Hepatic Transcriptome Analysis Reveals Genes, Polymorphisms, and Molecules Related to Lamb Tenderness
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Phenotype
2.2. Library Construction and Sequencing
2.3. Differential Gene Expression and Pathway Analysis
2.4. Network Enrichment Analysis
2.5. Analysis of Quantitative Real-Time PCR (qRT–PCR) Validation
2.6. Analysis of Gene Variation
2.7. SNP Validation and Association Study
3. Results
3.1. Phenotype of Meat Quality Traits in Sheep
3.2. Overview of the RNA Deep Sequencing Data
3.3. Differential Gene Expression Analysis
3.4. Functional Analysis
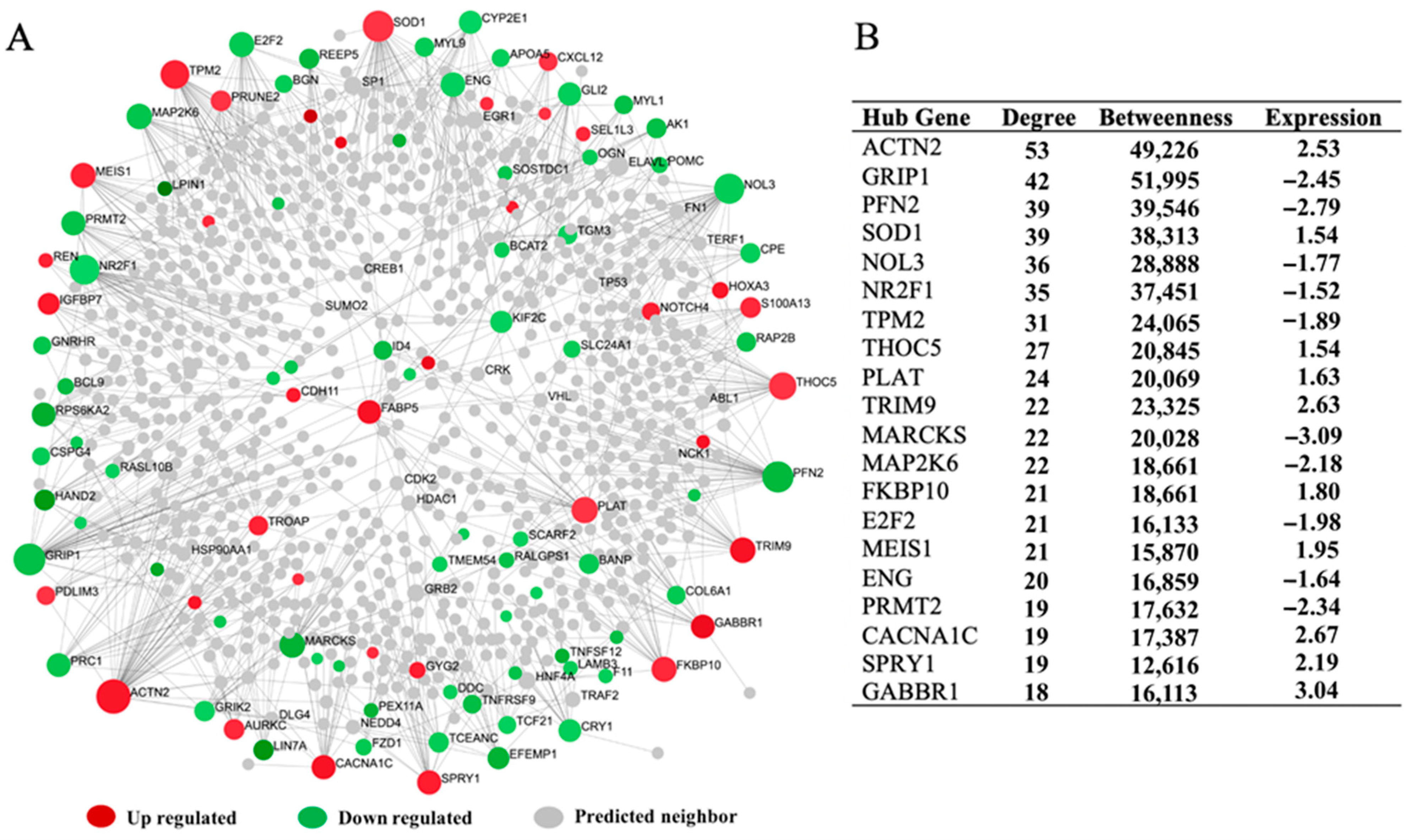
3.5. The Hepatic Transcriptome Network’s Regulatory Hub Genes
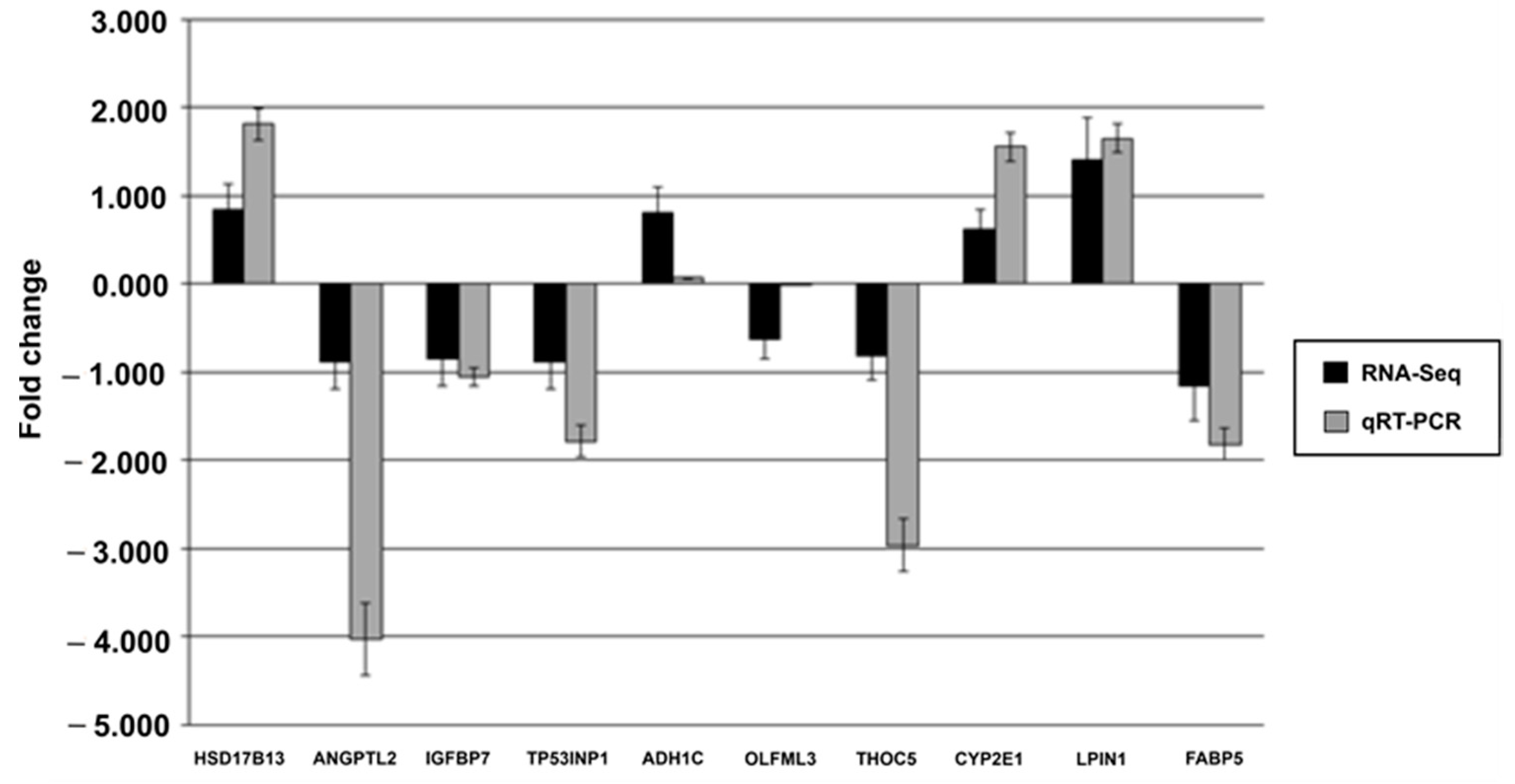
3.6. Quantitative Real-Time PCR Validation of Selected DEGs (qRT–PCR)
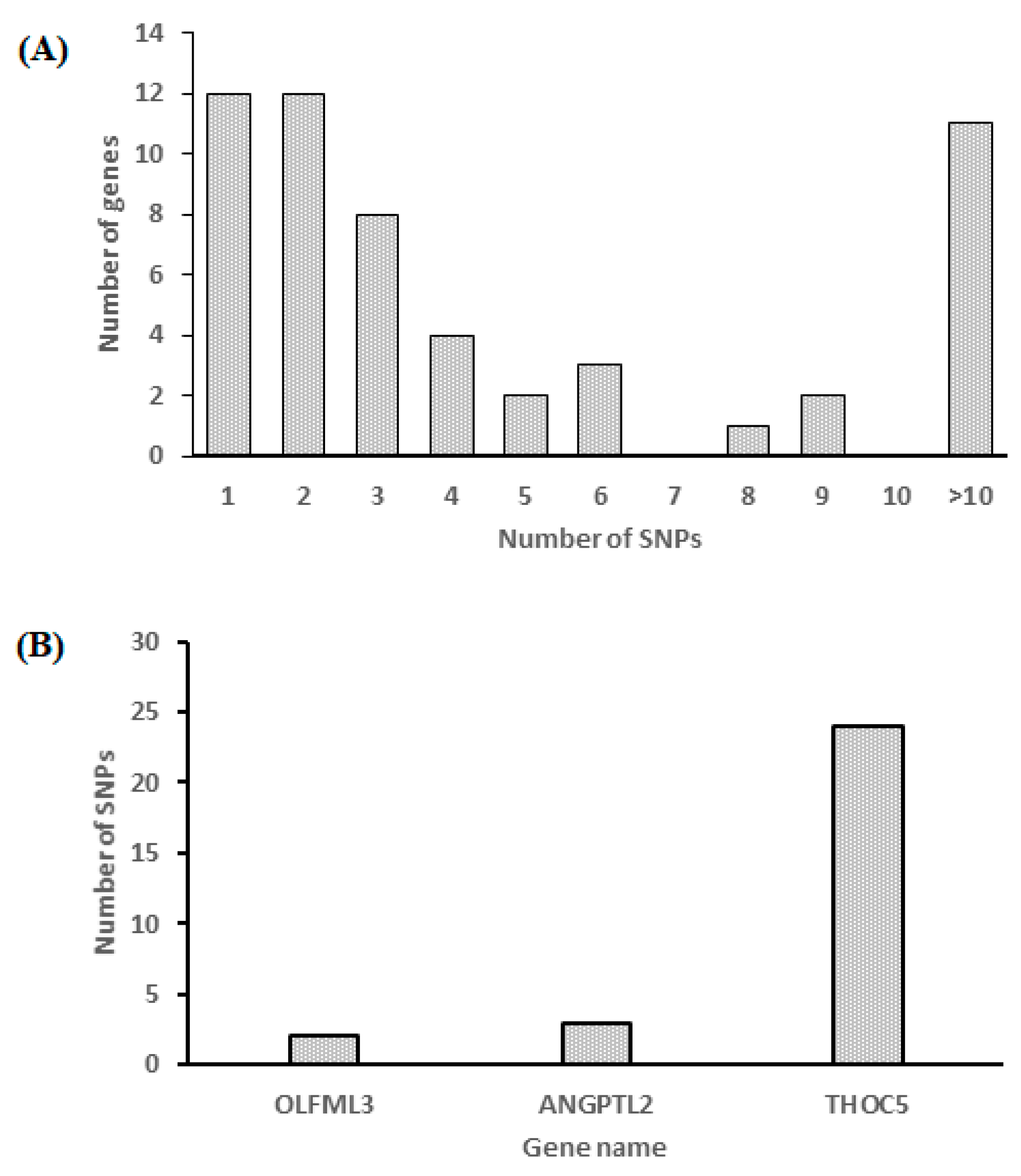
3.7. Analysis of Gene Variation and an Association Study
4. Discussion
4.1. Analysis of RNA Seq Data
4.2. Differentially Expressed Gene Analysis
4.3. Biological Function Analysis for DEGs
4.4. The Hepatic Transcriptome Network’s Regulatory Hub Genes
4.5. Association between Candidate Markers and Phenotypes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maltin, C.; Balcerzak, D.; Tilley, R.; Delday, M. Determinants of meat quality: Tenderness. Proc. Nutr. Soc. 2003, 62, 337–347. [Google Scholar] [CrossRef] [PubMed]
- FAO (Food and Agriculture Organization). Agriculture and Consumer Protection Department Animal Production and Health: Meat Quality. 2014. Available online: http://www.fao.org/ag/againfo/themes/en/meat/quality_meat.html (accessed on 1 February 2021).
- Thu, D.T.N. Meat quality: Understanding of meat tenderness and influence of fat content on meat flavor. Sci. Technol. Dev. 2006, 9, 65–70. [Google Scholar]
- Shackelford, S.D.; Morgan, J.B.; Cross, H.R.; Savell, J.W. Identification of threshold levels for warner-bratzler shear force in beef top loin steaks. J. Muscle Foods 1991, 2, 289–296. [Google Scholar] [CrossRef]
- Joo, S.T.; Kim, G.D.; Hwang, Y.H.; Ryu, Y.C. Control of fresh meat quality through manipulation of muscle fiber characteristics. Meat Sci. 2013, 95, 828–836. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.W.; Hwang, Y.H.; Joo, S.T. Meat tenderness characteristics of teh major muscles from Hanwoo steers according to quality grades of carcasses. Korean J. Food Sci. Anim. Resour. 2017, 37, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, S.I.; van der Werf, J.H.J.; Jacob, R.H.; Hopkins, D.L.; Pannier, L.; Pearce, K.L.; Gardner, G.E.; Warner, R.D.; Geesink, G.H.; Hocking Edwards, J.E.; et al. Genetic parameters for meat quality traits of Australian lamb meat. Meat Sci. 2014, 96, 1016–1024. [Google Scholar] [CrossRef]
- Koch, R.M.; Cundiff, L.V.; Gregory, K.E. Heritabilities and genetic, environmental and phenotypic correlations of carcass traits in a population of diverse biological types and their implications in selection programs. J. Anim. Sci. 1982, 55, 1319–1329. [Google Scholar] [CrossRef]
- Listyarini, K.; Jakaria, J.; Uddin, M.J.; Sumantri, C.; Gunawan, A. Association and expression of CYP2A6 and KIF12 genes related to lamb flavour and odour. Trop. Anim. Sci. J. 2018, 41, 100–107. [Google Scholar] [CrossRef]
- Gunawan, A.; Anggrela, D.; Listyarini, K.; Abuzahra, M.A.; Jakaria; Yamin, M.; Inounu, I.; Sumantri, C. Identification of single nucleotide polymorphism and pathway analysis of Apolipoprotein A5 (APOA5) related to fatty acid traits in Indonesian sheep. Trop. Anim. Sci. J. 2018, 41, 165–173. [Google Scholar] [CrossRef]
- Bagatoli, A.; Gasparino, E.; Soares, M.A.M.; Amaral, R.M.; Macedo, F.A.F.; Voltolini, D.M.; Del Vesco, A.P. Expression of calpastatin and myostatin genes associated with lamb meat quality. Genet. Mol. Res. 2013, 12, 6168–6175. [Google Scholar] [CrossRef]
- Lambe, N.R.; Haresign, W.; Macfarlane, J.; Richardson, R.I.; Matika, O.; Bunger, L. The effect of conditioning period on loin muscle tenderness in crossbred lambs with or without the Texel muscling QTL (TM-QTL). Meat Sci. 2010, 85, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Matika, O.; Riggio, V.; Anselme-Moizan, M.; Law, A.S.; Pong-Wong, R.; Archibald, A.L.; Bishop, S.C. Genome-wide association reveals QTL for growth, bone, and in vivo carcass traits as assessed by computed tomography in Scottish Blackface lambs. Genet. Sel. Evol. 2016, 48, 11. [Google Scholar] [CrossRef] [PubMed]
- Bolormaa, S.; van der Werf, J.H.J.; Hayes, B.J.; Goddard, M.E.; Daetwyler, H.D. Pleiotropic multi-trait genome-wide association reveals putative candidate gene for fatty acid composition in Australian sheep. Proc. Assoc. Advmt. Breed Genet. 2015, 21, 49–52. [Google Scholar]
- Armstrong, E.; Iriarte, A.; Nicolini, P.P.; De Los Santos, J.; Ithurraide, J.; Bielli, A.; Bianchi, G.; Penagaricano, F. Comparison of transcriptomic landscapes of different lamb muscles using RNA-Seq. PLoS ONE 2018, 13, e0200732. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Gunawan, A.; Jakaria; Listyarini, K.; Furqon, A.; Sumantri, C.; Akter, S.H.; Uddin, M.J. Transcriptome signature of liver tissue with divergent mutton odour and flavour using RNA deep sequencing. Gene 2018, 15, 86–94. [Google Scholar] [CrossRef]
- Gunawan, A.; Listyarini, K.; Harahap, R.S.; Jakaria; Roosita, K.; Sumantri, C.; Inounu, I.; Akter, S.H.; Islam, M.A.; Uddin, M.J. Hepatic transcriptome analysis identifies genes, polymorphisms and pathways involved in the fatty acids metabolism in sheep. PLoS ONE 2021, 16, e0260514. [Google Scholar] [CrossRef]
- Cheng, S.; Wang, X.; Zhang, Q.; He, Y.; Zhang, X.; Yang, L.; Shi, J. Comparative transcriptome analysis identifying the different molecular genetic markers related to production performance and meat quality in longissimus dorsi tissues of MG × STH and STH sheep. Genes 2020, 11, 183. [Google Scholar] [CrossRef]
- Raza, S.H.A.; Kaster, N.; Khan, R.; Abdelnour, S.A.; El-Hack, M.E.A.; Khafaga, A.F.; Taha, A.; Ohran, H.; Swelum, A.A.; Schreurs, N.M.; et al. The role of microRNAs in muscle tissue development in beef cattle. Genes 2020, 11, 295. [Google Scholar] [CrossRef]
- Bergman, E.N.; Brockman, R.P.; Kaufman, C.F. Glucose metabolism in ruminants: Comparison of whole-body turnover with production by gut, liver, and kidneys. Fed. Proc. 1974, 33, 1849–1854. [Google Scholar]
- Murray, R.; Granner, D.; Mayes, P.; Rodwell, V. Harpers Biochemistry, 24th ed.; Prentice Hall International, Inc.: Upper Saddle River, NY, USA, 1996. [Google Scholar]
- Warris, P.D.; Bevis, E.A. Liver glycogen in slaughtered pigs and estimated time of fasting before slaughter. Br. Vet. J. 1987, 143, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Warris, P.D.; Bevis, E.A.; Ekins, P.J. The relationships between glycogen stores and muscle ultimate pH in commercially slaughtered pigs. Br. Vet. J. 1989, 145, 378. [Google Scholar] [CrossRef] [PubMed]
- Onopiuk, A.; Poltorak, A.; Wierzbicka, A. Influence of post-mortem muscle glycogen content on the quality of beef during aging. J. Vet. Res. 2016, 60, 301–307. [Google Scholar] [CrossRef]
- Immonen, K.; Ruusunen, M.; Puolanne, E. Some effects of residual glycogen concentration on the physical and sensory quality of normal pH beef. Meat Sci. 2000, 55, 33–38. [Google Scholar] [CrossRef]
- Choe, J.H.; Choi, Y.M.; Lee, S.H.; Shin, H.G.; Ryu, Y.C.; Hong, K.C.; Kim, B.C. The relation between glycogen, lactate content and muscle fiber type composition, and their influence on postmortem glycolytic rate and pork quality. Meat Sci. 2008, 80, 355–362. [Google Scholar] [CrossRef] [PubMed]
- England, E.M.; Matarneh, S.K.; Oliver, E.M.; Apaoblaza, A.; Scheffler, T.L.; Shi, H.; Gerrard, D.E. Excess glycogen does not resolve high ultimate pH of oxidative muscle. Meat Sci. 2016, 114, 95–102. [Google Scholar] [CrossRef]
- Inounu, I.; Hidayati, N.; Subandriyo; Tiesnamurti, B.; Nafiu, L.O. Relative superiority analysis of Garut lamb and its crossbred. Indones. J. Anim. Vet. Sci. 2003, 8, 170–182. [Google Scholar]
- Dagong, M.I.A.; Herman, R.; Sumantri, C.; Noor, R.R.; Yamin, M. Carcass and physical meat characteristics of thin tail sheep (TTS) based on calpastatin gene (CAST) (Locus intron 5-exon 6) genotypes variation. Indones. J. Anim. Vet. Sci. 2012, 17, 13–24. [Google Scholar]
- Cinar, M.U.; Kayan, A.; Uddin, M.J.; Jonas, E.; Tesfaye, D.; Phatsara, C.; Ponsuksili, S.; Wimmers, K.; Tholen, E.; Looft, C.; et al. Association and expression quantitative trait loci (eQTL) analysis of porcine AMBP, GC and PPP1R3B genes with meat quality traits. Mol. Biol. Rep. 2012, 39, 4809–4821. [Google Scholar] [CrossRef]
- Robinson, M.D.; Mccarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, X.; Zhu, J.; Gu, Y.; Zhao, W.; Zou, J.; Guo, Z. GO-function: Deriving biologically relevant functions from statistically significant functions. Brief. Bioinform. 2012, 13, 216–227. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, 480–484. [Google Scholar] [CrossRef]
- Xia, J.; Benner, M.J.; Hancock, R.E. NetworkAnalyst—Integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014, 42, W167–W174. [Google Scholar] [CrossRef]
- Basha, O.; Shpringer, R.; Argov, C.M.; Yeger-Lotem, E. The DifferentialNet database of differential protein-protein interactions in human tissues. Nucleic Acids Res. 2018, 46, D522–D526. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Zhang, C.; Arif, M.; Liu, Z.; Benfeitas, R.; Bidkhori, G.; Deshmukh, S.; Al Sobky, M.; Lovric, A.; Boren, J.; et al. TCSBN: A database of tissue and cancer specific biological networks. Nucleic Acids Res. 2018, 46, D595–D600. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [CrossRef]
- Gunawan, A.; Sahadavan, S.; Cinar, M.U.; Neuhoff, C.; Große-Brinkhaus, C.; Frieden, L.; Tesfaye, D.; Tholen, E.; Looft, C.; Wondim, D.S.; et al. Identification of the novel candidate genes and variants in boar liver tissues with divergent skatole levels using RNA deep sequencing. PLoS ONE 2013, 8, e72298. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and Genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szczesniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Kim, J.R.; Moon, J.K.; Choi, B.H.; Kim, T.H.; Kim, K.S.; Kim, J.J.; Lee, C.K. Transcriptional Alteration of p53 Related Processes as a Key Factor for Skeletal Muscle Characteristics in Sus scrofa. Mol. Cells 2009, 28, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Morlein, D.; Lungershausen, M.; Steinke, K.; Sharifi, A.R.; Knorr, C. A single nucleotide polymorphism in the CYP2E1 gen promoter affects skatole content in backfat of boars of two commercial Duroc-sired crossbred populations. Meat Sci. 2012, 92, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Jo, J.L.; Hwang, J.H.; Kwon, S.G.; Park, D.H.; Kim, T.W.; Kang, D.G.; Yu, G.E.; Kim, I.S.; Ha, J.G.; Kim, C.W. Association between a non-synonymous HSD17β4 single nucleotide polymorphism and meat-quality traits in Berkshire pigs. Gent. Mol. Res. 2016, 15, 1–9. [Google Scholar] [CrossRef]
- Peng, D.Q.; Jung, U.S.; Lee, J.S.; Kim, W.S.; Jo, Y.H.; Kim, M.J.; Oh, Y.K.; Baek, Y.C.; Hwang, S.G.; Lee, H.G. Effect of alcohol dehydrogenase 1C (ADH1C) genotype on vitamin A restriction and marbling in Korean native steers. Asian-Australas. J. Anim. Sci. 2017, 30, 1099–1104. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Kwon, S.; Ham, S.; Lee, D.; Park, H.E.H.; Yamaoka, Y.; Jeong, D.E.; Artan, M.; Altintas, O.; Park, S.; et al. Caenorhabditis elegans Lipin 1 moderates the lifespan-shortening effects of dietary glucose by maintaining ω-6 polyunsaturated fatty acids. Aging Cell. 2020, 19, e13150. [Google Scholar] [CrossRef]
- Duan, X.; An, B.; Du, L.; Chang, T.; Liang, M.; Yang, B.G.; Xu, L.; Zhang, L.; Li, J.; Guangxin, E.; et al. Genome-wide association analysis of growth curve parameters in chinese simmental beef cattle. Animals 2021, 11, 192. [Google Scholar] [CrossRef]
- Óvilo, C.; Benítez, R.; Fernández, A.; Núñez, Y.; Ayuso, M.; Fernández, A.I.; Rodriguez, C.; Isabel, B.; Rey, A.I.; Lopez-Bote, C.; et al. Longissimus dorsi transcriptome analysis of purebred and crossbred Iberian pigs differing in muscle characteristics. BMC Genom. 2014, 15, 413. [Google Scholar] [CrossRef]
- Zhongchang, H.; Jian, W.; Qin, L.; Haigou, J.; Yang, C.; Yumin, Z. IGFBP7 downregulation or overexpression effect on bovine preadipocyte differentiation. Anim. Biotechnol. 2021, 32, 21–30. [Google Scholar] [CrossRef]
- Shu, L.; Hoo, R.L.; Wu, X.; Pan, Y.; Lee, I.P.; Cheong, L.Y.; Bornstein, S.R.; Rong, X.; Guo, J.; Xu, A. A-FABP mediates adaptive thermogenesis by promoting intracellular activation of thyroid hormonesin brown adipocytes. Nat. Commun. 2017, 8, 14147. [Google Scholar] [CrossRef]
- Holmes, R.S.; Vandeberg, J.L.; Cox, L.A. Genomics and Proteomics of Vertebrate Cholesterol Ester Lipase (LIPA) and Cholesterol 25-Hydroxylase (CH25H). 3 Biotech. 2011, 1, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, R.; Liu, Z.; Hou, C.; Zong, W.; Zhang, A.; Sun, X.; Gao, J. Loss of lysyl oxidase-like 3 causes cleft palate and spinal deformity in mice. Hum. Mol. Genet. 2015, 24, 6174–6185. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, J.; Hou, X.; Zan, L.; Wang, N.; Tang, Z.; Li, K. OLFML3 expression is decreased during prenatal muscle development and regulated by microRNA-155 in pigs. Int. J. Biol. Sci. 2012, 8, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Braz, C.U.; Taylor, J.F.; Bresolin, T.; Espigolan, R.; Feitosa, F.L.B.; Carvalheiro, R.; Baldi, F.; de Albuquerque, L.G.; de Oliveira, H.N. Sliding window haplotype approaches overcome single SNP analysis limitations in identifying genes for meat tenderness in Nelore cattle. BMC Genet. 2019, 20, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Picard, B.; Gagaoua, M. Chapter 11—Proteomic Investigations of Beef Tenderness. In Proteomics in Food Science: From Farm to Fork; Colgrave, M.L., Ed.; Academic Press: London, UK, 2017; pp. 177–197. [Google Scholar]
- Ouali, A.; Gagaoua, M.; Boudida, Y.; Becila, S.; Boudjellal, A.; Herrera-Mendez, C.H.; Sentandreu, M.A. Biomarkers of meat tenderness: Present knowledge and perspectives in regards to our current understanding of the mechanisms involved. Meat Sci. 2013, 95, 854–870. [Google Scholar] [CrossRef]
- Picard, B.; Gagaoua, M. Meta-proteomics for the discovery of protein biomarkers of beef tenderness: An overview of integrated studies. Food Res. Int. 2020, 127, 108739. [Google Scholar] [CrossRef]
- Nishimura, T. Role of extracellular matrix in development of skeletal muscle and postmortem aging of meat. Meat Sci. 2015, 109, 48–55. [Google Scholar] [CrossRef]
- Cho, J.H.; Lee, R.H.; Jeon, Y.-J.; Park, S.-M.; Shin, J.-C.; Kim, S.-H.; Jeong, J.Y.; Kang, H.-S.; Choi, N.-J.; Seo, K.S.; et al. Proteomic assessment of the relevant factors affecting pork meat quality associated with longissimus dorsi muscles in Duroc pig. Asian-Australas. J. Anim. Sci. 2016, 29, 1653–1663. [Google Scholar] [CrossRef]
- Markey, C.M.; Coombs, M.A.; Sonnenschein, C.; Soto, A.M. Mammalian development in a changing environment: Exposure to endocrine disruptors reveals the developmental plasticity of steroid-hormone target organs. Evol. Dev. 2003, 5, 67–75. [Google Scholar] [CrossRef]
- McEwen, B. Steroid hormones: Effect on brain development and function. Horm. Res. 1992, 37, 1–10. [Google Scholar] [CrossRef]
- Berger, J.; Moller, D.E. The mechanisms of action of PPARs. Annu. Rev. Med. 2002, 53, 409–435. [Google Scholar] [CrossRef]
- Lim, D.; Chai, H.H.; Lee, S.H.; Cho, Y.M.; Choi, J.W.; Kim, N.K. Gene expression patterns associated with peroxisome proliferator-activated receptor (PPAR) signaling in the Longissimus dorsi of Hanwoo (Korean Cattle). Asian-Australas. J. Anim. Sci. 2015, 28, 1075–1082. [Google Scholar] [CrossRef] [PubMed]
- Macneil, L.T.; Walhout, A.J. Gene regulatory networks and the role of robustness and stochasticity in the control of gene expression. Genome Res. 2011, 21, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Siddaraju, N.K.; Manjunatha, S.S.; Sudarshan, S.; Fairoze, M.N.; Kumar, A.; Chhabra, P.; Kaur, M.; Sreesujatha, R.M.; Ahlawat, S.; et al. Muscle transcriptome provides the first insight into the dynamics of gene expression with progression of age in sheep. Sci. Rep. 2021, 11, 22360. [Google Scholar] [CrossRef] [PubMed]
- Noce, A.; Cardoso, T.F.; Manunza, A.; Martínez, A.; Cánovas, A.; Pons, A.; Bermejo, L.A.; Landi, V.; Sanchez, A.; Jordana, J.; et al. Expression patterns and genetic variation of the ovine skeletal muscle transcriptome of sheep from five Spanish meat breeds. Sci. Rep. 2018, 8, 10486. [Google Scholar] [CrossRef]
- Leal-Gutierrez, J.D.; Elzo, M.A.; Johnson, D.D.; Hamblen, H.; Mateescu, R.G. Genome wide association and gene enrichment analysis reveal membrane anchoring and structural proteins associated with meat quality in beef. BMC Genom. 2019, 20, 151. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.; Kim, Y.; Kim, C.; Kim, J.; Lee, C. Association of bovine carcass phenotypes with genes in an adaptive thermogenesis pathway. Mol. Biol. Rep. 2012, 39, 1441–1445. [Google Scholar] [CrossRef]
- Remy, G.; Risco, A.M.; Inesta-Vaguera, F.A.; Gonzalez-Teran, B.; Sabio, G.; Davis, R.J.; Cuenda, A. Differential activation of p38MAPK isoforms by MKK6 and MKK3. Cell Signal 2010, 22, 660–667. [Google Scholar] [CrossRef]
- Guang-kai, W.; Tao, Z.; Hui-hua, W.; Shu-zhen, Z.; Li, Z.; Cai-hong, W.; Fu-Ping, Z.; Li-Xin, D. Genome-wide detection of selection signature on sunite sheep. Sci. Agric. Sin. 2014, 47, 1190–1199. [Google Scholar] [CrossRef]
- Jin-Ping, S.; Gui-Shan, M.A.; Quan-Wei, Z.; Ting, L.; Jian-Wei, T.; Jian-Hong, S.; Yu-Bing, W.; Lei, Y.; Shu-Ru, C. Identify the genes related to muscle traits in crossbred sheep populations (Ovis aries) by RNA-seq. J. Agric. Biotechnol. 2021, 29, 288–297. [Google Scholar] [CrossRef]
- Salehian-Dehkordi, H.; Xu, Y.-X.; Xu, S.-S.; Li, X.; Luo, L.-Y.; Liu, Y.-J.; Wang, D.-F.; Cao, Y.-H.; Shen, M.; Gao, L.; et al. Genome-wide detection of copy number variations and their association with distinct phenotypes in the world’s sheep. Front. Genet. 2021, 12, 670582. [Google Scholar] [CrossRef] [PubMed]
- Suryati, T.; Arief, I.I.; Polii, B.N. Correlation and categories of meat tenderness based on equipment and panelist test. Anim. Prod. 2008, 10, 188–193. [Google Scholar]
- Suman, S.P.; Joseph, P. Proteomic technologies and their applications in the meat industry. Encycl. Meat Sci. 2014, 3, 155–158. [Google Scholar] [CrossRef]
- Listyarini, K.; Sumantri, C.; Rahayu, S.; Uddin, M.J.; Gunawan, A. Association study and expression analysis of olfactomedin like 3 gene related to meat quality, carcass characteristics, retail meat cut, and fatty acid composition in sheep. Anim. Biosci. 2022, 35, 1489–1498. [Google Scholar] [CrossRef] [PubMed]



| Meat Quality Composition | Mean | SD | Low (n = 5) | High (n = 5) | ||
|---|---|---|---|---|---|---|
| n = 140 | n = 140 | Mean | SD | Mean | SD | |
| pH | 5.98 | 0.57 | 6.11 | 0.11 | 5.95 | 0.22 |
| Tenderness * | 3.66 | 0.76 | 4.69 | 0.67 | 3.14 | 0.09 |
| Cooking loss (%) | 46.46 | 8.09 | 47.91 | 6.30 | 49.40 | 2.90 |
| Water holding capacity (%) | 28.09 | 3.22 | 26.22 | 2.00 | 26.68 | 3.17 |
| Gene Name | Accession Number | Primer sequence | Tm (°C) | Application | Enzymes | Size (bp) | Cutting Size (bp) |
|---|---|---|---|---|---|---|---|
| HSD17B13 | XM_004009979.5 | F: 5′-CCC ATC AAC ACC TAG AAT GC-3′ R: 5′-CAG CAG TGA TTC CAA GTA GG-3′ | 61 | qRT–PCR | - | 178 | - |
| ANGPTL2 | XM_027966435.2 | F: 5′-TTA ATG AAT AAC CAG GGG CC-3′ R: 5′-CTG CTG AGG TAA TAG GCA CA-3′ | 53 | qRT–PCR | - | 215 | - |
| IGFBP7 | NM_001145181.1 | F: 5′-CTG TCC TCA TCT GGA ACA AG-3′ R: 5′-TCT CCA GCA TCT TCC TTA CT-3′ | 56 | qRT–PCR | - | 169 | - |
| TP53INP1 | XM_042254467.1 | F: 5′-GTG CAG TCT GAA GTT CTC CT-3′ R: 5′-TTT CCA AAA CCT GTC TTC GG-3′ | 52 | qRT–PCR | - | 181 | - |
| ADH1C | XM_004009680.4 | F: 5′-GAA TCT GTC GCT CAG ATG AC-3′ R: 5′-GCT CAT TCA GGT CGT GTT TC-3′ | 52 | qRT–PCR | - | 225 | - |
| OLFML3 | XM_004002351.5 | F: 5′-TCC AGA GTA GTG AGA GAG AC-3′ R: 5′-ACA AAA GGA ACA AGA TCA GC-3′ | 53 | qRT–PCR | - | 182 | - |
| THOC5 | XM_042234811.1 | F: 5′-ATT GGC CCA CAT CAG GTT GA-3′ R: 5′-TCT CCC ATG GTG ACT TCT GC-3′ | 53 | qRT–PCR | - | 237 | - |
| CYP2E1 | NM_001245972.1 | F: 5′-ATT CCC AAG TCC TTC ACC AG-3′ R: 5′-GTT GTT TTT GTG CAC CTG GA-3′ | 61 | qRT–PCR | - | 180 | - |
| LPIN1 | NM_001280700.1 | F: 5′-CTC AGA CCA TGA ACT ACG TC-3′ R: 5′-AGT TTC ATG TGC AAA TCC AC-3′ | 57 | qRT–PCR | - | 247 | - |
| FABP5 | NM_001145180.1 | F: 5′-GTC TGC AAC TTT ACG GAT GG-3′ R: 5′-CAG CAG TAT GGA GAT TTG CT-3′ | 61 | qRT–PCR | - | 233 | - |
| GAPDH | NC_019460.2 | F: 5′-GAG AAA CCT GCC AAG TAT GA-3′ R: 5′-TAC CAG GAA ATG AGC TTG AC-3′ | 62 | qRT–PCR | - | 203 | - |
| β-Actin | NC_019471.2 | F: 5′-GAA AAC GAG ATG AGA TTG GC-3′ R: 5′-CCA TCA TAG AGT GGA GTT CG-3′ | 62 | qRT–PCR | - | 194 | - |
| ANGPTL2 | NC_040254.1 | F: 5′-ACA GCT CTG CTC TTA GGA GA-3′ R: 5′-AGA AGC TAG GGA ATC TTG CC-3′ | 62 | Genotyping | NsbI | 454 | GG: 154, 300 bp AA: 454 bp GA: 154, 300, 454 bp |
| OLFML3 | NC_019458.2 | F: 5′-ATG ATG GCT ACC AGA TTG TC-3′ R: 5′-AGT CTG CAG TAC AGA AGG AG-3′ | 59 | Genotyping | MspI | 498 | CC = 195, 303 bp TT = 498 bp CT = 195, 303, 498 bp |
| THOC5 | NC_019474.2 | F: 5′-CCC AGG AAG GTT TGA TTC TC-3′ R: 5′-AGG ACT ACA TGG TAG GTG TG-3′ | 60 | Genotyping | TaiI | 322 | CC = 129, 193 bp TT = 322 bp CT = 129, 193, 322 bp |
| Group | Sample | Total Number of Reads (Million) | Unmapped Reads (Million) | Mapped Reads (Million) | Percentage of Unmapped Reads (%) | Percentage of Mapped Reads (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|---|---|---|
| Low Tenderness | LT1 | 20.95 | 2.67 | 18.28 | 12.74 | 87.26 | 96.48 | 92.68 |
| LT2 | 21.90 | 2.62 | 19.28 | 11.96 | 88.04 | 96.52 | 92.80 | |
| LT3 | 20.06 | 2.40 | 17.66 | 11.96 | 88.04 | 96.06 | 91.95 | |
| LT4 | 21.04 | 2.36 | 18.68 | 11.22 | 88.78 | 96.32 | 92.45 | |
| LT5 | 20.84 | 2.48 | 18.36 | 11.90 | 88.10 | 96.45 | 92.67 | |
| High Tenderness | HT1 | 21.29 | 2.62 | 18.67 | 12.31 | 87.69 | 96.40 | 92.58 |
| HT2 | 20.00 | 3.23 | 16.77 | 16.15 | 83.85 | 96.50 | 92.70 | |
| HT3 | 20.18 | 2.46 | 17.72 | 12.19 | 87.81 | 96.65 | 93.05 | |
| HT4 | 20.02 | 3.12 | 16.90 | 15.58 | 84.42 | 96.41 | 92.56 | |
| HT5 | 21.17 | 2.37 | 18.80 | 11.20 | 88.80 | 96.65 | 93.12 |
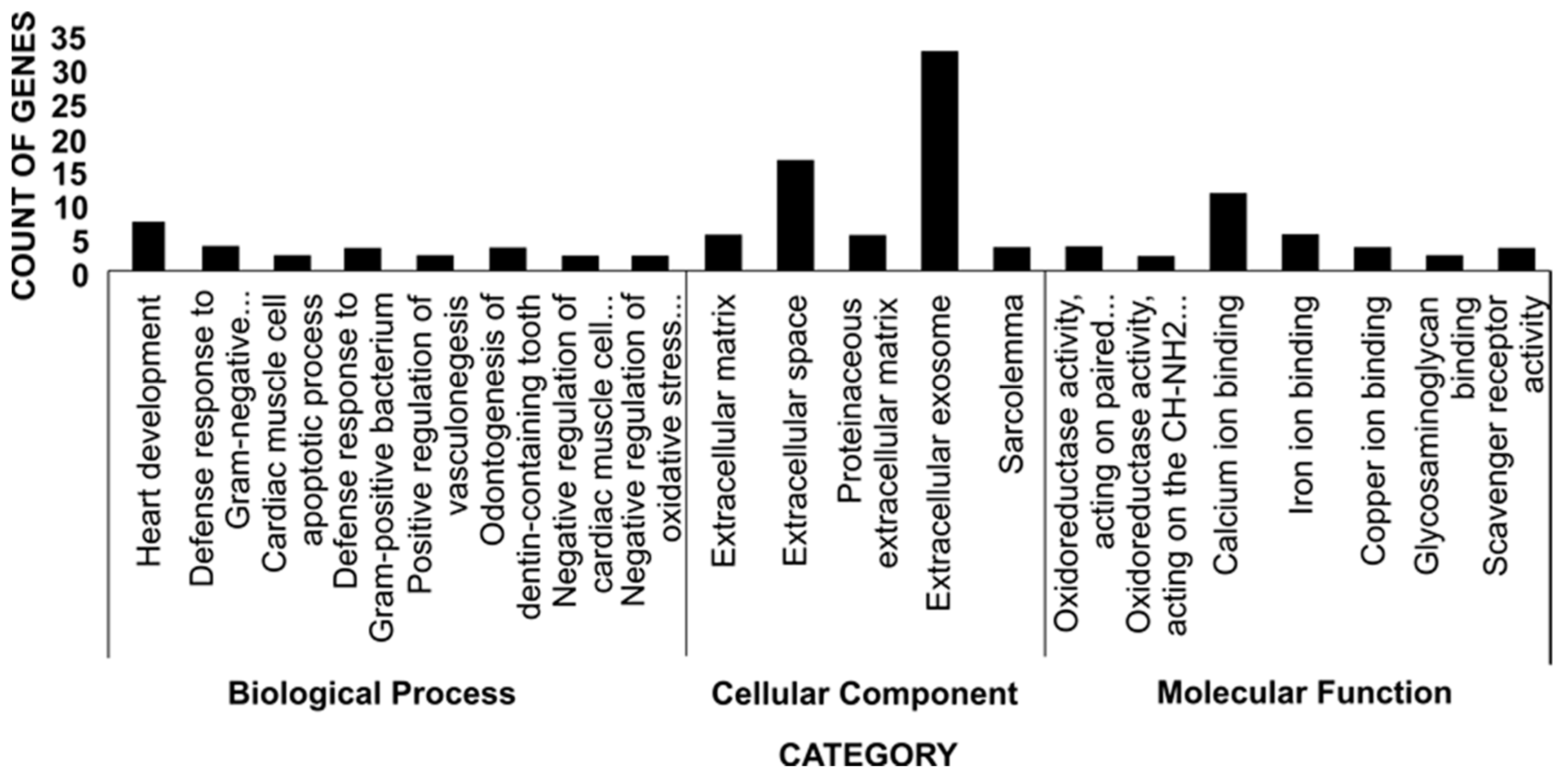
| Category | Term | Count of Genes | Genes |
|---|---|---|---|
| Biological Process | Heart development | 7 | ADM, KCNJ8, RPS6KA2, PDLIM3, GLI2, CACNA1C, DNAH5 |
| Defense response to Gram-negative bacterium | 3 | ADM, HMGB2, SSC5D | |
| Cardiac muscle cell apoptotic process | 2 | NOL3, RPS6KA2 | |
| Defense response to Gram-positive bacterium | 3 | ADM, HMGB2, SSC5D | |
| Positive regulation of vasculogenesis | 2 | ADM, TMEM100 | |
| Odontogenesis of dentin-containing teeth | 3 | HAND2, SOSTDC1, GLI2 | |
| Negative regulation of cardiac muscle cell apoptotic process | 2 | NOL3, HAND2 | |
| Negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 2 | NOL3, VNN1 | |
| Cellular Component | Extracellular matrix | 5 | OGN, AEBP1, EFEMP1, SSC5D, LOXL1 |
| Extracellular space | 16 | F11, PLAT, AEBP1, ADAMTS13, EFEMP1, HMGB2, POMC, S100A13, TNFRSF9, OGN, ADM, SOSTDC1, REN, GDF10, ANGPTL1, SSC5D | |
| Proteinaceous extracellular matrix | 5 | BGN, ADAMTS13, COL6A1, PRELP, WNT2B | |
| Extracellular exosome | 32 | AEBP1, CSPG4, ALDH1L2, EXTL2, CXCL12, OGN, ASPA, TGM3, COL6A1, VNN1, ANGPTL1, ANGPTL2, RHOF, RAP2B, PLAT, F11, DDC, FAM26E, AK1, EFEMP1, ACTN2, REEP2, S100A13, LIN7A, PRELP, REEP5, BGN, CPE, FBLN7, ZNF114, PCYOX1, CDH11 | |
| Sarcolemma | 3 | BGN, COL6A1, CCDC78 | |
| Molecular Function | Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen | 3 | CYP2D14, CYP2D14-like, CYP2E1 |
| Oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor | 2 | LOXL3, LOXL1 | |
| Calcium ion binding | 11 | NOL3, SCUBE2, NOTCH4, EFEMP1, MYL1, FBLN7, TGM3, ACTN2, FKBP10, S100A13, CDH11 | |
| Iron ion binding | 5 | P3H3, CH25H, CYP2D14, CYP2D14-like, CYP2E1 | |
| Copper ion binding | 3 | LOXL3, LOXL1, S100A13 | |
| Glycosaminoglycan binding | 2 | BGN, ENG | |
| Scavenger receptor activity | 3 | LOXL3, SSC5D, SCARA5 |
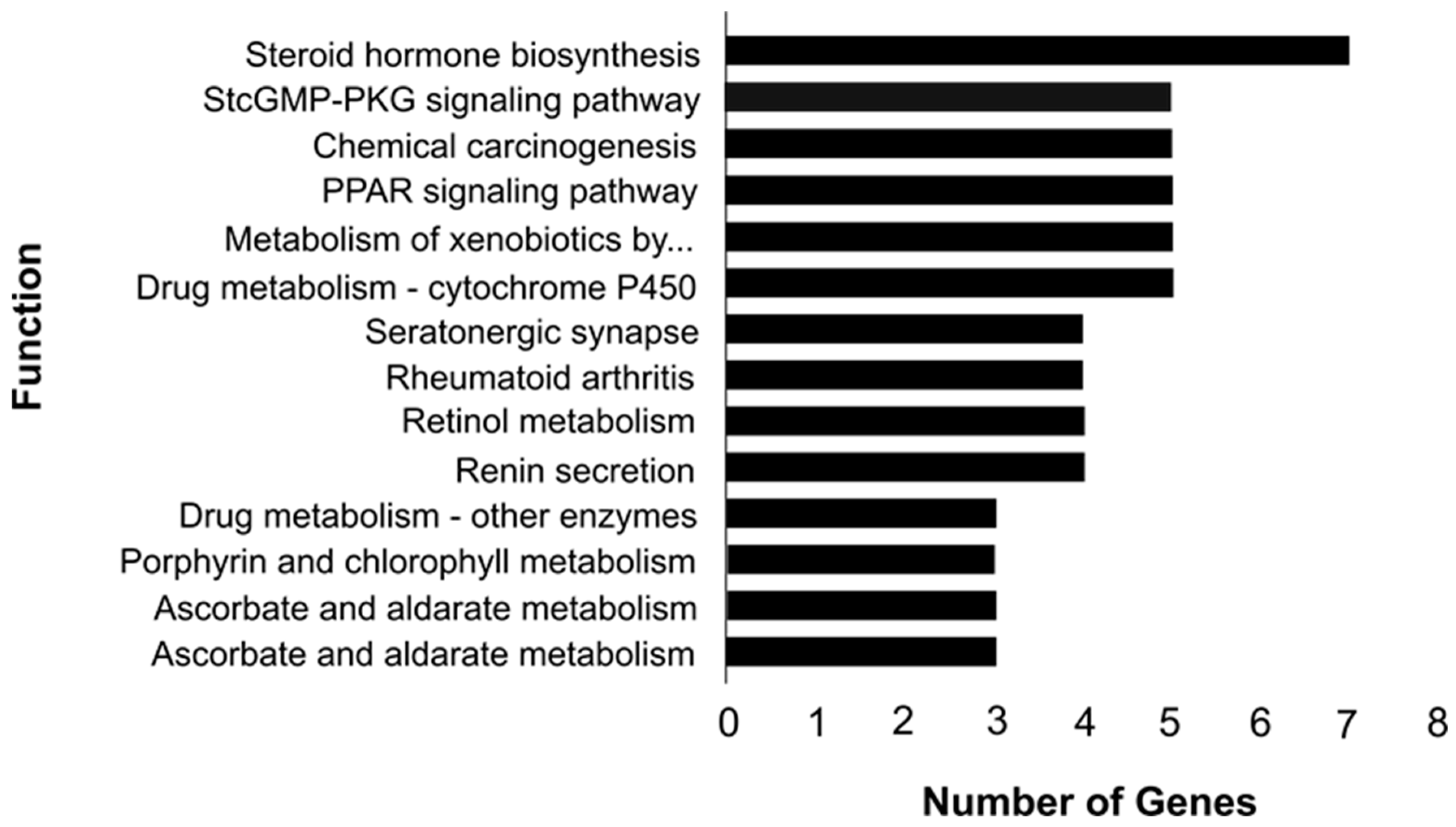
| Function | Number of Genes | Benjamini-Hochberg p-Value | Genes |
|---|---|---|---|
| Ascorbate and aldarate metabolism | 3 | 0.025004 | UGT2B18-like, UGT2B31-like, UGT2A1-like |
| Pentose and glucuronate interconversions | 3 | 0.045079 | UGT2B18-like, UGT2B31-like, UGT2A1-like |
| Porphyrin and chlorophyll metabolism | 3 | 0.066439 | UGT2B18-like, UGT2B31-like, UGT2A1-like |
| Drug metabolism—other enzymes | 3 | 0.066439 | UGT2B18-like, UGT2B31-like, UGT2A1-like |
| Renin secretion | 4 | 0.029158 | REN, GUCY1B2, CACNA1C, LOC101116002 |
| Retinol metabolism | 4 | 0.030325 | UGT2B18-like, UGT2B31-like, ADH1C, UGT2A1-like |
| Rheumatoid arthritis | 4 | 0.089781 | MMP1, DQA, CXCL12 |
| Serotonergic synapse | 4 | 0.095866 | DDC, CYP2D14, CYP2D14-like, CACNA1C |
| Drug metabolism—cytochrome P450 | 5 | 0.003294 | UGT2B18, UGT2B31-like, ADH1C, UGT2A1-like, CYP2E1 |
| Metabolism of xenobiotics by cytochrome P450 | 5 | 0.004904 | UGT2B18-like, UGT2B31-like, ADH1C, UGT2A1-like, CYP2E1 |
| PPAR signaling pathway | 5 | 0.005732 | MMP1, PLIN1, APOA5, ACSL6, FABP5 |
| Chemical carcinogenesis | 5 | 0.007657 | UGT2B18-like, UGT2B31-like, ADH1C, UGT2A1-like, CYP2E1 |
| cGMP-PKG signaling pathway | 5 | 0.099557 | KCNJ8, GUCY1B2, MRVI1, CACNA1C, MYL9 |
| Steroid hormone biosynthesis | 7 | 8.33 × 10−5 | UGT2B18, UGT2B31, DHD3-like, UGT2A1, CYP2D14, CYP2D14-like, CYP2E1 |
| Meat Quality | OLFML3 C > T | ANGPTL2 G > A | THOC5 C > T | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype (µ ± S.D) | Genotype (µ ± S.D) | Genotype (µ ± S.D) | |||||||
| CC (n = 57) | CT (n = 62) | TT (n = 21) | GG (n = 21) | GA (n = 69) | AA (n = 50) | CC (n = 135) | CT (n = 3) | TT (n = 2) | |
| pH value | 6.08 ± 0.58 | 5.93 ± 0.56 | 5.83 ± 0.54 | 6.09 ± 0.75 | 5.98 ± 0.61 | 6.06 ± 0.55 | 6.01 ± 0.62 | 5.97 ± 0.10 | 5.90 ± 0.41 |
| Tenderness (shear force, kg/cm2) | 3.63 ± 0.91 ab | 3.79 ± 0.67 a | 3.35 ± 0.44 b | 3.09 ± 0.51 a | 3.61 ± 0.74 a | 3.75 ± 0.86 b | 3.55 ± 0.70 b | 4.97 ± 0.53 a | 3.45 ± 1.20 b |
| Cooking loss (%) | 45.31 ± 8.53 b | 46.47 ± 7.76 ab | 49.54 ± 7.35 a | 49.47 ± 5.90 | 46.36 ± 8.09 | 46.85 ± 8.01 | 46.44 ± 8.05 | 48.42 ± 3.00 | 49.69 ± 3.88 |
| WHC (mgH2O) | 84.80 ± 12.00 | 83.88 ± 8.02 | 84.07 ± 6.98 | 84.04 ± 7.36 | 84.59 ± 9.37 | 84.16 ± 10.48 | 84.18 ± 9.53 | 77.95 ± 11.30 | 84.87 ± 4.32 |
| WHC (% mgH2O) | 28.26 ± 4.00 | 27.96 ± 2.67 | 28.02 ± 2.32 | 28.01 ± 2.45 | 28.19 ± 3.12 | 28.05 ± 3.49 | 28.06 ± 3.17 | 25.98 ± 3.76 | 28.29 ± 1.44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Listyarini, K.; Sumantri, C.; Rahayu, S.; Islam, M.A.; Akter, S.H.; Uddin, M.J.; Gunawan, A. Hepatic Transcriptome Analysis Reveals Genes, Polymorphisms, and Molecules Related to Lamb Tenderness. Animals 2023, 13, 674. https://doi.org/10.3390/ani13040674
Listyarini K, Sumantri C, Rahayu S, Islam MA, Akter SH, Uddin MJ, Gunawan A. Hepatic Transcriptome Analysis Reveals Genes, Polymorphisms, and Molecules Related to Lamb Tenderness. Animals. 2023; 13(4):674. https://doi.org/10.3390/ani13040674
Chicago/Turabian StyleListyarini, Kasita, Cece Sumantri, Sri Rahayu, Md. Aminul Islam, Syeda Hasina Akter, Muhammad Jasim Uddin, and Asep Gunawan. 2023. "Hepatic Transcriptome Analysis Reveals Genes, Polymorphisms, and Molecules Related to Lamb Tenderness" Animals 13, no. 4: 674. https://doi.org/10.3390/ani13040674
APA StyleListyarini, K., Sumantri, C., Rahayu, S., Islam, M. A., Akter, S. H., Uddin, M. J., & Gunawan, A. (2023). Hepatic Transcriptome Analysis Reveals Genes, Polymorphisms, and Molecules Related to Lamb Tenderness. Animals, 13(4), 674. https://doi.org/10.3390/ani13040674








