and dissemination, data gaps, and future directions. J. Wildl. Dis. 2020, 56, 1–15. [Google Scholar] [CrossRef]
- Torres, R.T.; Carvalho, J.; Cunha, M.V.; Serrano, E.; Palmeira, J.D.; Fonseca, C. Temporal and geographical research trends of antimicrobial resistance in wildlife—A bibliometric analysis. One Health 2020, 11, 100198. [Google Scholar] [CrossRef]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell. Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef]
- Smoglica, C.; Vergara, A.; Angelucci, S.; Festino, A.R.; Antonucci, A.; Marsilio, F.; Di Francesco, C.E. Evidence of Linezolid Resistance and Virulence Factors in Enterococcus spp. Isolates from Wild and Domestic Ruminants, Italy. Antibiotics 2022, 11, 223. [Google Scholar] [CrossRef]
- Smoglica, C.; Vergara, A.; Angelucci, S.; Festino, A.R.; Antonucci, A.; Moschetti, L.; Farooq, M.; Marsilio, F.; Di Francesco, C.E. Resistance Patterns, mcr-4 and OXA-48 Genes, and Virulence Factors of Escherichia coli from Apennine Chamois Living in Sympatry with Domestic Species, Italy. Animals 2022, 12, 129. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 11.0. 2021. Available online: https://www.eucast.org/clinical_breakpoints/ (accessed on 27 November 2022).
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- Stata Statistical Software; Release 16; StataCorp LLC.: College Station, TX, USA, 2019.
- Rothman, K. Episheet, Spreadsheets for the Analysis of Epidemiological Data, Version of 21 May 2007. Available online: http://www.drugepi.org/dope-downloads/#Epishee (accessed on 27 November 2022).
- Graham, D.W.; Bergeron, G.; Bourassa, M.W.; Dickson, J.; Gomes, F.; Howe, A.; Kahn, L.H.; Morley, P.S.; Scott, H.M.; Simjee, S.; et al. Complexities in understanding antimicrobial resistance across domesticated animal, human, and environmental systems. Ann. N. Y. Acad. Sci. 2019, 1441, 17–30. [Google Scholar] [CrossRef]
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, R.; et al. Antibiotic Resistance Is the Quintessential One Health Issue. Trans. R Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef]
- Stępień-Pyśniak, D.; Hauschild, T.; Nowaczek, A.; Marek, A.; Dec, M. Wild birds as a potential source of known and novel multilocus sequence types of antibiotic-resistant enterococcus faecalis. J. Wildl. Dis. 2018, 54, 219–228. [Google Scholar] [CrossRef]
- Turchi, B.; Dec, M.; Bertelloni, F.; Winiarczyk, S.; Gnat, S.; Bresciani, F.; Viviani, F.; Cerri, D.; Fratini, F. Antibiotic Susceptibility and Virulence Factors in Escherichia coli from Sympatric Wildlife of the Apuan Alps Regional Park (Tuscany, Italy). Microb. Drug Resist. 2019, 25, 772–780. [Google Scholar] [CrossRef]
- Dec, M.; Stępień-Pyśniak, D.; Gnat, S.; Fratini, F.; Urban-Chmiel, R.; Cerri, D.; Winiarczyk, S.; Turchi, B. Antibiotic Susceptibility and Virulence Genes in Enterococcus Isolates from Wild Mammals Living in Tuscany, Italy. Microb. Drug Resist. 2020, 26, 505–519. [Google Scholar] [CrossRef]
- Massella, E.; Reid, C.J.; Cummins, M.L.; Anantanawat, K.; Zingali, T.; Serraino, A.; Piva, S.; Giacometti, F.; Djordjevic, S.P. Snapshot Study of Whole Genome Sequences of Escherichia coli from Healthy Companion Animals, Livestock, Wildlife, Humans and Food in Italy. Antibiotics 2020, 9, 782. [Google Scholar] [CrossRef] [PubMed]
- Torres, R.T.; Cunha, M.V.; Araujo, D.; Ferreira, H.; Fonseca, C.; Palmeira, J.D. Emergence of colistin resistance genes (mcr-1) in Escherichia coli among widely distributed wild ungulates. Environ. Pollut. 2021, 291, 118136. [Google Scholar] [CrossRef] [PubMed]
- Katakweba, A.A.; Møller, K.S.; Muumba, J.; Muhairwa, A.P.; Damborg, P.; Rosenkrantz, J.T.; Minga, U.M.; Mtambo, M.M.; Olsen, J.E. Antimicrobial resistance in faecal samples from buffalo, wildebeest and zebra grazing together with and without cattle in Tanzania. J. Appl. Microbiol. 2015, 118, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Farnleitner, A.H.; Ryzinska-Paier, G.; Reischer, G.H.; Burtscher, M.M.; Knetsch, S.; Kirschner, A.K.; Dirnböck, T.; Kuschnig, G.; Mach, R.L.; Sommer, R. Escherichia coli and enterococci are sensitive and reliable indicators for human, livestock and wildlife faecal pollution in alpine mountainous water resources. J. Appl. Microbiol. 2010, 109, 1599–1608. [Google Scholar] [CrossRef]
- Hassell, J.M.; Ward, M.J.; Muloi, D.; Bettridge, J.M.; Robinson, T.P.; Kariuki, S.; Ogendo, A.; Kiiru, J.; Imboma, T.; Kang’ethe, E.K.; et al. Clinically relevant antimicrobial resistance at the wildlife-livestock-human interface in Nairobi: An epidemiological study. Lancet Planet Health 2019, 3, e259–e269. [Google Scholar] [CrossRef]
- Mubita, C.; Syakalima, M.; Chisenga, C.; Munyeme, M.; Muma, B.; Chifumpa, G.; Hangombe, B.; Sinkala, P.; Simuunza, M.; Fukushi, H.; et al. Antimicrobial resistance of faecal Escherichia coli and Enterococci spp. from Apparently Health Pastoralist Cattle in the Interface Areas of the Kafue basin of Zambia-short communication. Vet. Arh. 2008, 78, 179–185. [Google Scholar]
- Kabali, E.; Pandey, G.S.; Munyeme, M.; Kapila, P.; Mukubesa, A.N.; Ndebe, J.; Muma, J.B.; Mubita, C.; Muleya, W.; Muonga, E.M.; et al. Identification of Escherichia coli and Related Enterobacteriaceae and Examination of Their Phenotypic Antimicrobial Resistance Patterns: A Pilot Study at A Wildlife-Livestock Interface in Lusaka, Zambia. Antibiotics 2021, 10, 238. [Google Scholar] [CrossRef]
- Sulzner, K.; Kelly, T.; Smith, W.; Johnson, C.K. Enteric pathogens and antimicrobial resistance in turkey vultures (Cathartes aura) feeding at the wildlife-livestock interface. J. Zoo Wildl. Med. 2014, 45, 931–934. [Google Scholar] [CrossRef]
- Espunyes, J.; Cabezón, O.; Dias-Alves, A.; Miralles, P.; Ayats, T.; Cerdà-Cuéllar, M. Assessing the role of livestock and sympatric wild ruminants in spreading antimicrobial resistant Campylobacter and Salmonella in alpine ecosystems. BMC Vet. Res. 2021, 17, 79. [Google Scholar] [CrossRef]
- Luo, X.; Zhai, Y.; He, D.; Cui, X.; Yang, Y.; Yuan, L.; Liu, J.; Hu, G. Molecular characterization of a novel blaCTX-M-3-carrying Tn6741 transposon in Morganella morganii isolated from swine. J. Med. Microbiol. 2020, 69, 1089–1094. [Google Scholar] [CrossRef]
- Palmieri, N.; Hess, C.; Hess, M.; Alispahic, M. Sequencing of five poultry strains elucidates phylogenetic relationships and divergence in virulence genes in Morganella morganii. BMC Genom. 2020, 21, 579. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Lee, K.; Cho, Y.; Lim, S.R.; Kwon, H.; Han, J.E.; Kim, J.H. Emergence of Third-Generation Cephalosporin-Resistant Morganella morganii in a Captive Breeding Dolphin in South Korea. Animals 2020, 10, 2052. [Google Scholar] [CrossRef] [PubMed]
- Ryser, L.T.; Arias-Roth, E.; Perreten, V.; Irmler, S.; Bruggmann, R. Genetic and Phenotypic Diversity of Morganella morganii Isolated from Cheese. Front. Microbiol. 2021, 12, 738492. [Google Scholar] [CrossRef] [PubMed]
- Trotta, A.; Marinaro, M.; Sposato, A.; Galgano, M.; Ciccarelli, S.; Paci, S.; Corrente, M. Antimicrobial Resistance in Loggerhead Sea Turtles (Caretta caretta): A Comparison between Clinical and Commensal Bacterial Isolates. Animals 2021, 11, 2435. [Google Scholar] [CrossRef]
- Shi, H.; Chen, X.; Yao, Y.; Xu, J. Morganella morganii: An unusual analysis of 11 cases of pediatric urinary tract infections. J. Clin. Lab. Anal. 2022, 36, e24399. [Google Scholar] [CrossRef]
- Govender, R.; Amoah, I.D.; Adegoke, A.A.; Singh, G.; Kumari, S.; Swalaha, F.M.; Bux, F.; Stenström, T.A. Identification, antibiotic resistance, and virulence profiling of Aeromonas and Pseudomonas species from wastewater and surface water. Environ. Monit. Assess. 2021, 193, 294. [Google Scholar] [CrossRef]
- Smoglica, C.; Evangelisti, G.; Fani, C.; Marsilio, F.; Trotta, M.; Messina, F.; Di Francesco, C.E. Antimicrobial Resistance Profile of Bacterial Isolates from Urinary Tract Infections in Companion Animals in Central Italy. Antibiotics 2022, 11, 1363. [Google Scholar] [CrossRef]
- Oliveira, A.F.de.; Arrais, B.R.; Bannwart, P.F.; Pinto, J.F.N.; Stella, A.E. Detection of beta-lactamase-producing Enterobacteriaceae in a veterinary hospital environment. Braz. J. Vet. Res. Anim. Sci. 2022, 59, e191724. [Google Scholar] [CrossRef]
- Iredell, J.; Brown, J.; Tagg, K. Antibiotic resistance in Enterobacteriaceae: Mechanisms and clinical implications. BMJ 2016, 352, h6420. [Google Scholar] [CrossRef]
- Martins, L.; Gonçalves, J.L.; Leite, R.F.; Tomazi, T.; Rall, V.L.M.; Santos, M.V. Association between antimicrobial use and antimicrobial resistance of Streptococcus uberis causing clinical mastitis. J. Dairy Sci. 2021, 104, 12030–12041. [Google Scholar] [CrossRef]
- Malaluang, P.; Wilén, E.; Frosth, S.; Lindahl, J.; Hansson, I.; Morrell, J.M. Vaginal Bacteria in Mares and the Occurrence of Antimicrobial Resistance. Microorganisms 2022, 10, 2204. [Google Scholar] [CrossRef] [PubMed]
- Hai, P.D.; Son, P.N.; Thi Thu Huong, N.; Thanh Binh, N.; Thi Viet Hoa, L.; Manh Dung, N. A Case of Streptococcus thoraltensis Bacteremia and Prosthetic Valve Endocarditis in a 68-Year-Old Vietnamese Man. Am. J. Case Rep. 2020, 21, e925752. [Google Scholar] [CrossRef] [PubMed]
- Wazir, M.; Grewal, M.; Jain, A.G.; Everett, G. Streptococcus thoraltensis Bacteremia: A Case of Pneumonia in a Postpartum Patient. Cureus 2019, 11, e5659. [Google Scholar] [CrossRef]
- Borø, S.; McCartney, C.A.; Snelling, T.J.; Worgan, H.J.; McEwan, N.R. Isolation of Streptococcus thoraltensis from rabbit faeces. Curr. Microbiol. 2010, 61, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Thanantong, N.; Edwards, S.; Sparagano, O.A. Characterization of lactic acid bacteria and other gut bacteria in pigs by a macroarraying method. Ann. N. Y. Acad. Sci. 2006, 1081, 276–279. [Google Scholar] [CrossRef] [PubMed]
- Devriese, L.A.; Pot, B.; Vandamme, P.; Kersters, K.; Collins, M.D.; Alvarez, N.; Haesebrouck, F.; Hommez, J. Streptococcus hyovaginalis sp. nov. and Streptococcus thoraltensis sp. nov., from the genital tract of sows. Int. J. Syst. Bacteriol. 1997, 47, 1073–1077. [Google Scholar] [CrossRef]
- Nomura, R.; Otsugu, M.; Hamada, M.; Matayoshi, S.; Teramoto, N.; Iwashita, N.; Naka, S.; Matsumoto-Nakano, M.; Nakano, K. Potential involvement of Streptococcus mutans possessing collagen binding protein Cnm in infective endocarditis. Sci. Rep. 2020, 10, 19118. [Google Scholar] [CrossRef]
- World Health Organization. WHO List of Critically Important Antimicrobials for Human Medicine (WHO CIA List). Available online: https://www.who.int/publications/i/item/9789241515528 (accessed on 27 November 2022).
- Sadowy, E. Linezolid resistance genes and genetic elements enhancing their dissemination in enterococci and streptococci. Plasmid 2018, 99, 89–98. [Google Scholar] [CrossRef]
- Ranjan, R.; Thatikonda, S. β-Lactam Resistance Gene NDM-1 in the Aquatic Environment: A Review. Curr. Microbiol. 2021, 78, 3634–3643. [Google Scholar] [CrossRef]
- Silva, N.; Igrejas, G.; Figueiredo, N.; Gonçalves, A.; Radhouani, H.; Rodrigues, J.; Poeta, P. Molecular characterization of antimicrobial resistance in enterococci and Escherichia coli isolates from European wild rabbit (Oryctolagus cuniculus). Sci. Total Environ. 2010, 408, 4871–4876. [Google Scholar] [CrossRef]
- Radhouani, H.; Silva, N.; Poeta, P.; Torres, C.; Correia, S.; Igrejas, G. Potential impact of antimicrobial resistance in wildlife, environment and human health. Front Microbiol. 2014, 5, 23. [Google Scholar] [CrossRef]
- Mo, S.S.; Urdahl, A.M.; Madslien, K.; Sunde, M.; Nesse, L.L.; Slettemeås, J.S.; Norström, M. What does the fox say? Monitoring antimicrobial resistance in the environment using wild red foxes as an indicator. PLoS ONE 2018, 13, e0198019. [Google Scholar] [CrossRef]
- Gonçalves, A.; Igrejas, G.; Radhouani, H.; Santos, T.; Monteiro, R.; Pacheco, R.; Alcaide, E.; Zorrilla, I.; Serra, R.; Torres, C.; et al. Detection of antibiotic resistant enterococci and Escherichia coli in free range Iberian Lynx (Lynx pardinus). Sci. Total Environ. 2013, 456-457, 115–119. [Google Scholar] [CrossRef]
- Mengistu, T.S.; Garcias, B.; Castellanos, G.; Seminati, C.; Molina-López, R.A.; Darwich, L. Occurrence of multidrug resistant Gram-negative bacteria and resistance genes in semi-aquatic wildlife—Trachemys scripta, Neovison vison and Lutra lutra—As sentinels of environmental health. Sci. Total Environ. 2022, 830, 154814. [Google Scholar] [CrossRef]
- Ballash, G.A.; Dennis, P.M.; Mollenkopf, D.F.; Albers, A.L.; Robison, T.L.; Adams, R.J.; Li, C.; Tyson, G.H.; Wittum, T.E. Colonization of White-Tailed Deer (Odocoileus virginianus) from Urban and Suburban Environments with Cephalosporinase- and Carbapenemase-Producing Enterobacterales. Appl. Environ. Microbiol. 2022, 88, e0046522. [Google Scholar] [CrossRef]
- Neila-Ibáñez, C.; Pintado, E.; Velarde, R.; Fernández Aguilar, X.; Vidal, E.; Aragon, V.; Abarca, M.L. First Report of Streptococcus ruminantium in Wildlife: Phenotypic Differences with a Spanish Domestic Ruminant Isolate. Microbiol. Res. 2022, 13, 102–113. [Google Scholar] [CrossRef]
- Smoglica, C.; Angelucci, S.; Farooq, M.; Antonucci, A.; Marsilio, F.; Di Francesco, C.E. Microbial community and antimicrobial resistance in fecal samples from wild and domestic ruminants in Maiella National Park, Italy. One Health 2022, 15, 100403. [Google Scholar] [CrossRef]
- Lee, S.; Fan, P.; Liu, T.; Yang, A.; Boughton, R.K.; Pepin, K.M.; Miller, R.S.; Jeong, K.C. Transmission of antibiotic resistance at the wildlife-livestock interface. Comm. Biol. 2022, 5, 585. [Google Scholar] [CrossRef]
- Forslund, K.; Sunagawa, S.; Kultima, J.R.; Mende, D.R.; Arumugam, M.; Typas, A.; Bork, P. Country-Specific Antibiotic Use Practices Impact the Human Gut Resistome. Genome Res. 2013, 23, 1163–1169. [Google Scholar] [CrossRef]
- World Health Organization. 2017. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 27 November 2022).
| Bacterial Species | Sample | Sympatric Animals Area | Non-Sympatric Animals Area | Total |
|---|---|---|---|---|
| Acinetobacter baumannii | W | 0 | 1 | 1 |
| Aeromonas sobria | W | 1 | 0 | 1 |
| Citrobacter freundii | D | 0 | 1 | 1 |
| Enterobacter cloacae complex | W | 0 | 2 | 2 |
| Enterococcus casseliflavus | AC, C, S | 2 | 2 | 4 |
| Enterococcus faecalis | AC, D, S | 5 | 6 | 11 |
| Enterococcus faecium | AC, C, D, G, S | 4 | 8 | 12 |
| Enterococcus gallinarum | C, D, G, S | 3 | 12 | 15 |
| Enterococcus hirae | AC, G, S | 3 | 3 | 6 |
| Escherichia coli | AC, C, D, G, S | 14 | 26 | 40 |
| Klebsiella oxytoca | W | 1 | 0 | 1 |
| Klebsiella pneumoniae | D | 0 | 1 | 1 |
| Morganella morganii | C | 0 | 1 | 1 |
| Pseudomonas mendocina | W | 0 | 2 | 2 |
| Streptococcus alactolyticus | S | 1 | 0 | 1 |
| Streptococcus gallolyticus | D | 0 | 2 | 2 |
| Streprococcus mutans | D | 0 | 1 | 1 |
| Streptococcus thoraltensis | C | 0 | 1 | 1 |
| Streptococcus sanguinis | C | 0 | 2 | 2 |
| Streptococcus uberis | AC | 0 | 1 | 1 |
| Total | 34 | 72 | 106 |
| Strain | Source | Area | Multidrug Resistant Phenotypes | Number of Isolates |
|---|---|---|---|---|
| Enterococcus gallinarum | D, G | sympatric | LNZ QD TET | 3 |
| Enterococcus faecium | AC | sympatric | LNZ QD TEIC TET VAN | 1 |
| Enterobacter cloacae complex | W | non-sympatric | AK CTZ MER | 1 |
| Escherichia coli | G | non-sympatric | AMP SXT TET | 1 |
| Pseudomonas mendocina | W | non-sympatric | AK CTX ETP | 1 |
| Pseudomonas mendocina | W | non-sympatric | AK CTZ ETP | 1 |
| Streptococcus mutans | D | non-sympatric | AMP BEN CLIN CRO CTX LIN VAN TGC | 1 |
| Streptococcus uberis | AC | non-sympatric | AMP BEN CLIN CRO CTX ERY LIN TGC | 1 |
| Streptococcus thoraltensis | C | non-sympatric | CLIN CTX CRO VAN | 1 |
| Bacteria | Sample | Antibiotics | Resistant Isolates | Gene Detected by PCR | |
|---|---|---|---|---|---|
| Resistance Genes | Isolates | ||||
| Aeromon sobria | W | MER, TZP/TAZ | 1 | armA, rmtF | - |
| Escherichia coli | AC, S | AMP | 3 | blaCMY-2 | 1 |
| tetB | 1 | ||||
| - | 1 | ||||
| AC, S, D, C | CS | 7 | mcr-4, blaCMY-2 | 1 | |
| mcr-4, blaTEM, blaCMY2 | 1 | ||||
| mcr-4, tetB | 1 | ||||
| mcr-4, blaTEM | 1 | ||||
| mcr-4, blaCMY1, blaCMY2 | 3 | ||||
| AC | TET | 2 | tetB, blaTEM, blaCMY2 | 1 | |
| tetB | 1 | ||||
| AC, D | CS, TET | 2 | tetB, blaCMY2, mcr-4 | 1 | |
| tetB, mcr-4 | 1 | ||||
| AC | CS, CAZ, MRP | 1 | mcr-4, blaOXA-48, blaTEM, blaCMY1 | 1 | |
| D | AMP, TET, SXT | 1 | - | - | |
| G | TET, SXT | 1 | - | - | |
| Enterococcus faecium | D | QD | 1 | - | - |
| AC | QD, LNZ, TEIC, VAN, TET | 1 | msrC, TetB, cfrD | 1 | |
| Enterococcus faecalis | S | LNZ | 1 | cfrD | 1 |
| Enterococcus gallinarum | D, S | QD, LNZ, TET | 3 | cfrD, TetM | 1 |
| tetB, tetM, msrC, cfrD | 1 | ||||
| tetB, tetM, tetL,msrC,cfrD | 1 | ||||
| G | QD, LNZ | 3 | tetB, tetM, msrC, cfrD | 1 | |
| msrC, cfrD | 1 | ||||
| cfrD | 1 | ||||
| D | QD, TET | 1 | tetM, msrC | 1 | |
| Streptococcus alactolyticus | S | CTX, CRO, CLIN | 1 | gyrA | 1 |
| Streptococcus mutans | D | AMP, BEN, CTX, CRO, CLIN, LIN, TGC, VAN | 1 | ermA, gyrA | - |
| Streptococcus thoraltensis | C | CTX, CRO, CLIN, ERY, VAN | 1 | vanC2, vanG | 1 |
| Streptococcus uberis | AC | CTX, CRO, CLIN, LIN, AMP, BEN, ERY, TGC | 1 | ermB, gyrA, parC, poxtA, PBP2b | |
| Enterobacter cloacae complex | W | CTX, ETP, MER, TZP/TAZ | 1 | blaCTX-M, blaTEM, blaNDM, aaC1, armA, aphA6, rmtF | 1 |
| W | AK, CTZ, MER | 1 | blaCTX-M, blaTEM, blaNDM, rmtB, armA, aphA6 | 1 | |
| Pseudomonas mendocina | W | AK, CTX, ETP | 1 | blaCTX-M | 1 |
| W | AK, CTZ, ETP | 1 | blaCTX-M, blaCMY-1 | 1 | |
| Morganella morganii | C | NIT TGC | 1 | aaCA4, nfsA, rmtF, rmtB | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smoglica, C.; Vergara, A.; Angelucci, S.; Festino, A.R.; Antonucci, A.; Marsilio, F.; Di Francesco, C.E. Antibiotic-Resistant Bacteria Dissemination in the Wildlife, Livestock, and Water of Maiella National Park, Italy. Animals 2023, 13, 432. https://doi.org/10.3390/ani13030432
Smoglica C, Vergara A, Angelucci S, Festino AR, Antonucci A, Marsilio F, Di Francesco CE. Antibiotic-Resistant Bacteria Dissemination in the Wildlife, Livestock, and Water of Maiella National Park, Italy. Animals. 2023; 13(3):432. https://doi.org/10.3390/ani13030432
Chicago/Turabian StyleSmoglica, Camilla, Alberto Vergara, Simone Angelucci, Anna Rita Festino, Antonio Antonucci, Fulvio Marsilio, and Cristina Esmeralda Di Francesco. 2023. "Antibiotic-Resistant Bacteria Dissemination in the Wildlife, Livestock, and Water of Maiella National Park, Italy" Animals 13, no. 3: 432. https://doi.org/10.3390/ani13030432
APA StyleSmoglica, C., Vergara, A., Angelucci, S., Festino, A. R., Antonucci, A., Marsilio, F., & Di Francesco, C. E. (2023). Antibiotic-Resistant Bacteria Dissemination in the Wildlife, Livestock, and Water of Maiella National Park, Italy. Animals, 13(3), 432. https://doi.org/10.3390/ani13030432






