Regulatory Role of microRNA of Milk Exosomes in Mastitis of Dairy Cows
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Housing
2.2. Extracellular Vesicles Isolation and RNA Extraction
2.3. miRNA Sequencing
2.4. Data Mining and Bioinformatic and Statistical Analysis
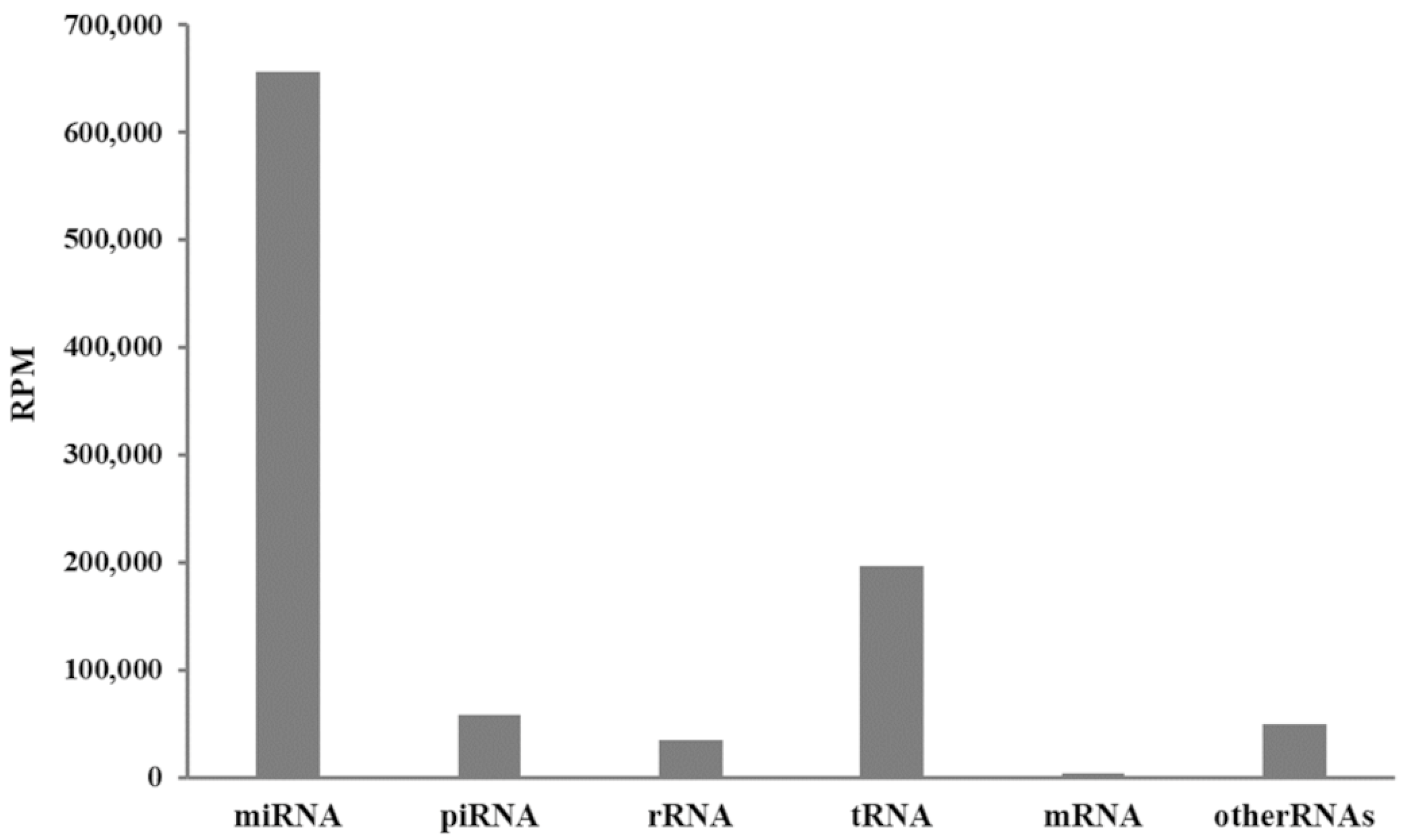
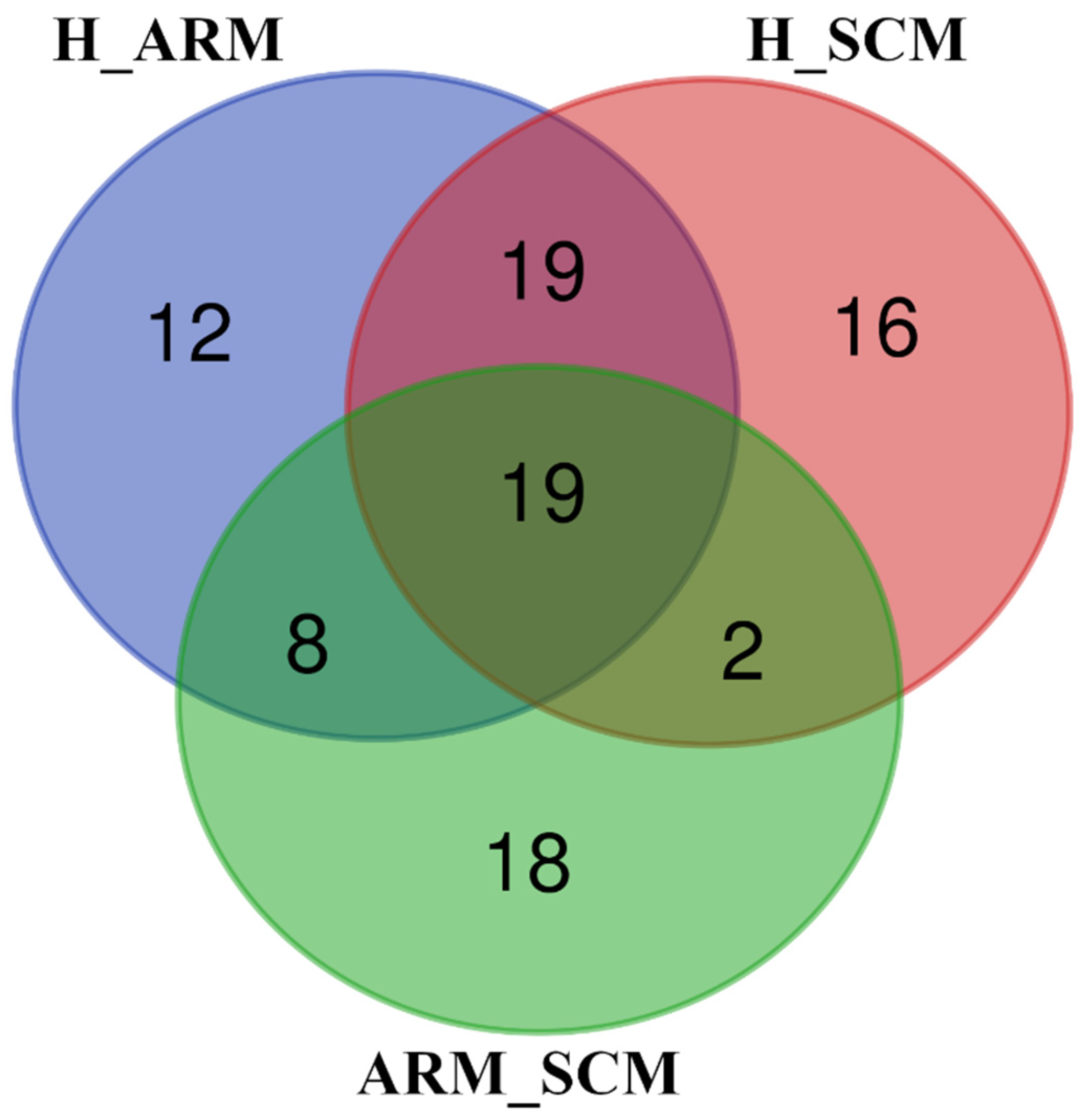
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van Hooijdonk, A.C.; Kussendrager, K.D.; Steijns, J.M. In vivo antimicrobial and antiviral activity of components in bovine milk and colostrum involved in non-specific defense. Br. J. Nutr. 2000, 84, S127–S134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sgorlon, S.; Fanzago, M.; Guiatti, D.; Gabai, G.; Stradaioli, G.; Stefanon, B. Factors affecting milk cortisol in mid lactating dairy cows. BMC Vet. Res. 2015, 11, 259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreas, N.J.; Kampmann, B.; Mehring Le-Doare, K. Human breast milk: A review on its composition and bioactivity. Early Hum. Dev. 2015, 91, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Verhasselt, V.; Milcent, V.; Cazareth, J.; Kanda, A.; Fleury, S.; Dombrowicz, D.; Glaichenhaus, N.; Julia, V. Breast milk-mediated transfer of an antigen induces tolerance and protection from allergic asthma. Nat. Med. 2008, 14, 170–175. [Google Scholar] [CrossRef]
- Kosaka, N.; Izumi, H.; Sekine, K.; Ochiya, T. MicroRNA as a new immune-regulatory agent in breast milk. Silence 2010, 1, 7. [Google Scholar] [CrossRef] [Green Version]
- Benmoussa, A.; Ly, S.; Shan, S.T.; Laugier, J.; Boilard, E.; Gilbert, C.; Provost, P. A subset of extracellular vesicles carries the bulk of microRNAs in commercial dairy cow’s milk. J. Extracell. Vesicles 2017, 6, 1401897. [Google Scholar] [CrossRef] [Green Version]
- Benmoussa, A.; Laugier, J.; Beauparlant, C.J.; Lambert, M.; Droit, A.; Provost, P. Complexity of the microRNA transcriptome of cow milk and milk-derived extracellular vesicles isolated via differential ultracentrifugation. J. Dairy Sci. 2020, 103, 16–29. [Google Scholar] [CrossRef]
- Srikok, S.; Patchaneeb, P.; Boonyayatra, S.; Chuammitria, P. Potential role of MicroRNA as a diagnostic tool in the detection of bovine mastitis. Prev. Vet. Med. 2020, 182, 105101. [Google Scholar] [CrossRef]
- Tucker, A.R.; Salazar, N.A.; Ayoola, A.O.; Memili, E.; Thomas, B.N.; Morenikeji, O.B. Regulatory network of miRNA, lncRNA, transcription factor and target immune response genes in bovine mastitis. Sci. Rep. 2021, 11, 21899. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Benmoussa, A.; Provost, P. Milk microRNAs in health and disease. Compr. Rev. Food Sci. Food Saf. 2019, 18, 703–722. [Google Scholar] [CrossRef] [Green Version]
- Laurent, L.C. MicroRNAs in embryonic stem cells and early embryonic development. J. Cell Mol. Med. 2008, 12, 2181–2188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krömker, V.; Leimbach, S. Mastitis treatment-Reduction in antibiotic usage in dairy cows. Reprod. Domest. Anim. 2017, 52 (Suppl. S3), 21–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weersink, A.; VanLeeuwen, J.A.; Chi, J.; Keefe, G.P. Direct Production Losses and Treatment Costs due to Four Dairy Cattle Diseases. Adv. Dairy Technol. 2002, 14, 55–75. [Google Scholar]
- Hommels, N.M.C.; Ferreira, F.C.; van den Borne, B.H.P.; Hogeveen, H. Antibiotic use and potential economic impact of implementing selective dry cow therapy in large US dairies. J. Dairy Sci. 2021, 104, 8931–8946. [Google Scholar] [CrossRef]
- Trevisi, E.; Zecconi, A.; Cogrossi, S.; Razzuoli, E.; Grossi, P.; Amadori, M. Strategies for reduced antibiotic usage in dairy cattle farms. Res. Vet. Sci. 2014, 96, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Kuipers, A.; Koops, W.J.; Wemmenhove, H. Antibiotic use in dairy herds in the Netherlands from 2005 to 2012. J. Dairy Sci. 2016, 99, 1632–1648. [Google Scholar] [CrossRef] [Green Version]
- Farre, M.; Zecconi, A.; Kelton, D. International Dairy Federation. Guidelines for defining quarter and udder health status and cured clinical and subclinical mastitis cases. Bull. IDF 2022, 515, 34. [Google Scholar] [CrossRef]
- Ebrahimie, E.; Ebrahimi, F.; Ebrahimi, M.; Tomlinson, S.; Petrovski, K.R. A large-scale study of indicators of sub-clinical mastitis in dairy cattle by attribute weighting analysis of milk composition features: Highlighting the predictive power of lactose and electrical conductivity. J. Dairy Res. 2018, 85, 193–200. [Google Scholar] [CrossRef]
- Schwarz, D.; Lipkens, Z.; Piepers, S.; De Vliegher, S. Investigation of differential somatic cell count as a potential new supplementary indicator to somatic cell count for identification of intramammary infection in dairy cows at the end of the lactation period. Prev. Vet. Med. 2019, 172, 104803. [Google Scholar] [CrossRef]
- Zecconi, A.; Zanini, L.; Cipolla, M.; Stefanon, B. Factors Affecting the Patterns of Total Amount and Proportions of Leukocytes in Bovine Milk. Animals 2020, 10, 992. [Google Scholar] [CrossRef] [PubMed]
- Zecconi, A.; Vairani, D.; Cipolla, M.; Rizzi, N.; Zanini, L. Assessment of Subclinical Mastitis Diagnostic Accuracy by Differential Cell Count in Individual Cow Milk. Ital. J. Anim. Sci. 2018, 18, 435–440. [Google Scholar] [CrossRef] [Green Version]
- Mariani, E.; Cipolat-Gotet, C.; Stefanon, B.; Zecconi, A.; Stocco, G.; Sandri, M.; Ablondi, M.; Mountricha, M.; Summer, A. Effect of total and differential somatic cell count on yield, composition and predicted coagulation properties from individual dairy cows. Int. J. Dairy Technol. 2022, 75, 298–307. [Google Scholar] [CrossRef]
- Zecconi, A.; Dell’Orco, F.; Vairani, D.; Rizzi, N.; Cipolla, M.; Zanini, L. Differential cell count as a marker for changes of milk composition in cows very low somatic cell counts. Animals 2020, 10, 604. [Google Scholar] [CrossRef] [Green Version]
- O’Connell, R.M.; Rao, D.S.; Baltimore, D. microRNA regulation of inflammatory responses. Ann. Rev. Immunol. 2012, 30, 295–312. [Google Scholar] [CrossRef]
- Colitti, M.; Sgorlon, S.; Licastro, D.; Stefanon, B. Differential expression of miRNAs in milk exosomes of cows subjected to group relocation. Res. Vet. Sci. 2018, 122, 148–155. [Google Scholar] [CrossRef]
- Colitti, M.; Sgorlon, S.; Stefanon, B. Exosome cargo in milk as a potential marker of cow health. J. Dairy Res. 2020, 87 (Suppl. S1), 79–83. [Google Scholar] [CrossRef]
- Wang, Y.; Fang, J.; Zeng, H.F.; Zhong, J.F.; Li, H.X.; Chen, K.L. Identification and bioinformatics analysis of differentially expressed milk exosomal microRNAs in milk exosomes of heat-stressed Holstein cows. Funct. Integr. Genom. 2022, 22, 77–87. [Google Scholar] [CrossRef]
- Zheng, Y.; Chen, K.L.; Zheng, X.M.; Li, H.X.; Wang, G.L. Identification and bioinformatics analysis of microRNAs associated with stress and immune response in serum of heat-stressed and normal Holstein cows. Cell Stress Chaperones 2014, 19, 973–981. [Google Scholar] [CrossRef] [Green Version]
- Cai, M.; He, H.; Jia, X.; Chen, S.; Wang, J.; Shi, Y.; Liu, B.; Xiao, W.; Lai, S. Genome wide microRNA profiling of bovine milk-derived exosomes infected with Staphylococcus aureus. Cell Stress Chaperones 2018, 23, 663–672. [Google Scholar] [CrossRef]
- Saenz-de-Juano, M.D.; Silvestrelli, G.; Bauersachs, S.; Ulbrich, S.E. Determining extracellular vesicles properties and miRNA cargo variability in bovine milk from healthy cows and cows undergoing subclinical mastitis. BMC Genom. 2022, 23, 189. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, T.A.; Sacco, R.E.; Nonnecke, B.J.; Lippolis, J.D. Bovine milk proteome: Quantitative changes in normal milk exosomes, milk fat globule membranes and whey proteomes resulting from Staphylococcus aureus mastitis. J. Proteom. 2013, 82, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Aswath, K.; Schroeder, S.G.; Lippolis, J.D.; Reinhardt, T.A.; Sonstegard, T.S. MicroRNA expression profiles of bovine milk exosomes in response to Staphylococcus aureus infection. BMC Genom. 2015, 16, 806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Passe Pereira, H.; Lima Verardo, L.; Morena Del Cambre Amaral Weller, M.; Paula Sbardella, A.; Prado Munari, D.; Morais de Paiva Daibert, R.; Araújo Carvalho, W.; Antonio Machado, M.; Fonseca Martins, M. Going further post-RNA-seq: In silico functional analyses revealing candidate genes and regulatory elements related to mastitis in dairy cattle. J. Dairy Res. 2021, 88, 286–292. [Google Scholar] [CrossRef]
- Van Herwijnen, M.J.C.; Driedonks, T.A.P.; Snoek, B.L.; Kroon, A.M.T.; Kleinjan, M.; Jorritsma, R.; Pieterse, C.M.J.; Hoen, E.N.M.N.; Wauben, M.H.M. Abundantly present miRNAs in milk-derived extracellular vesicles are conserved between mammals. Front. Nutr. 2018, 5, 81. [Google Scholar] [CrossRef]
- Izumi, H.; Kosaka, N.; Shimizu, T.; Sekine, K.; Ochiya, T.; Takase, M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J. Dairy Sci. 2012, 95, 4831–4841. [Google Scholar] [CrossRef] [Green Version]
- Lonnerdal, B.; Du, X.; Liao, Y.; Li, J. Human milk exosomes resist digestion in vitro and are internalized by human intestinal cells. FASEB J. 2015, 29, 121–123. [Google Scholar] [CrossRef]
- Shandilya, S.; Rani, P.; Onteru, S.K.; Singh, D. Small interfering RNA in milk exosomes is resistant to digestion and crosses the intestinal barrier in vitro. J. Agric. Food Chem. 2017, 65, 9506–9513. [Google Scholar] [CrossRef]
- Kahn, S.; Liao, Y.; Du, X.; Xu, W.; Li, J.; Lonnerdal, B. Exosomal microRNAs in milk from mothers delivering preterm infants survive in vitro digestion and are taken up by human intestinal cells. Mol. Nutr. Food Res. 2018, 62, e1701050. [Google Scholar] [CrossRef]
- Liao, Y.; Du, X.; Li, J.; Lonnerdal, B. Human milk exosomes and their microRNAs survive digestion in vitro and are taken up by human intestinal cells. Mol. Nutr. Food Res. 2017, 61, 11. [Google Scholar] [CrossRef]
- Baier, S.R.; Nguyen, C.; Xie, F.; Wood, J.R.; Zempleni, J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow’s milk and affect gene expression in peripheral blood mononuclear cells, HEK- 293 kidney cell cultures, and mouse livers. J. Nutr. 2014, 144, 1495–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izumi, H.; Tsuda, M.; Sato, Y.; Kosaka, N.; Ochiya, T.; Iwamoto, H.; Namba, K.; Takeda, Y. Bovine milk exosomes contain microRNA and mRNA and are taken up by human macrophages. J. Dairy Sci. 2015, 98, 2920–2933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Pozo-Acebo, L.; Hazas, M.L.L.; Tomé-Carneiro, J.; Gil-Cabrerizo, P.; San-Cristobal, R.; Busto, R.; García-Ruiz, A.; Dávalos, A. Bovine Milk-Derived Exosomes as a Drug Delivery Vehicle for miRNA-Based Therapy. Int. J. Mol. Sci. 2021, 22, 1105. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; Schmitz, G. Exosomes of pasteurized milk: Potential pathogens of Western diseases. J. Transl. Med. 2019, 17, 3. [Google Scholar] [CrossRef] [Green Version]
- Cintio, M.; Polacchini, G.; Scarsella, E.; Montanari, T.; Stefanon, B.; Colitti, M. MicroRNA Milk Exosomes: From Cellular Regulator to Genomic Marker. Animals 2020, 10, 1126. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 14 December 2022).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef] [Green Version]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.Y.; Lin, Y.C.; Li, J.; Huang, K.Y.; Shrestha, S.; Hong, H.C.; Tang, Y.; Chen, Y.G.; Jin, C.N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betel, D.; Wilson, M.; Gabow, A.; Marks, D.S.; Sander, C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008, 36, D149–D153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, S.; Tong, C.; Ibeagha-Awemu, E.M.; Zhao, X. Identification and characterization of differentially expressed exosomal microRNAs in bovine milk infected with Staphylococcus aureus. BMC Genom. 2019, 20, 934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, C.; Lv, H.; Peng, Z.; Yang, D.; Shen, P.; Yu, J.; Tong, C.; Wang, X. Analysis of miRNA expression changes in bovine endometrial stromal cells treated with lipopolysaccharide. Theriogenolgy 2021, 167, 85–93. [Google Scholar] [CrossRef]
- Muroya, S.M.; Hagi, T.; Kimura, A.; Aso, H.; Matsuzaki, M.; Nomura, M. Lactogenic hormones alter cellular and extracellular microRNA expression in bovine mammary epithelial cell culture. J. Anim. Sci. Biotechol. 2016, 7, 8. [Google Scholar] [CrossRef] [Green Version]
- Doherty, R.; Whiston, R.; Cormican, P.M.; Finlay, E.K.; Couldrey, C.; Brady, C.; O’Farrelly, C.; Meade, K.G. The CD4(+) T cell methylome contributes to a distinct CD4(+) T cell transcriptional signature in Mycobacterium bovis-infected cattle. Sci. Rep. 2016, 6, 31014. [Google Scholar] [CrossRef] [Green Version]
- Nata, T.; Fujiya, M.; Ueno, N.; Moriichi, K.; Konishi, H.; Tanabe, H.; Ohtake, T.; Ikuta, K.; Kohgo, Y. MicroRNA-146b improves intestinal injury in mouse colitis by activating nuclear factor-?B and improving epithelial barrier function. J. Gene Med. 2013, 15, 249–260. [Google Scholar] [CrossRef]
- Wang, X.P.; Luoreng, Z.M.; Zan, L.S.; Li, F.; Li, N. Bovine miR-146a regulates inflammatory cytokines of bovine mammary epithelial cells via targeting the TRAF6 gene. J. Dairy Sci. 2017, 100, 7648–7658. [Google Scholar] [CrossRef] [Green Version]
- Tzelos, T.; Ho, W.; Charmana, V.I.; Lee, S.; Donadeu, F.X. MiRNAs in milk can be used towards early prediction of mammary gland inflammation in cattle. Sci. Rep. 2022, 12, 5131. [Google Scholar] [CrossRef]
- Wollowski, L.; Heuwieser, W.; Kossatz, A.; Addis, M.F.; Puggioni, G.M.G.; Meriaux, L.; Bertulat, S. The value of the biomarkers cathelicidin, milk amyloid A, and haptoglobin to diagnose and classify clinical and subclinical mastitis. J. Dairy Sci. 2020, 104, 2106–2122. [Google Scholar] [CrossRef]
- Jiang, Q.; Zhao, H.; Li, R.; Zhang, Y.; Liu, Y.; Wang, J.; Wang, X.; Ju, Z.; Liu, W.; Hou, M.; et al. In silico genome-wide miRNA-QTL-SNPs analyses identify a functional SNP associated with mastitis in Holsteins. BMC Genet. 2019, 20, 46. [Google Scholar] [CrossRef] [PubMed]
- Billa, P.A.; Faulconnier, Y.; Ye, T.; Chervet, M.; Le Provost, F.; Pires, J.A.A.; Leroux, C. Deep RNA-Seq reveals miRNome differences in mammary tissue of lactating Holstein and Montbéliarde cows. BMC Genom. 2019, 20, 621. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, P.S.; Coutinho, L.L.; Cesar, A.S.M.; Diniz, W.J.D.S.; de Souza, M.M.; Andrade, B.G.; Koltes, J.E.; Mourão, G.B.; Zerlotini, A.; Reecy, J.M.; et al. Co-Expression Networks Reveal Potential Regulatory Roles of miRNAs in Fatty Acid Composition of Nelore Cattle. Front. Genet. 2019, 10, 651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzilli, M.; Zecconi, A. Assessment of epithelial cells’ immune and inflammatory response to Staphylococcus aureus when exposed to a macrolide. J. Dairy Res. 2010, 77, 404–410. [Google Scholar] [CrossRef]
- Riollet, C.; Rainard, P.; Poutrel, B. Cells and cytokines in inflammatory secretions of bovine mammary gland. Adv. Exp. Med. Biol. 2000, 480, 247–258. [Google Scholar] [PubMed]
- Glazov, E.A.; Kongsuwan, K.; Assavalapsakul, W.; Horwood, P.F.; Mitter, N.; Mahony, T.J. Repertoire of bovine miRNA and miRNA-like small regulatory RNAs expressed upon viral infection. PLoS ONE 2009, 4, e6349. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Ju, Z.; Li, Q.; Hou, Q.; Wang, C.; Li, J.; Li, R.; Wang, L.; Sun, T.; Hang, S.; et al. Solexa sequencing of novel and differentially expressed microRNAs in testicular and ovarian tissues in Holstein cattle. Int. J. Biol. Sci. 2011, 7, 1016–1026. [Google Scholar] [CrossRef]
- Strozzi, F.; Mazza, R.; Malinverni, R.; Williams, J.L. Annotation of 390 bovine miRNA genes by sequence similarity with other species. Anim. Genet. 2009, 40, 125. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stefanon, B.; Cintio, M.; Sgorlon, S.; Scarsella, E.; Licastro, D.; Zecconi, A.; Colitti, M. Regulatory Role of microRNA of Milk Exosomes in Mastitis of Dairy Cows. Animals 2023, 13, 821. https://doi.org/10.3390/ani13050821
Stefanon B, Cintio M, Sgorlon S, Scarsella E, Licastro D, Zecconi A, Colitti M. Regulatory Role of microRNA of Milk Exosomes in Mastitis of Dairy Cows. Animals. 2023; 13(5):821. https://doi.org/10.3390/ani13050821
Chicago/Turabian StyleStefanon, Bruno, Michela Cintio, Sandy Sgorlon, Elisa Scarsella, Danilo Licastro, Alfonso Zecconi, and Monica Colitti. 2023. "Regulatory Role of microRNA of Milk Exosomes in Mastitis of Dairy Cows" Animals 13, no. 5: 821. https://doi.org/10.3390/ani13050821




