Insights into Early Ontogenesis of Salmo salar: RNA Extraction, Housekeeping Gene Validation and Transcriptional Expression of Important Primordial Germ Cell and Sex-Determination Genes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fertilization and Embryo Development of S. salar
2.2. Sampling
2.3. Cell Extraction from the Embryo during Different Developmental Stages
2.4. RNA Extraction and cDNA Synthesis
2.5. Primers
2.6. Quantitative PCR (qPCR) Analysis
2.7. Validation of HKGs
2.8. Data Analysis
3. Results
3.1. Embryonic Development
3.2. RNA Quality
3.3. Validation of HKG
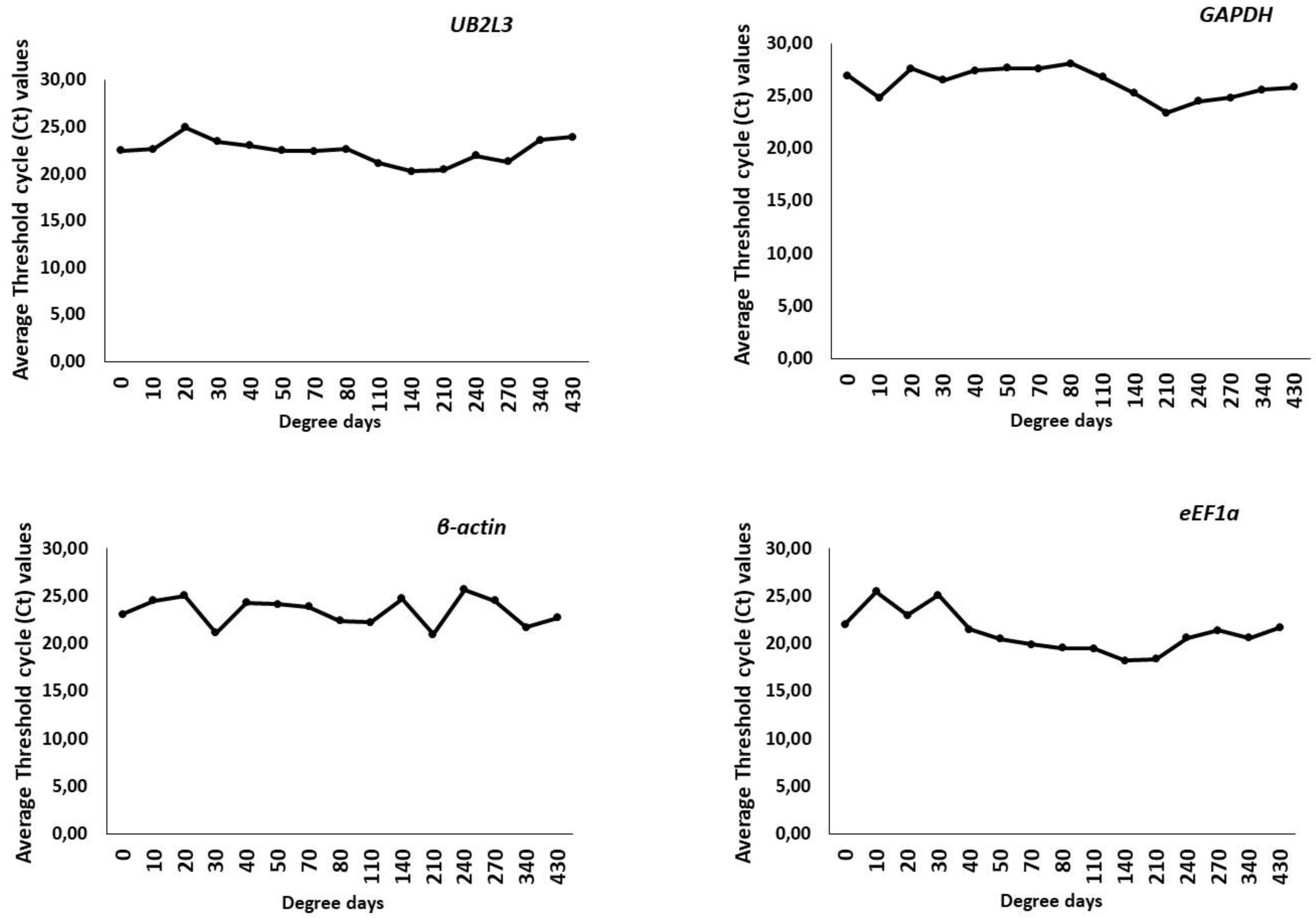
3.3.1. Expression Level of Selected Candidate Reference Genes by Cycle Threshold (Ct) Value Analysis
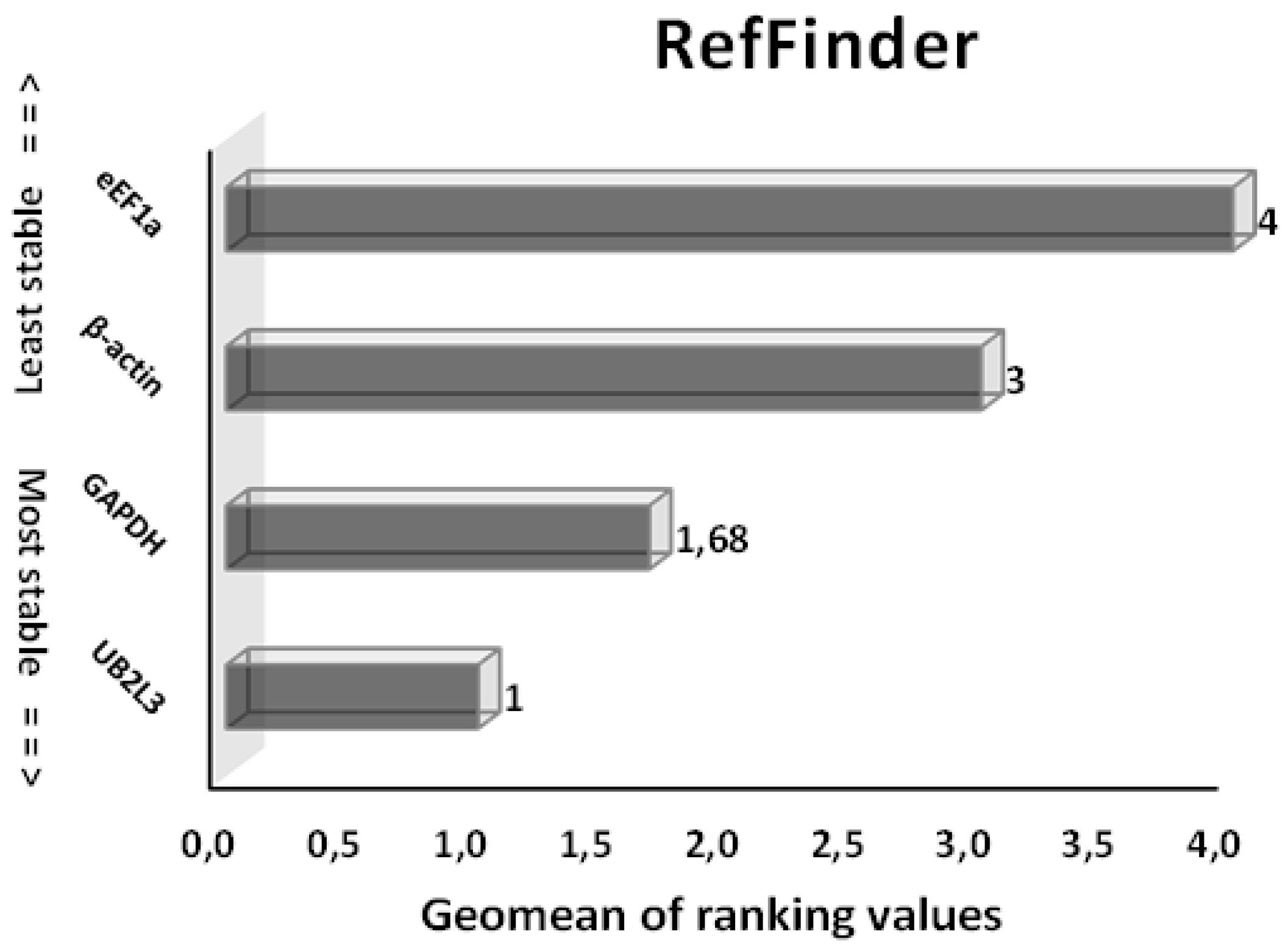
3.3.2. Gene Stability Analysis of HKGs
3.4. mRNA Expression of Genes Involved in PGC Development of S. salar Collected from Fertilization to Hatching Stages
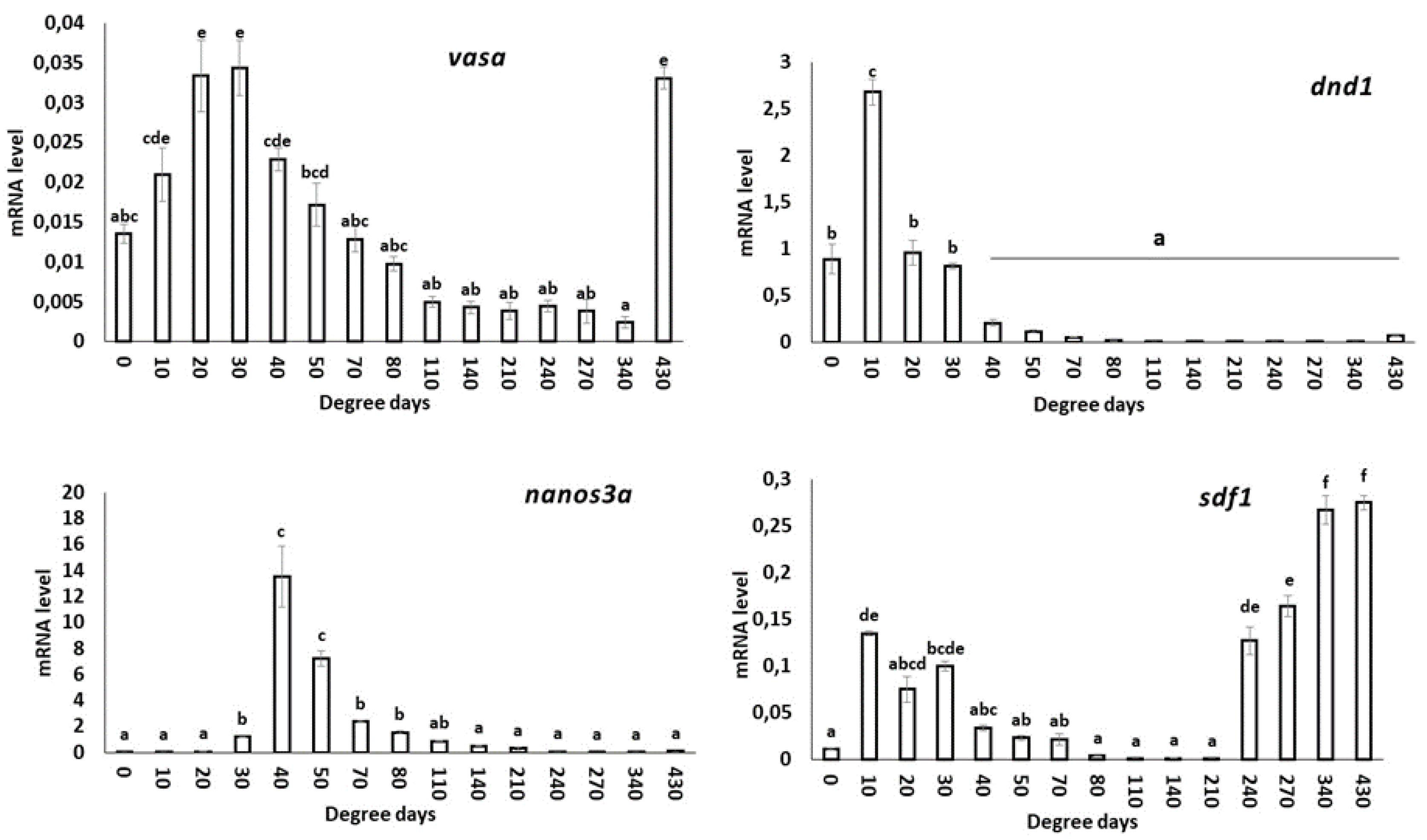
3.4.1. vasa
3.4.2. dnd1
3.4.3. nanos3a
3.4.4. sdf1
3.5. Expression of SD Genes in S. salar during Embryonic Developmental Stages
3.5.1. gsdf
3.5.2. amh
3.5.3. cyp19a
3.5.4. dmrt1
3.5.5. foxl2
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Claytor, R.R.; MacCrimmon, H.R. Partitioning Size from Morphometric Data: A Comparison of Five Statistical Procedures Used in Fisheries Stock Identification Research; Fisheries and Oceans: Ottawa, On, Canada, 1987. [Google Scholar]
- International Council for the Exploration of the Sea (ICES). Working Group on North Atlantic Salmon (WGNAS). ICES Scientific Reports. 2019. Available online: https://www.ices.dk/sites/pub/Publication%20Reports/Expert%20Group%20Report/Fisheries%20Resources%20Steering%20Group/2019/WGNAS/WGNAS_2019.pdf (accessed on 10 December 2020).
- Forseth, T.; Barlaup, B.T.; Finstad, B.; Fiske, P.; Gjøsæter, H.; Falkegård, M.; Hindar, A.; Mo, T.A.; Rikardsen, A.H.; Thorstad, E.B.; et al. The major threats to Atlantic salmon in Norway. ICES J. Mar. Sci. 2017, 74, 1496–1513. [Google Scholar] [CrossRef]
- Lorentzen, T. Modeling climate change and the effect on the Norwegian salmon farming industry. Nat. Resour. Model. 2008, 21, 416–435. [Google Scholar] [CrossRef]
- Bjarnason, A.; Magnusdottir, K.S. The Salmon Sea Fish Farming Industry in Iceland. A review. Fish Aqua J. 2021, 10, 2. [Google Scholar] [CrossRef]
- Thorstad, E.B.; Fleming, I.A.; McGinnity, P.; Soto, D.; Wennevik, V.; Whoriskey, F. Incidence and impacts of escaped farmed Atlantic salmon Salmo salar in nature. In NINA Special Report 36; NINA: Trondheim, Norway, 2008. [Google Scholar]
- Naylor, R.L.; Hindar, K.; Fleming, I.A.; Goldburg, R.J.; Williams, S.E.I.; Volpe, J.P.; Whoriskey, F.G.; Eagle, J.; Kelso, D.J.; Mangel, M. Fugitive salmon: Assessing the risks of escaped fish from net-pen aquaculture. BioScience 2005, 55, 427–437. [Google Scholar] [CrossRef]
- Johansen, L.H.; Jensen, I.; Mikkelsen, H.; Bjørn, P.A.; Jansen, P.A.; Bergh, Ø. Disease interaction and pathogens ex-change between wild and farmed fish populations with special reference to Norway. Aquaculture 2011, 315, 167–186. [Google Scholar] [CrossRef]
- NASCO. Guidelines on Containment of Farm Salmon. The Guidelines Are Attached as Annex 3 to the Williamsburg Resolution, Adopted at the Twentieth Annual Meeting of NASCO in June 2003. Available online: https://nasco.int/wpcontent/uploads/2020/09/slg_containmentguidelines.pdf (accessed on 13 December 2022).
- Bailey, J.L.; Eggereide, S.S. Indicating sustainable salmon farming: The case of the new Norwegian aquaculture management scheme. Mar. Policy 2020, 117, 103925. [Google Scholar] [CrossRef]
- Wong, A.C.; Van Eenennaam, A.L. Transgenic approaches for the reproductive containment of genetically engineered fish. Aquaculture 2008, 275, 1–12. [Google Scholar] [CrossRef]
- Devlin, R.H.; Donaldson, E.M. Containment of genetically altered fish with emphasis on salmonids. In Transgenic Fish; World Scientific press: Singapore, 1992; pp. 229–265. [Google Scholar]
- Ihssen, P.E.; McKay, L.R.; McMillan, I.; Phillips, R.B. Ploidy manipulation and gynogenesis in fishes: Cytogenetic and fisheries applications. Trans. Am. Fish. Soc. 1990, 119, 698–717. [Google Scholar] [CrossRef]
- McClure, C.A.; Hammell, K.L.; Moore, M.; Dohoo, I.R.; Burnley, H. Risk factors for early sexual maturation in Atlantic salmon in seawater farms in New Brunswick and Nova Scotia, Canada. Aquaculture 2007, 272, 370–379. [Google Scholar] [CrossRef]
- Jhingan, E.; Devlin, R.H.; Iwama, G.K. Disease resistance, stress response and effects of triploidy in growth hormone transgenic coho salmon. J. Fish Biol. 2003, 63, 806–823. [Google Scholar] [CrossRef]
- Sambraus, F.; Olsen, R.E.; Remen, M.; Hansen, T.J.; Torgersen, T.; Fjelldal, P.G. Water temperature and oxygen: The effect of triploidy on performance and metabolism in farmed Atlantic salmon (Salmo salar L.) post-smolts. Aquaculture 2017, 473, 1–12. [Google Scholar] [CrossRef]
- Wong, T.T.; Zohar, Y. Production of reproductively sterile fish by a non-transgenic gene silencing technology. Sci. Rep. 2015, 5, 15822. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, J.; Cui, X.; Luo, D.; Xia, H.; Dai, J.; Zhu, Z.; Hu, W. A controllable on-off strategy for the reproductive containment of fish. Sci. Rep. 2015, 5, 7614. [Google Scholar] [CrossRef] [PubMed]
- Wargelius, A.; Leininger, S.; Skaftnesmo, K.O.; Kleppe, L.; Andersson, E.; Taranger, G.L.; Schulz, R.W.; Edvardsen, R.B. Dnd1 knockout ablates germ cells and demonstrates germ cell independent sex differentiation in Atlantic salmon. Sci. Rep. 2016, 6, 21284. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.T.; Zohar, Y. Production of reproductively sterile fish: A mini-review of germ cell elimination technologies. Gen. Comp. Endocrinol. 2015, 221, 3–8. [Google Scholar] [CrossRef]
- Bonisławska, M.; Formicki, K.; Korzelecka-Orkisz, A.; Winnicki, A. Fish egg size variability: Biological significance. Differences 2001, 40, 55. [Google Scholar]
- Cirera, S. Highly efficient method for isolation of total RNA from adipose tissue. BMC Res. Notes 2013, 6, 472. [Google Scholar] [CrossRef]
- Ahi, E.P.; Guðbrandsson, J.; Kapralova, K.H.; Franzdóttir, S.R.; Snorrason, S.S.; Maier, V.H.; Jónsson, Z.O. Vali-dation of reference genes for expression studies during craniofacial development in arctic charr. PLoS ONE 2013, 8, e66389. [Google Scholar] [CrossRef]
- Köhsler, M.; Leitsch, D.; Müller, N.; Walochnik, J. Validation of reference genes for the normalization of RT-qPCR gene expression in Acanthamoeba spp. Sci. Rep. 2020, 10, 10362. [Google Scholar] [CrossRef]
- McCurley, A.T.; Callard, G.V. Characterization of housekeeping genes in zebrafish: Male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol. Biol. 2008, 9, 102. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Zakrajsek, B.A. Effect of experimental treatment on housekeeping gene expression: Validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods 2000, 46, 69–81. [Google Scholar] [CrossRef]
- Olsvik, P.A.; Lie, K.K.; Jordal, A.E.O.; Nilsen, T.O.; Hordvik, I. Evaluation of potential reference genes in real-time RT-PCR studies of Atlantic salmon. BMC Mol. Biol. 2005, 6, 21. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: M inimum I nformation for Publication of Quantitative Real-Time PCR E xperiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef]
- Jin, Y.H.; Davie, A.; Migaud, H. Expression pattern of nanos, piwil, dnd1, vasa and pum genes during ontogenic development in Nile tilapia Oreochromis niloticus. Gene 2019, 688, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Knaut, H.; Pelegri, F.; Bohmann, K.; Schwarz, H.; Nüsslein-Volhard, C. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J. Cell Biol. 2000, 149, 875–888. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, X.; Du, S.; Wang, Y.; Zhao, H.; Du, T.; Yu, J.; Wu, L.; Song, Z.; Liu, Q.; et al. Germline Specific Expression of a vasa Homologue Gene in the Viviparous Fish Black Rockfish (Sebastes schlegelii) and Functional Analysis of the vasa 3′ Untranslated Region. Front. Cell Dev. Biol. 2020, 8, 575788. [Google Scholar] [CrossRef]
- Kedde, M.; Strasser, M.J.; Boldajipour, B.; Vrielink, J.A.F.O.; Slanchev, K.; le Sage, C.; Nagel, R.; Voorhoeve, P.M.; van Duijse, J.; Ørom, U.A.; et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 2007, 131, 1273–1286. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, Q.; Xiao, Y.; Yang, Y.; Wang, Y.; Song, Z.; You, F.; An, H.; Xiao, Z.; Xu, S.; et al. The dnd RNA identifies germ cell origin and migration in olive flounder (Paralichthys olivaceus). BioMed Res. Int. 2015, 2015, 428591. [Google Scholar] [PubMed]
- Köprunner, M.; Thisse, C.; Thisse, B.; Raz, E. A zebrafish nanos-related gene is essential for the development of pri-mordial germ cells. Genes Dev. 2001, 15, 2877–2885. [Google Scholar] [CrossRef]
- Tsuda, M.; Sasaoka, Y.; Kiso, M.; Abe, K.; Haraguchi, S.; Kobayashi, S.; Saga, Y. Conserved role of nanos proteins in germ cell development. Science 2003, 301, 1239–1241. [Google Scholar] [CrossRef]
- Saga, Y. Function of Nanos2 in the male germ cell lineage in mice. Cell. Mol. Life Sci. 2010, 67, 3815–3822. [Google Scholar] [CrossRef]
- Han, K.; Chen, S.; Cai, M.; Jiang, Y.; Zhang, Z.; Wang, Y. Nanos3a not nanos1 and nanos2 is a germ cell marker gene in large yellow croaker during embryogenesis. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2018, 218, 13–22. [Google Scholar] [CrossRef]
- Saito, T.; Goto-Kazeto, R.; Kawakami, Y.; Nomura, K.; Tanaka, H.; Adachi, S.; Arai, K.; Yamaha, E. The mechanism for primordial germ-cell migration is conserved between Japanese eel and zebrafish. PLoS ONE 2011, 6, e24460. [Google Scholar] [CrossRef]
- Herpin, A.; Fischer, P.; Liedtke, D.; Kluever, N.; Neuner, C.; Raz, E.; Schartl, M. Sequential SDF1a and b-induced mobility guides Medaka PGC migration. Dev. Biol. 2008, 320, 319–327. [Google Scholar] [CrossRef]
- Guiguen, Y.; Fostier, A.; Piferrer, F.; Chang, C.F. Ovarian aromatase and estrogens: A pivotal role for gonadal sex differentiation and sex change in fish. Gen. Comp. Endocrinol. 2010, 165, 352–366. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.W.; Chung, B.C. Evolution, Expression, and Function of Gonadal Somatic Cell-Derived Factor. Front. Cell Dev. Biol. 2021, 9, 1600. [Google Scholar] [CrossRef]
- Bhat, I.A.; Dar, J.Y.; Ahmad, I.; Mir, I.N.; Bhat, H.; Bhat, R.A.; Ganie, P.A.; Sharma, R. Testicular development and sper-matogenesis in fish: Insights into molecular aspects and regulation of gene expression by different exogenous factors. Rev. Aquac. 2021, 13, 2142–2168. [Google Scholar] [CrossRef]
- Gunnes, K. Survival and development of Atlantic salmon eggs and fry at three different temperatures. Aquaculture 1979, 16, 211–218. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticu-locytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Trukhina, A.V.; Lukina, N.A.; Wackerow-Kouzova, N.D.; Smirnov, A.F. The variety of vertebrate mechanisms of sex determination. BioMed Res. Int. 2013, 2013, 587460. [Google Scholar] [CrossRef]
- Estermann, M.A.; Major, A.T.; Smith, C.A. Gonadal sex differentiation: Supporting versus steroidogenic cell lineage specification in mammals and birds. Front. Cell Dev. Biol. 2020, 8, 616387. [Google Scholar] [CrossRef]
- Cinalli, R.M.; Rangan, P.; Lehmann, R. Germ cells are forever. Cell 2008, 132, 559–562. [Google Scholar] [CrossRef]
- Robles, V.; Riesco, M.F.; Psenicka, M.; Saito, T.; Valcarce, D.G.; Cabrita, E.; Herraez, P. Biology of teleost primordial germ cells (PGCs) and spermatogonia: Biotechnological applications. Aquaculture 2017, 472, 4–20. [Google Scholar] [CrossRef]
- Kobayashi, T.; Kajiura-Kobayashi, H.; Guan, G.; Nagahama, Y. Sexual dimorphic expression of DMRT1 and Sox9a during gonadal differentiation and hormone-induced sex reversal in the teleost fish Nile tilapia (Oreochromis niloticus). Dev. -Ment. Dyn. Off. Publ. Am. Assoc. Anat. 2008, 237, 297–306. [Google Scholar]
- Li, M.; Yang, H.; Zhao, J.; Fang, L.; Shi, H.; Li, M.; Sun, Y.; Zhang, X.; Jiang, D.; Zhou, L.; et al. Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics 2014, 197, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Dai, S.; Wu, J.; Wei, X.; Zhou, X.; Chen, M.; Tan, D.; Pu, D.; Li, M.; Wang, D. Roles of anti-Müllerian hormone and its duplicates in sex determination and germ cell proliferation of Nile tilapia. Genetics 2022, 220, iyab237. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.L.; Medrano, J.F. Real-time PCR for mRNA quantitation. Biotechniques 2005, 39, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Pabinger, S.; Rödiger, S.; Kriegner, A.; Vierlinger, K.; Weinhäusel, A. A survey of tools for the analysis of quantitative PCR (qPCR) data. Biomol. Detect. Quantif. 2014, 1, 23–33. [Google Scholar] [CrossRef]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef] [PubMed]
- Szabo, A.; Perou, C.M.; Karaca, M.; Perreard, L.; Quackenbush, J.F.; Bernard, P.S. Statistical modeling for selecting housekeeper genes. Genome Biol. 2004, 5, R59. [Google Scholar] [CrossRef]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Asp. Med. 2006, 27, 126–139. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, F.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.M.; Mommens, M.; Hagen, Ø.; Babiak, I.; Solberg, C. Selection of suitable reference genes for real-time PCR studies of Atlantic halibut development. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2008, 150, 23–32. [Google Scholar] [CrossRef]
- Johnstone, K.A.; Lubieniecki, K.P.; Koop, B.F.; Davidson, W.S. Expression of olfactory receptors in different life stages and life histories of wild Atlantic salmon (Salmo salar). Mol. Ecol. 2011, 20, 4059–4069. [Google Scholar] [CrossRef] [PubMed]
- Lubieniecki, K.P.; Botwright, N.A.; Taylor, R.S.; Evans, B.S.; Cook, M.T.; Davidson, W.S. Expression analysis of sex-determining pathway genes during development in male and female Atlantic salmon (Salmo salar). Physiol. Genom. 2015, 47, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Løvoll, M.; Johnsen, H.; Boshra, H.; Bøgwald, J.; Sunyer, J.O.; Dalmo, R.A. The ontogeny and extrahepatic expression of complement factor C3 in Atlantic salmon (Salmo salar). Fish Shellfish. Immunol. 2007, 23, 542–552. [Google Scholar] [CrossRef]
- Kleppe, L.; Wargelius, A.; Johnsen, H.; Andersson, E.; Edvardsen, R.B. Gonad specific genes in Atlantic salmon (Salmon salar L.): Characterization of tdrd7-2, dazl-2, piwil1 and tdrd1 genes. Gene 2015, 560, 217–225. [Google Scholar] [CrossRef]
- Brubacher, J.L.; Secombes, C.J.; Zou, J.; Bols, N.C. Constitutive and LPS-induced gene expression in a macrophage-like cell line from the rainbow trout (Oncorhynchus mykiss). Dev. Comp. Immunol. 2000, 24, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Peddie, S.; Campos-Pérez, J.J.; Zou, J.; Secombes, C.J. The effect of intraperitoneally administered recom-binant IL-1β on immune parameters and resistance to Aeromonas salmonicida in the rainbow trout (Oncorhynchus mykiss). Dev. Comp. Immunol. 2003, 27, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Robertsen, B.; Bergan, V.; Røkenes, T.; Larsen, R.; Albuquerque, A. Atlantic salmon interferon genes: Cloning, sequence analysis, expression, and biological activity. J. Interferon Cytokine Res. 2003, 23, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Acosta, F.; Lockhart, K.; Gahlawat, S.K.; Real, F.; Ellis, A.E. Mx expression in Atlantic salmon (Salmo salar L.) parr in response to Listonella anguillarum bacterin, lipopolysaccharide and chromosomal DNA. Fish Shellfish. Immunol. 2004, 17, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Takle, H.; Baeverfjord, G.; Lunde, M.; Kolstad, K.; Andersen, Ø. The effect of heat and cold exposure on HSP70 ex-pression and development of deformities during embryogenesis of Atlantic salmon (Salmo salar). Aquaculture 2005, 249, 515–524. [Google Scholar] [CrossRef]
- Takle, H.; McLeod, A.; Andersen, O. Cloning and characterization of the executioner caspases 3, 6, 7 and Hsp70 in hyperthermic Atlantic salmon (Salmo salar) embryos. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2006, 144, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Julin, K.; Johansen, L.H.; Sommer, A.I. Reference genes evaluated for use in infectious pancreatic necrosis virus re-al-time RT-qPCR assay applied during different stages of an infection. J. Virol. Methods 2009, 162, 30–39. [Google Scholar] [CrossRef]
- Fodnes, J.O.; Kvaal, G. Variance in Relative Gene Expression RT-qPCR Analysis of Atlantic Salmon-before, during and after Delousing. Bachelor’s Thesis, Norwegian University of Science and Technology, Faculty of Natural Sciences, Trondheim Ålesund Gjøvik, Norway, 2020. [Google Scholar]
- Nagasawa, K.; Fernandes, J.M.; Yoshizaki, G.; Miwa, M.; Babiak, I. Identification and migration of primordial germ cells in Atlantic salmon, Salmo salar: Characterization of vasa, dead end, and lymphocyte antigen 75 genes. Mol. Reprod. Dev. 2013, 80, 118–131. [Google Scholar] [CrossRef]
- Du, S.; Zhou, L.; Wang, X.; Xu, S.; Li, J.; Song, Z.; Liu, Q. Characterization of vasa and dnd1 homologs in summer flounder, Paralichthys dentatus: Expression analysis and colocalization of PGCs during embryogenesis. Theriogenology 2022, 181, 180–189. [Google Scholar] [CrossRef]
- Weidinger, G.; Stebler, J.; Slanchev, K.; Dumstrei, K.; Wise, C.; Lovell-Badge, R.; Thisse, C.; Thisse, B.; Raz, E. Dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr. Biol. 2003, 13, 1429–1434. [Google Scholar] [CrossRef]
- Giraldez, A.J.; Mishima, Y.; Rihel, J.; Grocock, R.J.; Van Dongen, S.; Inoue, K.; Enright, A.J.; Schier, A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 2006, 312, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Stark, A.; Brennecke, J.; Cohen, S.M. Temporal reciprocity of miRNAs and their targets during the ma-ternal-to-zygotic transition in Drosophila. Curr. Biol. 2008, 18, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Liu, M.; Hartley, R.S.; Sheets, M.D.; Dahlberg, J.E. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. Rna 2009, 15, 2351–2363. [Google Scholar] [CrossRef] [PubMed]
- Kass, S.U.; Pruss, D.; Wolffe, A.P. How does DNA methylation repress transcription? Trends Genet. 1997, 13, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Pacchiarini, T.; Cross, I.; Leite, R.B.; Gavaia, P.; Ortiz-Delgado, J.B.; Pousão-Ferreira, P.; Rebordinos, L.; Sarasquete, C.; Cabrita, E. Solea senega-lensis vasa transcripts: Molecular characterisation, tissue distribution and developmental expression profiles. Reprod. Fertil. Dev. 2013, 25, 646–660. [Google Scholar] [CrossRef]
- Ricci, J.M.; Martinez, E.R.; Butzge, A.J.; Doretto, L.B.; Oliveira, M.A.; Bombardelli, R.A.; Bogerd, J.; Nóbrega, R.H. Char-acterization of vasa homolog in a neotropical catfish, Jundiá (Rhamdia quelen): Molecular cloning and expression analysis during embryonic and larval development. Gene 2018, 654, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Wang, T.; Zhao, C.; Wang, D.; Zhang, X.; Zhang, H.; Chi, M.; Yin, S.; Jia, Y. Evolutionary conservation and divergence of Vasa, Dazl and Nanos1 during embryogenesis and gametogenesis in dark sleeper (Odontobutis potamophila). Gene 2018, 672, 21–33. [Google Scholar] [CrossRef]
- Presslauer, C.; Nagasawa, K.; Fernandes, J.M.O.; Babiak, I. Expression of vasa and nanos3a during primordial germ cell formation and migration in Atlantic cod (Gadus morhua L.). Theriogenology 2012, 78, 1262–1277. [Google Scholar] [CrossRef]
- Svetic, V.; Hollway, G.E.; Elworthy, S.; Chipperfield, T.R.; Davison, C.; Adams, R.J.; Eisen, J.S.; Ingham, P.W.; Currie, P.D.; Kelsh, R.N. Sdf1a patterns zebrafish melanophores and links the somite and melanophore pattern defects in choker mutants. Development 2007, 134, 1011–1022. [Google Scholar] [CrossRef]
- Chong, S.W.; Nguyet, L.M.; Jiang, Y.J.; Korzh, V. The chemokine Sdf-1 and its receptor Cxcr4 are required for for-mation of muscle in zebrafish. BMC Dev. Biol. 2007, 7, 54. [Google Scholar] [CrossRef] [PubMed]
- Hayes, T.B. Sex determination and primary sex differentiation in amphibian: Genetic and developmental mechanisms. J. Exp. Zool. 1988, 281, 373–399. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, G.; Shao, C.; Huang, Q.; Liu, G.; Zhang, P.; Song, W.; An, N.; Chalopin, D.; Volff, J.-N.; et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014, 46, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Nagahama, Y.; Chakraborty, T.; Paul-Prasanth, B.; Ohta, K.; Nakamura, M. Sex determination, gonadal sex differen-tiation, and plasticity in vertebrate species. Physiol. Rev. 2021, 101, 1237–1308. [Google Scholar] [CrossRef]
- Heule, C.; Salzburger, W.; Böhne, A. Genetics of sexual development: An evolutionary playground for fish. Genetics 2014, 196, 579–591. [Google Scholar] [CrossRef]
- Martínez, P.; Viñas, A.M.; Sánchez, L.; Díaz, N.; Ribas, L.; Piferrer, F. Genetic architecture of sex determination in fish: Applications to sex ratio control in aquaculture. Front. Genet. 2014, 5, 340. [Google Scholar] [CrossRef] [PubMed]
- Horie, Y.; Myosho, T.; Sato, T.; Sakaizumi, M.; Hamaguchi, S.; Kobayashi, T. Androgen induces gonadal soma-derived factor, Gsdf, in XX gonads correlated to sex-reversal but not Dmrt1 directly, in the teleost fish, northern medaka (Oryzias sakaizumii). Mol. Cell. Endocrinol. 2016, 436, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, T.; Zhou, L.Y.; Chaudhari, A.; Iguchi, T.; Nagahama, Y. Dmy initiates masculinity by altering Gsdf/Sox9a2/Rspo1 expression in medaka (Oryzias latipes). Sci. Rep. 2016, 6, 19480. [Google Scholar] [CrossRef] [PubMed]
- Sawatari, E.; Shikina, S.; Takeuchi, T.; Yoshizaki, G. A novel transforming growth factor-β superfamily member ex-pressed in gonadal somatic cells enhances primordial germ cell and spermatogonial proliferation in rainbow trout (Oncorhynchus mykiss). Dev. Biol. 2007, 301, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Shibata, Y.; Paul-Prasanth, B.; Suzuki, A.; Usami, T.; Nakamoto, M.; Matsuda, M.; Nagahama, Y. Expression of gonadal soma derived factor (GSDF) is spatially and temporally correlated with early testicular differentiation in medaka. Gene Expr. Patterns 2010, 10, 283–289. [Google Scholar] [CrossRef]
- Yan, Y.L.; Batzel, P.; Titus, T.; Sydes, J.; Desvignes, T.; Bremiller, R.; Draper, B.; Postlethwait, J.H. The roles of Amh in zebrafish gonad development and sex determination. BioRxiv 2019, 650218. [Google Scholar]
- Gautier, A.; Sohm, F.; Joly, J.S.; Le Gac, F.; Lareyre, J.J. The proximal promoter region of the zebrafish gsdfgene is sufficient to mimic the spatio-temporal expression pattern of the endogenous gene in Sertoli and granulosa cells. Biol. Reprod. 2011, 85, 1240–1251. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Kobayashi, D.; Aoki, Y.; Yokoi, H.; Ebe, Y.; Wittbrodt, J.; Tanaka, M. Identification and lineage tracing of two populations of somatic gonadal precursors in medaka embryos. Dev. Biol. 2006, 295, 678–688. [Google Scholar] [CrossRef]
- Klüver, N.; Pfennig, F.; Pala, I.; Storch, K.; Schlieder, M.; Froschauer, A.; Gutzeit, H.O.; Schartl, M. Differential expression of anti-Müllerian hormone (amh) and anti-Müllerian hormone receptor type II (amhrII) in the teleost medaka. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2007, 236, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Simpson, E.R.; Mahendroo, M.S.; Means, G.D.; Kilgore, M.W.; Hinshelwood, M.M.; Graham-Lorence, S.; Amarneh, B.; Ito, Y.; Fisher, C.R.; Michael, M.D.; et al. Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr. Rev. 1994, 15, 342–355. [Google Scholar]
- Kwon, J.Y.; Haghpanah, V.; Kogson-Hurtado, L.M.; McAndrew, B.J.; Penman, D.J. Masculinization of genetic female Nile tilapia (Oreochromis niloticus) by dietary administration of an aromatase inhibitor during sexual differentiation. J. Exp. Zool. 2000, 287, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Lau, E.S.W.; Zhang, Z.; Qin, M.; Ge, W. Knockout of zebrafish ovarian aromatase gene (Cyp19a1a) by TALEN and CRISPR/Cas9 leads to all-male offspring due to failed ovarian differentiation. Sci. Rep. 2016, 6, 37357. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Willett, K.L. Local expression of CYP19A1 and CYP19A2 in developing and adult killifish (Fundulus heteroclitus). Gen. Comp. Endocrinol. 2008, 155, 307–317. [Google Scholar] [CrossRef][Green Version]
- Coumailleau, P.; Pellegrini, E.; Adrio, F.; Diotel, N.; Cano-Nicolau, J.; Nasri, A.; Vaillant, C.; Kah, O. Aromatase, estrogen receptors and brain development in fish and amphibians. Biochim. Et Biophys. Acta (BBA)-Gene Regul. Mech. 2015, 1849, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.S.; Evans, B.S.; Afonso, L.O. Developmental changes in gene expression and gonad morphology during sex differentiation in Atlantic salmon (Salmo salar). Gene 2022, 823, 146393. [Google Scholar] [CrossRef]
- Otake, H.; Shinomiya, A.; Matsuda, M.; Hamaguchi, S.; Sakaizumi, M. Wild-derived XY sex-reversal mutants in the medaka, Oryzias latipes. Genetics 2006, 173, 2083–2090. [Google Scholar] [CrossRef]
- Herpin, A.; Schartl, M. Dmrt1 genes at the crossroads: A widespread and central class of sexual development factors in fish. FEBS J. 2011, 278, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.C.; Carpenter, C.; Nebert, D.W.; Vasiliou, V. Update of human and mouse forkhead box (FOX) gene families. Hum. Genom. 2010, 4, 345. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, M.; Ma, H.; Liu, X.; Shi, H.; Li, M.; Wang, D. Mutation of Foxl2 or Cyp19a1a results in female to male sex reversal in XX Nile tilapia. Endocrinology 2017, 158, 2634–2647. [Google Scholar] [CrossRef]
- Lin, L.; Hongwei, L.; Zhong, L. Cloning and expression analysis of DMRT1 gene in Pelteobagrus fulvidraco. J. Huazhong Agric. University 2012, 31, 220–226. [Google Scholar]
- Jin-Hua, W.A.N.G.; Liang, M.I.A.O.; Ming-Yun, L.I.; Xiao-Fei, G.U.O.; Na, P.A.N.; Ying-Ying, C.H.E.N.; Liang, Z.H.A.O. Cloning the Dmrt1 and DmrtA2 genes of ayu (Plecoglossus altivelis) and mapping their expression in adult, larval, and embryonic stages. Zool. Res. 2014, 35, 99. [Google Scholar]
- Herpin, A.; Schindler, D.; Kraiss, A.; Hornung, U.; Winkler, C.; Schartl, M. Inhibition of primordial germ cell prolif-eration by the medaka male determining gene Dmrt1bY. BMC Dev. Biol. 2007, 7, 99. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Y.; Zhong, Y.; Guo, J.; Lu, M.; Gui, L.; Li, M. Molecular identification and expression analysis of Foxl2 and sox9b in Oryzias celebensis. Aquac. Fish. 2021, 6, 471–478. [Google Scholar] [CrossRef]



| Day\Days Post-Fertilization | Degree Days (d°C) | Pooled Number of Eggs/Embryos/Larvae per Sample |
|---|---|---|
| 0 | unfertilized egg | 10 |
| 1 | 10 | 10 |
| 2 | 20 | 10 |
| 3 | 30 | 10 |
| 4 | 40 | 10 |
| 5 | 50 | 6 |
| 7 | 70 | 6 |
| 8 | 80 | 6 |
| 11 | 110 | 6 |
| 14 | 140 | 6 |
| 21 | 210 | 6 |
| 24 | 240 | 6 |
| 27 | 270 | 6 |
| 34 | 340 | 6 |
| 43 | 430 (yolk sac fry or alevins) | 3 |
| Gene | Forward Primer Sequence (5′ to 3′) | Reverse Primer Sequence (5′ to 3′) | Accession Number and Reference |
|---|---|---|---|
| vasa | CGCTCCCTGGTCAAAGTCCTGTC | GCTAGTTGACTCGCCCCATCTCTC | JN712912 |
| dnd1 | TCTGTACAGGGCCTGATGGT | TAAAACAAAGTAGGGGATCTGTG | [19]. |
| nanos3a | ATGGAGTCCGAAAACAAGAGT | CGGTTCTGGGGTGAACTTGC | KC237283 |
| sdf1 | GTGTTGGTCCTACTGGCTGTGGC | GAGGGACGGTGTTGAGAGTGGAGC | NM_001140787 |
| gsdf | GACAAAGCAGTGGCTGTACC | GGCAGCATTTCAGACCACTA | XM_014138924 |
| amh | CAGTCACTCTCTGCAGCCTTACAA | CAACATTGAATCTCCATTTCAGTTTAC | NM_001123585 |
| cyp19a | TCAAACAGAACCCTGACGTAG | GCTCCCTTTCACCTATAGCAGTGT | AF436885 |
| dmrt1 | AGGAGGAGGAGATGGGGCTCTGTA | CCAGCAGAGGTGTTTCCACAGGTAG | XM_014172771 |
| foxl2 | GCGGTGATGGGTACGGCTACCTG | GACGGGACTCACGTTGCCACTGG | JX184084 |
| β-actin | CCAAAGCCAACAGGGAGAAG | AGGGACAACACTGCCTGGAT | BG933897 |
| GAPDH | AAGTGAAGCAGGAGGGTGGAA | CAGCCTCACCCCATTTGATG | AM230811 |
| eEf1a | CCCCTCCAGGACGTTTACAAA | CACACGGCCCACAGGTACA | BT058711 |
| UB2L3 | CGAGAAGGGACAGGTGTGTC | ACCAACGCAATCAGGGACT | NM_001141284 |
| UB2L3 | eEF1a | β-actin | GAPDH | |
|---|---|---|---|---|
| n * | 15 | 15 | 15 | 15 |
| geo Mean [CP] | 21.90 | 20.57 | 22.88 | 25.56 |
| AR Mean [CP] | 21.93 | 20.67 | 22.93 | 25.60 |
| min [CP] | 20.00 | 18.00 | 20.00 | 23.00 |
| max [CP] | 24.00 | 25.00 | 25.00 | 28.00 |
| std dev [±CP] | 0.89 | 1.64 | 1.28 | 1.23 |
| CV [% CP] | 4.05 | 7.96 | 5.58 | 4.79 |
| min [x-fold] | −3.74 | −5.92 | −7.38 | −5.90 |
| max [x-fold] | 4.27 | 21.61 | 4.33 | 5.42 |
| std dev [±x-fold] | 1.85 | 3.13 | 2.43 | 2.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhat, I.A.; Dubiel, M.M.; Rodriguez, E.; Jónsson, Z.O. Insights into Early Ontogenesis of Salmo salar: RNA Extraction, Housekeeping Gene Validation and Transcriptional Expression of Important Primordial Germ Cell and Sex-Determination Genes. Animals 2023, 13, 1094. https://doi.org/10.3390/ani13061094
Bhat IA, Dubiel MM, Rodriguez E, Jónsson ZO. Insights into Early Ontogenesis of Salmo salar: RNA Extraction, Housekeeping Gene Validation and Transcriptional Expression of Important Primordial Germ Cell and Sex-Determination Genes. Animals. 2023; 13(6):1094. https://doi.org/10.3390/ani13061094
Chicago/Turabian StyleBhat, Irfan Ahmad, Milena Malgorzata Dubiel, Eduardo Rodriguez, and Zophonías Oddur Jónsson. 2023. "Insights into Early Ontogenesis of Salmo salar: RNA Extraction, Housekeeping Gene Validation and Transcriptional Expression of Important Primordial Germ Cell and Sex-Determination Genes" Animals 13, no. 6: 1094. https://doi.org/10.3390/ani13061094
APA StyleBhat, I. A., Dubiel, M. M., Rodriguez, E., & Jónsson, Z. O. (2023). Insights into Early Ontogenesis of Salmo salar: RNA Extraction, Housekeeping Gene Validation and Transcriptional Expression of Important Primordial Germ Cell and Sex-Determination Genes. Animals, 13(6), 1094. https://doi.org/10.3390/ani13061094





