Phylogenetic and Spatiotemporal Analyses of Porcine Epidemic Diarrhea Virus in Guangxi, China during 2017–2022
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Detection of PEDVs in Guangxi, China between 2017 and 2022
2.2. Sequencing of the Partial S1 Genes
2.3. Sequence Alignment and Phylogenetic Analyses
2.4. Bayesian Temporal Dynamics Analysis
2.5. Bayesian Phylogeographic Analysis
2.6. Genome Recombination Events Analysis
3. Results
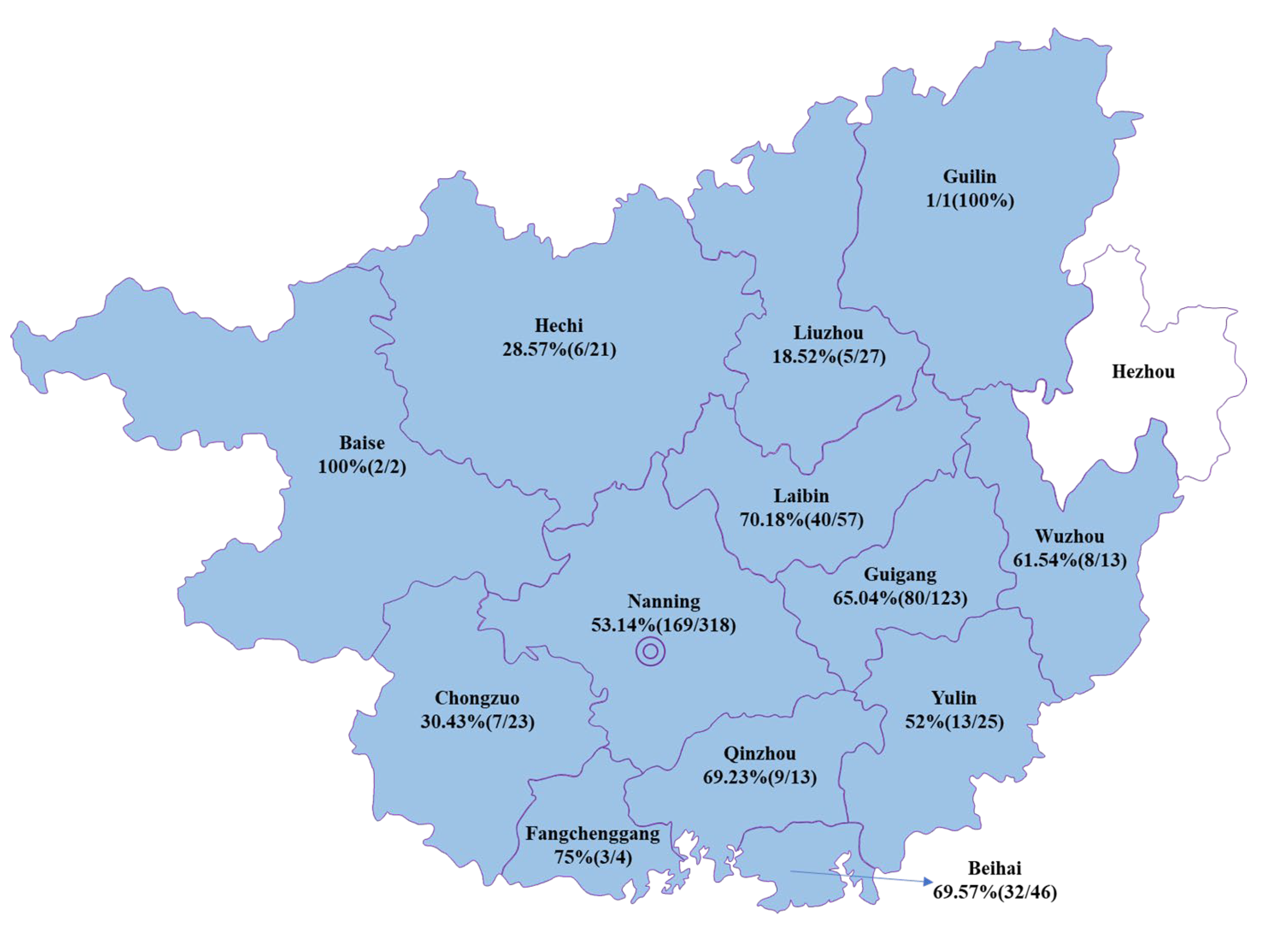
3.1. Prevalence of PEDV in Clinical Samples from Diarrheal Pigs
3.2. Phylogenetic Analysis of the PEDV Partial S1 Gene
3.3. Bayesian Temporal Dynamics Analysis
3.4. Bayesian Phylogeographic Analysis
3.5. Deduence Analysis of the Partial S1 Protein
3.6. Recombination Analysis of the PEDV Partial S1 Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, P.H.; Li, Y.Q.; Pan, Y.Q.; Guo, Y.Y.; Guo, F.; Shi, R.Z.; Xing, L. The spike glycoprotein genes of porcine epidemic diarrhea viruses isolated in China. Vet. Res. 2021, 52, 87. [Google Scholar] [CrossRef] [PubMed]
- Dee, S.; Neill, C.; Singrey, A.; Clement, T.; Cochrane, R.; Jones, C.; Patterson, G.; Spronk, G.; Christopher-Hennings, J.; Nelson, E. Modeling the transboundary risk of feed ingredients contaminated with porcine epidemic diarrhea virus. BMC Vet. Res. 2016, 12, 51. [Google Scholar] [CrossRef] [PubMed]
- Muckey, M.B.; Jones, C.K.; Woodworth, J.C.; Paulk, C.B.; Dritz, S.S.; Gebhardt, J.T. Using environmental sampling to evaluate the effectiveness of decontamination methods to reduce detection of porcine epidemic diarrhea virus RNA on feed manufacturing surfaces. Transl. Anim. Sci. 2021, 5, txab121. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.M.; Saif, L.J.; Marthaler, D.; Wang, Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016, 226, 20–39. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.; Lee, S.; Lee, C. Assessing the risk of recurrence of porcine epidemic diarrhea virus in affected farms on Jeju Island, South Korea. J. Vet. Sci. 2021, 22, e48. [Google Scholar] [CrossRef]
- Pensaert, M.B.; de Bouck, P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978, 58, 243–247. [Google Scholar] [CrossRef]
- Huang, B.; Gu, G.; Zhang, Y.; Chen, Z.; Tian, K. Molecular and Structural Evolution of Porcine Epidemic Diarrhea Virus. Animals 2022, 12, 3388. [Google Scholar] [CrossRef]
- Hu, X.; Lian, Y.; He, Y.; Liu, X.; Tian, Z.; Dai, Y.; Liu, M.; Fan, H.; Shi, Y.; Cong, F. Molecular Characterization and Phylogenetic Analysis of a Variant Recombinant Porcine Epidemic Diarrhea Virus Strain in China. Animals 2022, 12, 2189. [Google Scholar] [CrossRef]
- Yu, J.; Chai, X.; Cheng, Y.; Xing, G.; Liao, A.; Du, L.; Wang, Y.; Lei, J.; Gu, J.; Zhou, J. Molecular characteristics of the spike gene of porcine epidemic diarrhoea virus strains in Eastern China in 2016. Virus Res. 2018, 247, 47–54. [Google Scholar] [CrossRef]
- Chen, N.; Li, S.; Zhou, R.; Zhu, M.; He, S.; Ye, M.; Huang, Y.; Li, S.; Zhu, C.; Xia, P.; et al. Two novel porcine epidemic diarrhea virus (PEDV) recombinants from a natural recombinant and distinct subtypes of PEDV variants. Virus Res. 2017, 242, 90–95. [Google Scholar] [CrossRef]
- Jang, G.; Park, J.; Lee, C. Successful Eradication of Porcine Epidemic Diarrhea in an Enzootically Infected Farm: A Two-Year Follow-Up Study. Pathogens 2021, 10, 830. [Google Scholar] [CrossRef]
- Garcia-Hernandez, M.E.; Trujillo-Ortega, M.E.; Alcaraz-Estrada, S.L.; Lozano-Aguirre-Beltran, L.; Sandoval-Jaime, C.; Taboada-Ramirez, B.I.; Sarmiento-Silva, R.E. Molecular Detection and Characterization of Porcine Epidemic Diarrhea Virus and Porcine Aichivirus C Coinfection in Mexico. Viruses 2021, 13, 738. [Google Scholar] [CrossRef]
- Duarte, M.; Gelfi, J.; Lambert, P.; Rasschaert, D.; Laude, H. Genome organization of porcine epidemic diarrhoea virus. Adv. Exp. Med. Biol. 1993, 342, 55–60. [Google Scholar] [CrossRef]
- Lu, Y.; Huang, W.; Zhong, L.; Qin, Y.; Liu, X.; Yang, C.; Wang, R.; Su, X.; Du, C.; Mi, X.; et al. Comparative Characterization and Pathogenicity of a Novel Porcine Epidemic Diarrhea Virus (PEDV) with a Naturally Occurring Truncated ORF3 Gene Coinfected with PEDVs Possessing an Intact ORF3 Gene in Piglets. Viruses 2021, 13, 1562. [Google Scholar] [CrossRef]
- Salamat, S.E.A.; Collantes, T.M.A.; Lumbera, W.M.L.; Tablizo, F.A.; Mutia, C.T.M.; Ong, J.D.P.; Bandoy, D. Sequence analysis of new variants of porcine epidemic diarrhea virus in Luzon, Philippines, in 2017. Arch. Virol. 2021, 166, 1859–1867. [Google Scholar] [CrossRef]
- Sato, T.; Takeyama, N.; Katsumata, A.; Tuchiya, K.; Kodama, T.; Kusanagi, K. Mutations in the spike gene of porcine epidemic diarrhea virus associated with growth adaptation in vitro and attenuation of virulence in vivo. Virus Genes. 2011, 43, 72–78. [Google Scholar] [CrossRef]
- Lee, D.K.; Park, C.K.; Kim, S.H.; Lee, C. Heterogeneity in spike protein genes of porcine epidemic diarrhea viruses isolated in Korea. Virus Res. 2010, 149, 175–182. [Google Scholar] [CrossRef]
- Chen, P.; Wang, K.; Hou, Y.; Li, H.; Li, X.; Yu, L.; Jiang, Y.; Gao, F.; Tong, W.; Yu, H.; et al. Genetic evolution analysis and pathogenicity assessment of porcine epidemic diarrhea virus strains circulating in part of China during 2011–2017. Infect. Genet. Evol. 2019, 69, 153–165. [Google Scholar] [CrossRef]
- Ji, Z.; Shi, D.; Shi, H.; Wang, X.; Chen, J.; Liu, J.; Ye, D.; Jing, Z.; Liu, Q.; Fan, Q.; et al. A porcine epidemic diarrhea virus strain with distinct characteristics of four amino acid insertion in the COE region of spike protein. Vet. Microbiol. 2021, 253, 108955. [Google Scholar] [CrossRef]
- Chang, S.H.; Bae, J.L.; Kang, T.J.; Kim, J.; Chung, G.H.; Lim, C.W.; Laude, H.; Yang, M.S.; Jang, Y.S. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells 2002, 14, 295–299. [Google Scholar]
- Sun, D.; Feng, L.; Shi, H.; Chen, J.; Cui, X.; Chen, H.; Liu, S.; Tong, Y.; Wang, Y.; Tong, G. Identification of two novel B cell epitopes on porcine epidemic diarrhea virus spike protein. Vet. Microbiol. 2008, 131, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Cruz, D.J.; Kim, C.J.; Shin, H.J. Phage-displayed peptides having antigenic similarities with porcine epidemic diarrhea virus (PEDV) neutralizing epitopes. Virology 2006, 354, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Gerber, P.F.; Gong, Q.; Huang, Y.W.; Wang, C.; Holtkamp, D.; Opriessnig, T. Detection of antibodies against porcine epidemic diarrhea virus in serum and colostrum by indirect ELISA. Vet. J. 2014, 202, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Su, X.; Du, C.; Mo, L.; Ke, P.; Wang, R.; Zhong, L.; Yang, C.; Chen, Y.; Wei, Z.; et al. Genetic Diversity of Porcine Epidemic Diarrhea Virus With a Naturally Occurring Truncated ORF3 Gene Found in Guangxi, China. Front. Vet. Sci. 2020, 7, 435. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Mi, X.; Yang, C.; Lu, Y.; Wang, H.; Qin, Q.; Chen, R.; Chen, Z.; Luo, Y.; Chen, Y.; Wei, Z.; et al. Isolation, Identification, and Evaluation of the Pathogenicity of a Porcine Enterovirus G Isolated From China. Front. Vet. Sci. 2021, 8, 712679. [Google Scholar] [CrossRef]
- Chen, L.; Song, J.; Liu, H.; Cai, J.; Lin, Q.; Xu, C.; Ding, C.; Liao, M.; Ren, T.; Xiang, B. Phylodynamic analyses of class I Newcastle disease virus isolated in China. Transbound. Emerg. Dis. 2021, 68, 1294–1304. [Google Scholar] [CrossRef]
- Tsai, C.C.; Chou, C.H.; Wang, H.V.; Ko, Y.Z.; Chiang, T.Y.; Chiang, Y.C. Biogeography of the Phalaenopsis amabilis species complex inferred from nuclear and plastid DNAs. BMC Plant Biol. 2015, 15, 202. [Google Scholar] [CrossRef]
- Fan, W.; Chen, J.; Zhang, Y.; Deng, Q.; Wei, L.; Zhao, C.; Lv, D.; Lin, L.; Zhang, B.; Wei, T.; et al. Phylogenetic and Spatiotemporal Analyses of the Complete Genome Sequences of Avian Coronavirus Infectious Bronchitis Virus in China During 1985–2020: Revealing Coexistence of Multiple Transmission Chains and the Origin of LX4-Type Virus. Front. Microbiol. 2022, 13, 693196. [Google Scholar] [CrossRef]
- Siah, A.; Breyta, R.B.; Warheit, K.I.; Gagne, N.; Purcell, M.K.; Morrison, D.; Powell, J.F.F.; Johnson, S.C. Genomes reveal genetic diversity of Piscine orthoreovirus in farmed and free-ranging salmonids from Canada and USA. Virus Evol. 2020, 6, veaa054. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, H.; Zhao, J.; Zhong, Q.; Jin, J.H.; Zhang, G.Z. Evolution of infectious bronchitis virus in China over the past two decades. J. Gen. Virol. 2016, 97, 1566–1574. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Choi, G.K.Y.; Huang, Y.; Sivakumar, S.; Tsoi, H.W.; Yip, C.C.Y.; Jose, S.V.; Bai, R.; Wong, E.Y.M.; et al. Molecular epidemiology of canine picornavirus in Hong Kong and Dubai and proposal of a novel genus in Picornaviridae. Infect. Genet. Evol. 2016, 41, 191–200. [Google Scholar] [CrossRef]
- Zhang, C.; Ogilvie, H.A.; Drummond, A.J.; Stadler, T. Bayesian Inference of Species Networks from Multilocus Sequence Data. Mol. Biol. Evol. 2018, 35, 504–517. [Google Scholar] [CrossRef]
- Wang, K.; Hu, Z.; Fan, M.; Shao, Z.; Yu, Q.; Li, X. Development of an indirect ELISA to detect PEDV specific IgA antibody based on a PEDV epidemic strain. BMC Vet. Res. 2022, 18, 319. [Google Scholar] [CrossRef]
- Bielejec, F.; Baele, G.; Vrancken, B.; Suchard, M.A.; Rambaut, A.; Lemey, P. SpreaD3: Interactive Visualization of Spatiotemporal History and Trait Evolutionary Processes. Mol. Biol. Evol. 2016, 33, 2167–2169. [Google Scholar] [CrossRef]
- Shi, W.; Hao, H.; Li, M.; Niu, J.; Hu, Y.; Zhao, X.; Li, Q. Expression and Purification of a PEDV-Neutralizing Antibody and Its Functional Verification. Viruses 2021, 13, 472. [Google Scholar] [CrossRef]
- Beall, A.; Yount, B.; Lin, C.M.; Hou, Y.; Wang, Q.; Saif, L.; Baric, R. Characterization of a Pathogenic Full-Length cDNA Clone and Transmission Model for Porcine Epidemic Diarrhea Virus Strain PC22A. mBio 2016, 7, e01451-15. [Google Scholar] [CrossRef]
- Niu, X.; Wang, Q. Prevention and Control of Porcine Epidemic Diarrhea: The Development of Recombination-Resistant Live Attenuated Vaccines. Viruses 2022, 14, 1317. [Google Scholar] [CrossRef]
- Fan, B.; Jiao, D.; Zhao, X.; Pang, F.; Xiao, Q.; Yu, Z.; Mao, A.; Guo, R.; Yuan, W.; Zhao, P.; et al. Characterization of Chinese Porcine Epidemic Diarrhea Virus with Novel Insertions and Deletions in Genome. Sci. Rep. 2017, 7, 44209. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Gao, H.; Kuang, Q.Y.; Xing, J.B.; Wang, Z.Y.; Cao, X.Y.; Xu, S.J.; Liu, J.; Huang, Z.; Zheng, Z.Z.; et al. Clinical sequencing uncovers the genomic characteristics and mutation spectrum of the 2018 African swine fever virus in Guangdong, China. Front. Vet. Sci. 2022, 9, 978243. [Google Scholar] [CrossRef]
- Guo, J.; Fang, L.; Ye, X.; Chen, J.; Xu, S.; Zhu, X.; Miao, Y.; Wang, D.; Xiao, S. Evolutionary and genotypic analyses of global porcine epidemic diarrhea virus strains. Transbound. Emerg. Dis. 2019, 66, 111–118. [Google Scholar] [CrossRef] [PubMed]
- He, W.T.; Bollen, N.; Xu, Y.; Zhao, J.; Dellicour, S.; Yan, Z.; Gong, W.; Zhang, C.; Zhang, L.; Lu, M.; et al. Phylogeography Reveals Association between Swine Trade and the Spread of Porcine Epidemic Diarrhea Virus in China and across the World. Mol. Biol. Evol. 2022, 39, msab364. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Yang, X.; Li, H.; Ma, B.; Guan, R.; Yang, J.; Chen, D.; Han, X.; Zhou, L.; Song, Z.; et al. Molecular characterization of porcine epidemic diarrhea virus associated with outbreaks in southwest China during 2014–2018. Transbound. Emerg. Dis. 2021, 68, 3482–3497. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.; Yang, J.; Li, A.; Gong, Z.; Yang, L.; Cheng, Q.; Wang, C.; Zhao, M.; Yuan, S.; Chen, Y.; et al. Genetic characterization and phylogenetic analysis of porcine epidemic diarrhea virus in Guangdong, China, between 2018 and 2019. PLoS ONE 2021, 16, e0253622. [Google Scholar] [CrossRef]
- Zhou, C.J.; Chen, J.; Hou, J.B.; Zheng, Y.; Yu, Y.N.; He, H.; Zhang, Y.P.; Feng, X.L.; Zheng, Q.S. The Immunological Functions of Muramyl Dipeptide Compound Adjuvant on Humoral, Cellular-mediated and Mucosal Immune Responses to PEDV Inactivated Vaccine in Mice. Protein Pept. Lett. 2018, 25, 908–913. [Google Scholar] [CrossRef]
- Wen, F.; Yu, H.; Guo, J.; Li, Y.; Luo, K.; Huang, S. Identification of the hyper-variable genomic hotspot for the novel coronavirus SARS-CoV-2. J. Infect. 2020, 80, 671–693. [Google Scholar] [CrossRef]
- Li, W.; van Kuppeveld, F.J.M.; He, Q.; Rottier, P.J.M.; Bosch, B.J. Cellular entry of the porcine epidemic diarrhea virus. Virus Res. 2016, 226, 117–127. [Google Scholar] [CrossRef]
- Hou, Y.; Lin, C.M.; Yokoyama, M.; Yount, B.L.; Marthaler, D.; Douglas, A.L.; Ghimire, S.; Qin, Y.; Baric, R.S.; Saif, L.J.; et al. Deletion of a 197-Amino-Acid Region in the N-Terminal Domain of Spike Protein Attenuates Porcine Epidemic Diarrhea Virus in Piglets. J. Virol. 2017, 91, e00227-17. [Google Scholar] [CrossRef]
- Guo, X.; Hu, H.; Chen, F.; Li, Z.; Ye, S.; Cheng, S.; Zhang, M.; He, Q. iTRAQ-based comparative proteomic analysis of Vero cells infected with virulent and CV777 vaccine strain-like strains of porcine epidemic diarrhea virus. J. Proteom. 2016, 130, 65–75. [Google Scholar] [CrossRef]
- Zhang, H.; Han, F.; Yan, X.; Liu, L.; Shu, X.; Hu, H. Prevalence and phylogenetic analysis of spike gene of porcine epidemic diarrhea virus in Henan province, China in 2015–2019. Infect. Genet. Evol. 2021, 88, 104709. [Google Scholar] [CrossRef]






| Prevalence of PEDV Infection | Genotypes of PEDV Strains (n = 98) | |||||
|---|---|---|---|---|---|---|
| Year | Detection Rate | Positive Rate at Farm Level | G1b (n = 1) | G2a (n = 74) | G2b (n = 16) | G2c (n = 7) |
| 2017 | 59.12% (81/137) | 89.29% (25/28) | 1 | 11 | 2 | 0 |
| 2018 | 71.93% (123/171) | 89.19% (33/37) | 0 | 27 | 6 | 0 |
| 2019 | 53.49% (23/43) | 60.00% (9/15) | 0 | 3 | 0 | 1 |
| 2020 | 35.79% (34/95) | 66.67% (12/18) | 0 | 10 | 1 | 0 |
| 2021 | 46.08% (94/204) | 71.79% (28/39) | 0 | 22 | 7 | 4 |
| 2022 | 34.78% (8/23) | 50% (3/6) | 0 | 1 | 0 | 2 |
| Total | 53.94% (363/673) | 79.92% (110/143) | 1.02% (1/98) | 75.51% (74/98) | 16.33% (16/98) | 7.14% (7/98) |
| Guangxi PEDV Strains (n = 98) | Percentage of Nucleotide (Amino Acid) Identity (%) | |
|---|---|---|
| CV777 | AJ1102 | |
| G1b (n = 1) | 95.10 (81.50) | 93.50 (81.20) |
| G2a (n = 74) | 85.30–91.50 (82.30–89.10) | 96.40–99.50 (95.10–99.50) |
| G2b (n = 16) | 85.30–86.40 (82.00–83.90) | 97.10–99.50 (95.20–99.80) |
| G2c (n = 7) | 87.40–91.60 (87.50–91.60) | 84.30–87.10 (84.30–87.10) |
| Recombinant | Recombination Events | Putative Parental Virus | Detection Methods (Average p-Value) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Break Points † | Major | Minor | RDP | GENECONV | Bootscan | Maxchi | Chimaera | SiSscan | 3Seq | |
| 18-GXNN-6 | 1004–1106 | CH/SCAZ10 | CH/S | NS ‡ | 6.765 × 10−1 | 3.487 × 10−5 | NS | NS | 4.028 | 3.853 |
| 19-GXBH-2 | 54–703 | PEDV-7C | SC-L | 1.664 × 10−6 | NS | NS | 8.503 × 10−3 | 3.723 × 10−2 | 3.255 × 10−27 | 1.464 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, J.; Du, C.; Lu, Y.; Wang, R.; Su, X.; Yu, K.; Qin, Q.; Chen, Y.; Wei, Z.; Huang, W.; et al. Phylogenetic and Spatiotemporal Analyses of Porcine Epidemic Diarrhea Virus in Guangxi, China during 2017–2022. Animals 2023, 13, 1215. https://doi.org/10.3390/ani13071215
Bai J, Du C, Lu Y, Wang R, Su X, Yu K, Qin Q, Chen Y, Wei Z, Huang W, et al. Phylogenetic and Spatiotemporal Analyses of Porcine Epidemic Diarrhea Virus in Guangxi, China during 2017–2022. Animals. 2023; 13(7):1215. https://doi.org/10.3390/ani13071215
Chicago/Turabian StyleBai, Jiaguo, Chen Du, Ying Lu, Ruomu Wang, Xueli Su, Kechen Yu, Qiuying Qin, Ying Chen, Zuzhang Wei, Weijian Huang, and et al. 2023. "Phylogenetic and Spatiotemporal Analyses of Porcine Epidemic Diarrhea Virus in Guangxi, China during 2017–2022" Animals 13, no. 7: 1215. https://doi.org/10.3390/ani13071215
APA StyleBai, J., Du, C., Lu, Y., Wang, R., Su, X., Yu, K., Qin, Q., Chen, Y., Wei, Z., Huang, W., & Ouyang, K. (2023). Phylogenetic and Spatiotemporal Analyses of Porcine Epidemic Diarrhea Virus in Guangxi, China during 2017–2022. Animals, 13(7), 1215. https://doi.org/10.3390/ani13071215







