Comparison of Placental HSD17B1 Expression and Its Regulation in Various Mammalian Species
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Tissue Collection
2.2. Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
2.3. Cell Culture, Transfection, and Luciferase Assay
2.4. Methylation Analyses
2.5. Alignment Analysis and Plasmids
2.6. Western Blot Analysis
2.7. Statistical Analyses
3. Results
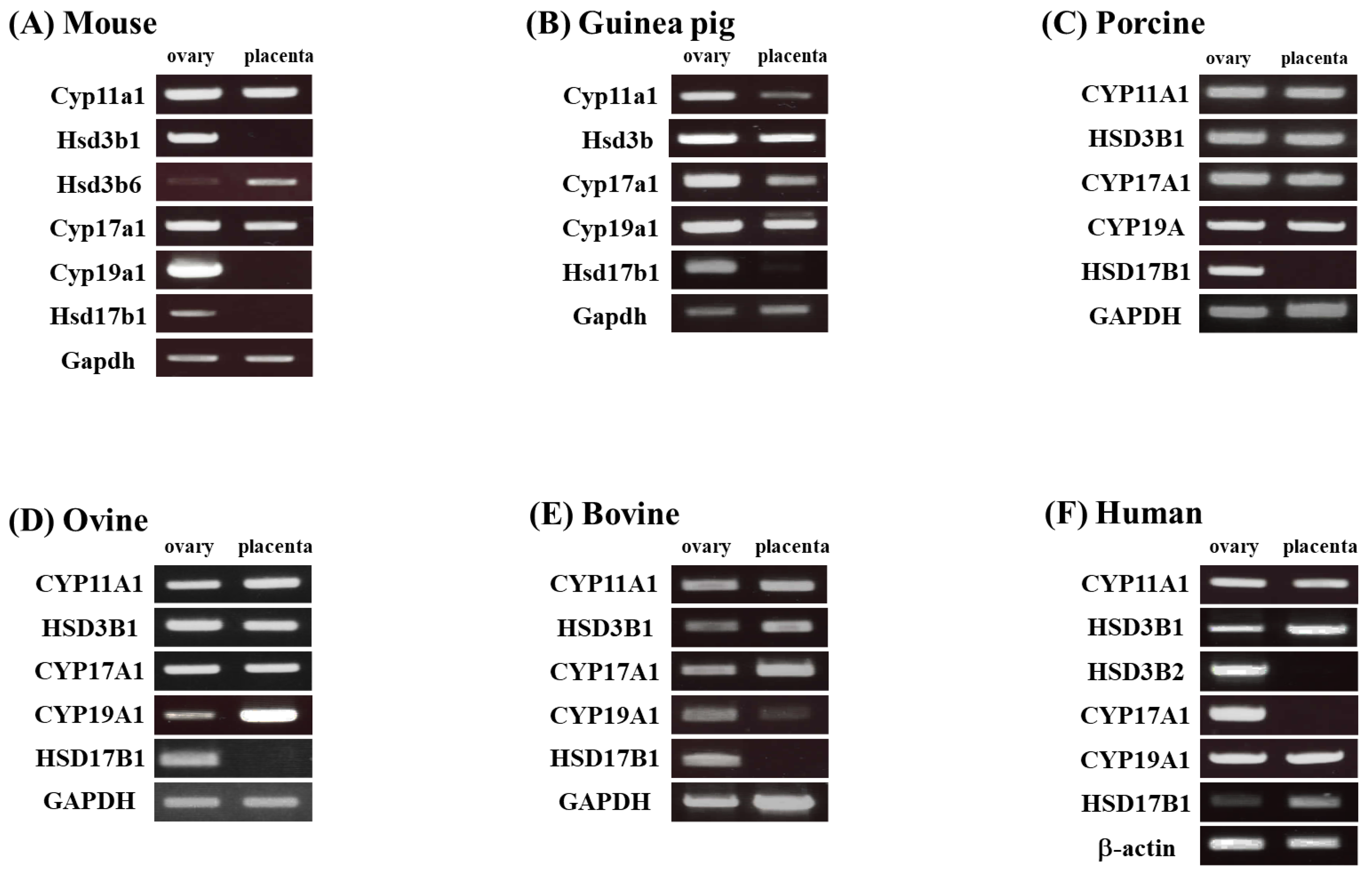
3.1. Comparison of the Placental Expression of Steroidogenic Genes in Various Mammals
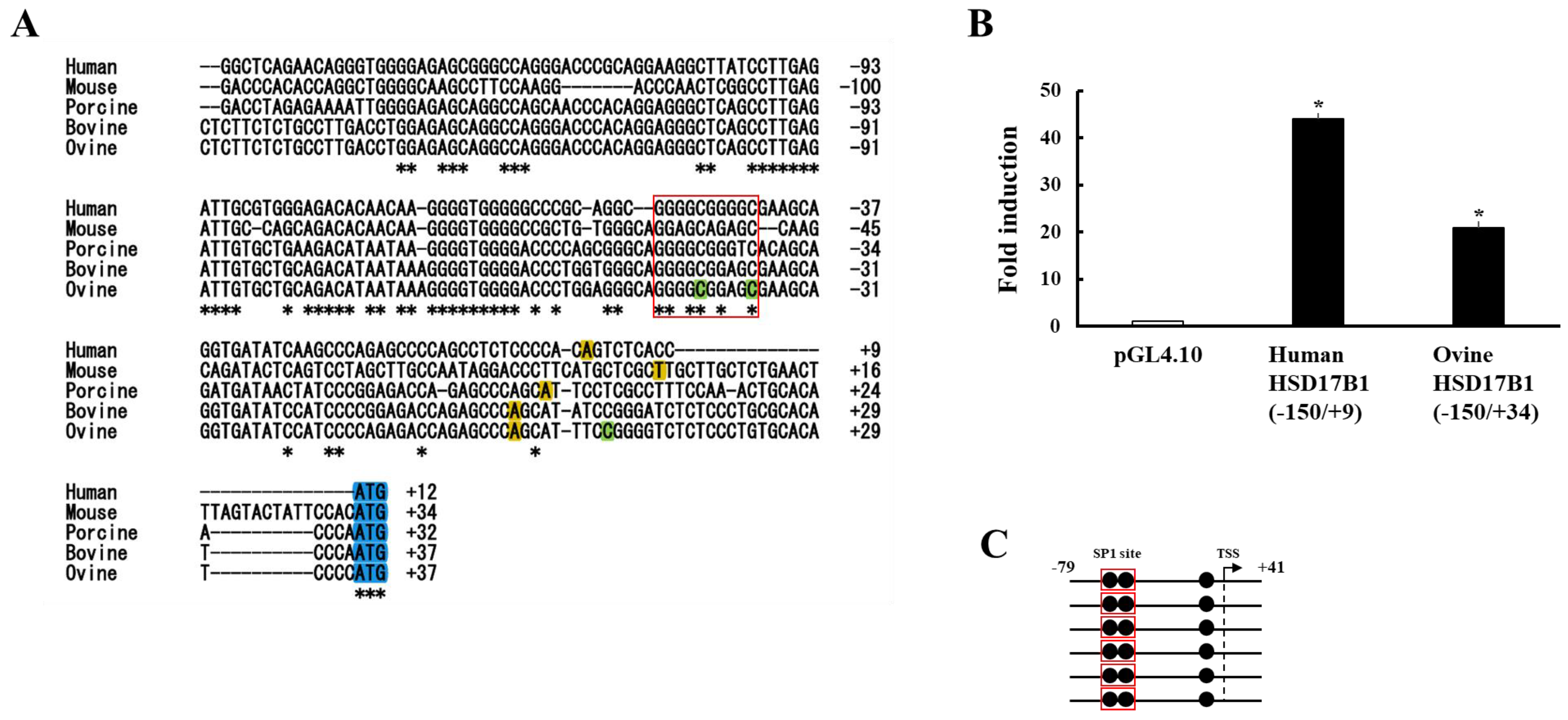
3.2. Analyses of Human and Ovine HSD17B1 Promoter Region
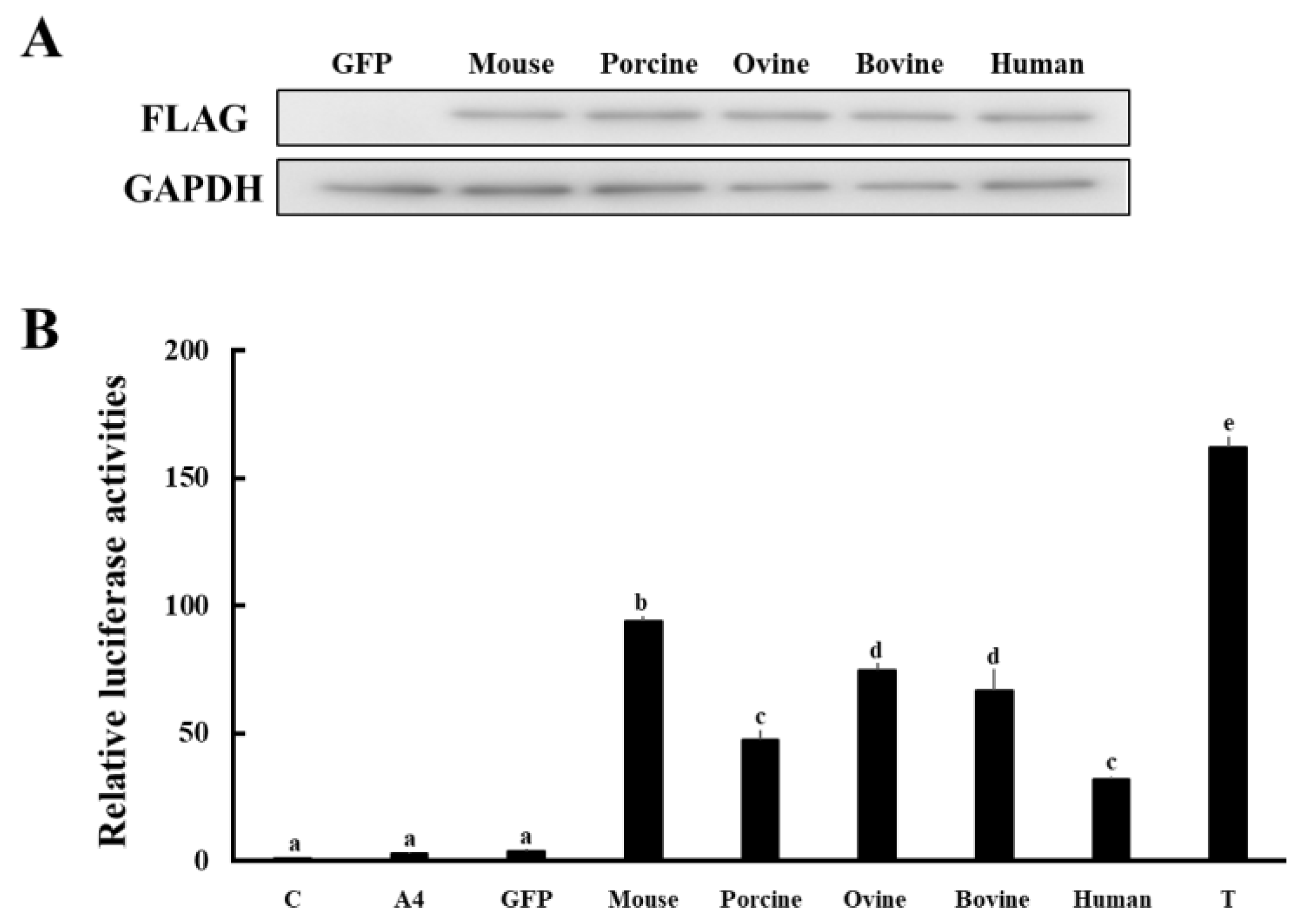
3.3. Comparison of Enzymatic Activities of HSD17B1/Hsd17b1 for Converting A4 to T
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tsachaki, M.; Odermatt, A. Subcellular localization and membrane topology of 17β-hydroxysteroid dehydrogenases. Mol. Cell. Endocrinol. 2019, 489, 98–106. [Google Scholar] [CrossRef]
- Heinosalo, T.; Saarinen, N.; Poutanen, M. Role of hydroxysteroid (17beta) dehydrogenase type 1 in reproductive tissues and hormone-dependent diseases. Mol. Cell. Endocrinol. 2019, 489, 9–31. [Google Scholar] [CrossRef]
- Puranen, T.; Poutanen, M.; Ghosh, D.; Vihko, P.; Vihko, R. Characterization of structural and functional properties of human 17 beta-hydroxysteroid dehydrogenase type 1 using recombinant enzymes and site-directed mutagenesis. Mol. Endocrinol. 1997, 11, 77–86. [Google Scholar] [CrossRef]
- Luu-The, V.; Zhang, Y.; Poirier, D.; Labrie, F. Characteristics of human types 1, 2 and 3 17 beta-hydroxysteroid dehydrogenase activities: Oxidation/reduction and inhibition. J. Steroid Biochem. Mol. Biol. 1995, 55, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Yazawa, T.; Imamichi, Y.; Uwada, J.; Sekiguchi, T.; Mikami, D.; Kitano, T.; Ida, T.; Sato, T.; Nemoto, T.; Nagata, S.; et al. Evaluation of 17β-hydroxysteroid dehydrogenase activity using androgen receptor-mediated transactivation. J. Steroid Biochem. Mol. Biol. 2020, 196, 105493. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.M. Evolution of Placental Hormones: Implications for Animal Models. Front. Endocrinol. 2022, 13, 891927. [Google Scholar] [CrossRef] [PubMed]
- Arck, P.; Hansen, P.J.; Mulac Jericevic, B.; Piccinni, M.P.; Szekeres-Bartho, J. Progesterone during pregnancy: Endocrine-immune cross talk in mammalian species and the role of stress. Am. J. Reprod. Immunol. 2007, 58, 268–279. [Google Scholar] [CrossRef]
- Morel, Y.; Roucher, F.; Plotton, I.; Goursaud, C.; Tardy, V.; Mallet, D. Evolution of steroids during pregnancy: Maternal, placental and fetal synthesis. Ann. D’endocrinologie 2016, 77, 82–89. [Google Scholar] [CrossRef]
- Adessi, G.L.; Nhuan, T.Q.; Prost, O. The in vitro metabolism of estrone and estradiol-17 beta and their 3-sulfates by the renal tissues from pregnant and female fetal guinea-pigs. J. Steroid Biochem. 1981, 15, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Bosu, W.T.; Edqvist, L.E.; Gustafsson, B. Peripheral plasma levels of estrone and progesterone in pregnant pigs treated with dexamethasone. Acta Vet. Scand. 1974, 15, 111–119. [Google Scholar] [CrossRef]
- Miura, H.; Yamazaki, T.; Kikuchi, M.; Sakaguchi, M. Plasma steroid hormone concentrations and their relationships in Suffolk ewes during gestation and parturition. Anim. Sci. J. = Nihon Chikusan Gakkaiho 2019, 90, 1426–1431. [Google Scholar] [CrossRef]
- Martins-Júnior, H.A.; Pinaffi, F.L.; Simas, R.C.; Tarouco, A.K.; Ferreira, C.R.; Silva, L.A.; Nogueira, G.P.; Meirelles, F.V.; Eberlin, M.N.; Perecin, F. Plasma steroid dynamics in late- and near-term naturally and artificially conceived bovine pregnancies as elucidated by multihormone high-resolution LC-MS/MS. Endocrinology 2014, 155, 5011–5023. [Google Scholar] [CrossRef]
- Tulchinsky, D.; Hobel, C.J.; Yeager, E.; Marshall, J.R. Plasma estrone, estradiol, estriol, progesterone, and 17-hydroxyprogesterone in human pregnancy. I. Normal pregnancy. Am. J. Obstet. Gynecol. 1972, 112, 1095–1100. [Google Scholar] [CrossRef] [PubMed]
- McCormack, J.T.; Greenwald, G.S. Progesterone and oestradiol-17beta concentrations in the peripheral plasma during pregnancy in the mouse. J. Endocrinol. 1974, 62, 101–107. [Google Scholar] [CrossRef]
- Gáspár, R.; Ducza, E.; Mihályi, A.; Márki, A.; Kolarovszki-Sipiczki, Z.; Páldy, E.; Benyhe, S.; Borsodi, A.; Földesi, I.; Falkay, G. Pregnancy-induced decrease in the relaxant effect of terbutaline in the late-pregnant rat myometrium: Role of G-protein activation and progesterone. Reproduction 2005, 130, 113–122. [Google Scholar] [CrossRef]
- Renaud, S.J.; Karim Rumi, M.A.; Soares, M.J. Review: Genetic manipulation of the rodent placenta. Placenta 2011, 32 (Suppl. 2), S130–S135. [Google Scholar] [CrossRef]
- Terakawa, J.; Watanabe, T.; Obara, R.; Sugiyama, M.; Inoue, N.; Ohmori, Y.; Hosaka, Y.Z.; Hondo, E. The complete control of murine pregnancy from embryo implantation to parturition. Reproduction 2012, 143, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Mindnich, R.; Deluca, D.; Adamski, J. Identification and characterization of 17 beta-hydroxysteroid dehydrogenases in the zebrafish, Danio rerio. Mol. Cell. Endocrinol. 2004, 215, 19–30. [Google Scholar] [CrossRef]
- Hakkarainen, J.; Jokela, H.; Pakarinen, P.; Heikelä, H.; Kätkänaho, L.; Vandenput, L.; Ohlsson, C.; Zhang, F.P.; Poutanen, M. Hydroxysteroid (17β)-dehydrogenase 1-deficient female mice present with normal puberty onset but are severely subfertile due to a defect in luteinization and progesterone production. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2015, 29, 3806–3816. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Uwada, J.; Hayashi, J.; Kikuya, K.I.; Muranishi, Y.; Watanabe, H.; Yaegashi, K.; Hasegawa, K.; Ida, T.; Sato, T.; et al. Analyses of Molecular Characteristics and Enzymatic Activities of Ovine HSD17B3. Animal 2021, 11, 2876. [Google Scholar] [CrossRef]
- Yazawa, T.; Inaba, H.; Imamichi, Y.; Sekiguchi, T.; Uwada, J.; Islam, M.S.; Orisaka, M.; Mikami, D.; Ida, T.; Sato, T.; et al. Profiles of 5α-Reduced Androgens in Humans and Eels: 5α-Dihydrotestosterone and 11-Ketodihydrotestosterone Are Active Androgens Produced in Eel Gonads. Front. Endocrinol. 2021, 12, 657360. [Google Scholar] [CrossRef] [PubMed]
- Yazawa, T.; Sato, T.; Nemoto, T.; Nagata, S.; Imamichi, Y.; Kitano, T.; Sekiguchi, T.; Uwada, J.; Islam, M.S.; Mikami, D.; et al. 11-Ketotestosterone is a major androgen produced in porcine adrenal glands and testes. J. Steroid Biochem. Mol. Biol. 2021, 210, 105847. [Google Scholar] [CrossRef]
- Imamichi, Y.; Yuhki, K.I.; Orisaka, M.; Kitano, T.; Mukai, K.; Ushikubi, F.; Taniguchi, T.; Umezawa, A.; Miyamoto, K.; Yazawa, T. 11-Ketotestosterone Is a Major Androgen Produced in Human Gonads. J. Clin. Endocrinol. Metab. 2016, 101, 3582–3591. [Google Scholar] [CrossRef]
- Shih, M.C.; Chiu, Y.N.; Hu, M.C.; Guo, I.C.; Chung, B.C. Regulation of steroid production: Analysis of Cyp11a1 promoter. Mol. Cell. Endocrinol. 2011, 336, 80–84. [Google Scholar] [CrossRef]
- Li, J.N.; Ge, Y.C.; Yang, Z.; Guo, C.M.; Duan, T.; Myatt, L.; Guan, H.; Yang, K.; Sun, K. The Sp1 transcription factor is crucial for the expression of 11beta-hydroxysteroid dehydrogenase type 2 in human placental trophoblasts. J. Clin. Endocrinol. Metab. 2011, 96, E899–E907. [Google Scholar] [CrossRef] [PubMed]
- Piao, Y.S.; Peltoketo, H.; Vihko, P.; Vihko, R. The proximal promoter region of the gene encoding human 17beta-hydroxysteroid dehydrogenase type 1 contains GATA, AP-2, and Sp1 response elements: Analysis of promoter function in choriocarcinoma cells. Endocrinology 1997, 138, 3417–3425. [Google Scholar] [CrossRef]
- Ezashi, T.; Matsuyama, H.; Telugu, B.P.; Roberts, R.M. Generation of colonies of induced trophoblast cells during standard reprogramming of porcine fibroblasts to induced pluripotent stem cells. Biol. Reprod. 2011, 85, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Suasnavas, E.A.; Heywood, S.; Ward, A.; Cox, L.; O’Grady, M.; Zhao, Y.; Gilbert, L.; Isom, S.C. Isolation and characterization of trophoblast-derived stem-like cells from peri-implantation porcine embryos. Anim. Reprod. Sci. 2015, 154, 128–141. [Google Scholar] [CrossRef]
- Huang, X.; Han, X.; Uyunbilig, B.; Zhang, M.; Duo, S.; Zuo, Y.; Zhao, Y.; Yun, T.; Tai, D.; Wang, C.; et al. Establishment of bovine trophoblast stem-like cells from in vitro-produced blastocyst-stage embryos using two inhibitors. Stem Cells Dev. 2014, 23, 1501–1514. [Google Scholar] [CrossRef]
- Matamoros, R.A.; Caamano, L.; Lamb, S.V.; Reimers, T.J. Estrogen production of bovine binuclate and mononuclate trophoblastic cells in vitro. Biol. Reprod. 1994, 51, 486–492. [Google Scholar] [CrossRef] [Green Version]
- Chu, S. Transcriptional regulation by post-transcriptional modification--role of phosphorylation in Sp1 transcriptional activity. Gene 2012, 508, 1–8. [Google Scholar] [CrossRef]
- Yuan, X.; Zhou, X.; Qiao, X.; Wu, Q.; Yao, Z.; Jiang, Y.; Zhang, H.; Zhang, Z.; Wang, X.; Li, J. FoxA2 and p53 regulate the transcription of HSD17B1 in ovarian granulosa cells of pigs. Reprod. Domest. Anim. 2021, 56, 74–82. [Google Scholar] [CrossRef]
- Zhu, H.; Huang, L.; He, Z.; Zou, Z.; Luo, Y. Estrogen-related receptor γ regulates expression of 17β-hydroxysteroid dehydrogenase type 1 in fetal growth restriction. Placenta 2018, 67, 38–44. [Google Scholar] [CrossRef]
- Luo, Y.; Kumar, P.; Chen, C.C.; Latham, J.; Wang, L.; Tudela, C.; Alexander, J.M.; Shelton, J.M.; McKown, L.; Mendelson, C.R. Estrogen-related receptor γ serves a role in blood pressure homeostasis during pregnancy. Mol. Endocrinol. 2014, 28, 965–975. [Google Scholar] [CrossRef]
- Poidatz, D.; Dos Santos, E.; Duval, F.; Moindjie, H.; Serazin, V.; Vialard, F.; De Mazancourt, P.; Dieudonné, M.N. Involvement of estrogen-related receptor-γ and mitochondrial content in intrauterine growth restriction and preeclampsia. Fertil. Steril. 2015, 104, 483–490. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Gauri, M.; Li, T.; Wang, R.; Lin, S.X. Current knowledge of the multifunctional 17β-hydroxysteroid dehydrogenase type 1 (HSD17B1). Gene 2016, 588, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Drzewiecka, H.; Gałęcki, B.; Jarmołowska-Jurczyszyn, D.; Kluk, A.; Dyszkiewicz, W.; Jagodziński, P.P. Increased expression of 17-beta-hydroxysteroid dehydrogenase type 1 in non-small cell lung cancer. Lung Cancer 2015, 87, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Rawłuszko, A.A.; Horbacka, K.; Krokowicz, P.; Jagodziński, P.P. Decreased expression of 17β-hydroxysteroid dehydrogenase type 1 is associated with DNA hypermethylation in colorectal cancer located in the proximal colon. BMC Cancer 2011, 11, 522. [Google Scholar] [CrossRef] [PubMed]
- Puranen, T.; Poutanen, M.; Ghosh, D.; Vihko, R.; Vihko, P. Origin of substrate specificity of human and rat 17beta-hydroxysteroid dehydrogenase type 1, using chimeric enzymes and site-directed substitutions. Endocrinology 1997, 138, 3532–3539. [Google Scholar] [CrossRef] [PubMed]
- Saloniemi, T.; Welsh, M.; Lamminen, T.; Saunders, P.; Mäkelä, S.; Streng, T.; Poutanen, M. Human HSD17B1 expression masculinizes transgenic female mice. Mol. Cell. Endocrinol. 2009, 301, 163–168. [Google Scholar] [CrossRef]
- Shozu, M.; Akasofu, K.; Harada, T.; Kubota, Y. A new cause of female pseudohermaphroditism: Placental aromatase deficiency. J. Clin. Endocrinol. Metab. 1991, 72, 560–566. [Google Scholar] [CrossRef]
- Belgorosky, A.; Guercio, G.; Pepe, C.; Saraco, N.; Rivarola, M.A. Genetic and clinical spectrum of aromatase deficiency in infancy, childhood and adolescence. Horm. Res. 2009, 72, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Makieva, S.; Saunders, P.T.; Norman, J.E. Androgens in pregnancy: Roles in parturition. Hum. Reprod. Update 2014, 20, 542–559. [Google Scholar] [CrossRef]
- Kumar, S.; Gordon, G.H.; Abbott, D.H.; Mishra, J.S. Androgens in maternal vascular and placental function: Implications for preeclampsia pathogenesis. Reproduction 2018, 156, R155–R167. [Google Scholar] [CrossRef]
- Abbott, D.H.; Kraynak, M.; Dumesic, D.A.; Levine, J.E. In utero Androgen Excess: A Developmental Commonality Preceding Polycystic Ovary Syndrome? Front. Horm. Res. 2019, 53, 1–17. [Google Scholar] [CrossRef]
- Padmanabhan, V.; Manikkam, M.; Recabarren, S.; Foster, D. Prenatal testosterone excess programs reproductive and metabolic dysfunction in the female. Mol. Cell. Endocrinol. 2006, 246, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Dapas, M.; Dunaif, A. Deconstructing a Syndrome: Genomic Insights into PCOS Causal Mechanisms and Classification. Endocr. Rev. 2022, 43, 927–965. [Google Scholar] [CrossRef] [PubMed]
- Stener-Victorin, E.; Padmanabhan, V.; Walters, K.A.; Campbell, R.E.; Benrick, A.; Giacobini, P.; Dumesic, D.A.; Abbott, D.H. Animal Models to Understand the Etiology and Pathophysiology of Polycystic Ovary Syndrome. Endocr. Rev. 2020, 41, bnaa010. [Google Scholar] [CrossRef] [PubMed]
- Risal, S.; Pei, Y.; Lu, H.; Manti, M.; Fornes, R.; Pui, H.P.; Zhao, Z.; Massart, J.; Ohlsson, C.; Lindgren, E.; et al. Prenatal androgen exposure and transgenerational susceptibility to polycystic ovary syndrome. Nat. Med. 2019, 25, 1894–1904. [Google Scholar] [CrossRef]
- Flück, C.E.; Miller, W.L. GATA-4 and GATA-6 modulate tissue-specific transcription of the human gene for P450c17 by direct interaction with Sp1. Mol. Endocrinol. 2004, 18, 1144–1157. [Google Scholar] [CrossRef] [Green Version]
- Escobar, J.C.; Patel, S.S.; Beshay, V.E.; Suzuki, T.; Carr, B.R. The human placenta expresses CYP17 and generates androgens de novo. J. Clin. Endocrinol. Metab. 2011, 96, 1385–1392. [Google Scholar] [CrossRef] [PubMed]
- Dudley, J.S.; Murphy, C.R.; Thompson, M.B.; McAllan, B.M. Uterine cellular changes during mammalian pregnancy and the evolution of placentation. Biol. Reprod. 2021, 105, 1381–1400. [Google Scholar] [CrossRef] [PubMed]
- Bigler, N.A.; Bruckmaier, R.M.; Gross, J.J. Implications of placentation type on species-specific colostrum properties in mammals. J. Anim. Sci. 2022, 100, skac287. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yazawa, T.; Islam, M.S.; Imamichi, Y.; Watanabe, H.; Yaegashi, K.; Ida, T.; Sato, T.; Kitano, T.; Matsuzaki, S.; Umezawa, A.; et al. Comparison of Placental HSD17B1 Expression and Its Regulation in Various Mammalian Species. Animals 2023, 13, 622. https://doi.org/10.3390/ani13040622
Yazawa T, Islam MS, Imamichi Y, Watanabe H, Yaegashi K, Ida T, Sato T, Kitano T, Matsuzaki S, Umezawa A, et al. Comparison of Placental HSD17B1 Expression and Its Regulation in Various Mammalian Species. Animals. 2023; 13(4):622. https://doi.org/10.3390/ani13040622
Chicago/Turabian StyleYazawa, Takashi, Mohammad Sayful Islam, Yoshitaka Imamichi, Hiroyuki Watanabe, Kazuhide Yaegashi, Takanori Ida, Takahiro Sato, Takeshi Kitano, Shigenori Matsuzaki, Akihiro Umezawa, and et al. 2023. "Comparison of Placental HSD17B1 Expression and Its Regulation in Various Mammalian Species" Animals 13, no. 4: 622. https://doi.org/10.3390/ani13040622
APA StyleYazawa, T., Islam, M. S., Imamichi, Y., Watanabe, H., Yaegashi, K., Ida, T., Sato, T., Kitano, T., Matsuzaki, S., Umezawa, A., & Muranishi, Y. (2023). Comparison of Placental HSD17B1 Expression and Its Regulation in Various Mammalian Species. Animals, 13(4), 622. https://doi.org/10.3390/ani13040622






