Can We Reliably Detect Respiratory Diseases through Precision Farming? A Systematic Review
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
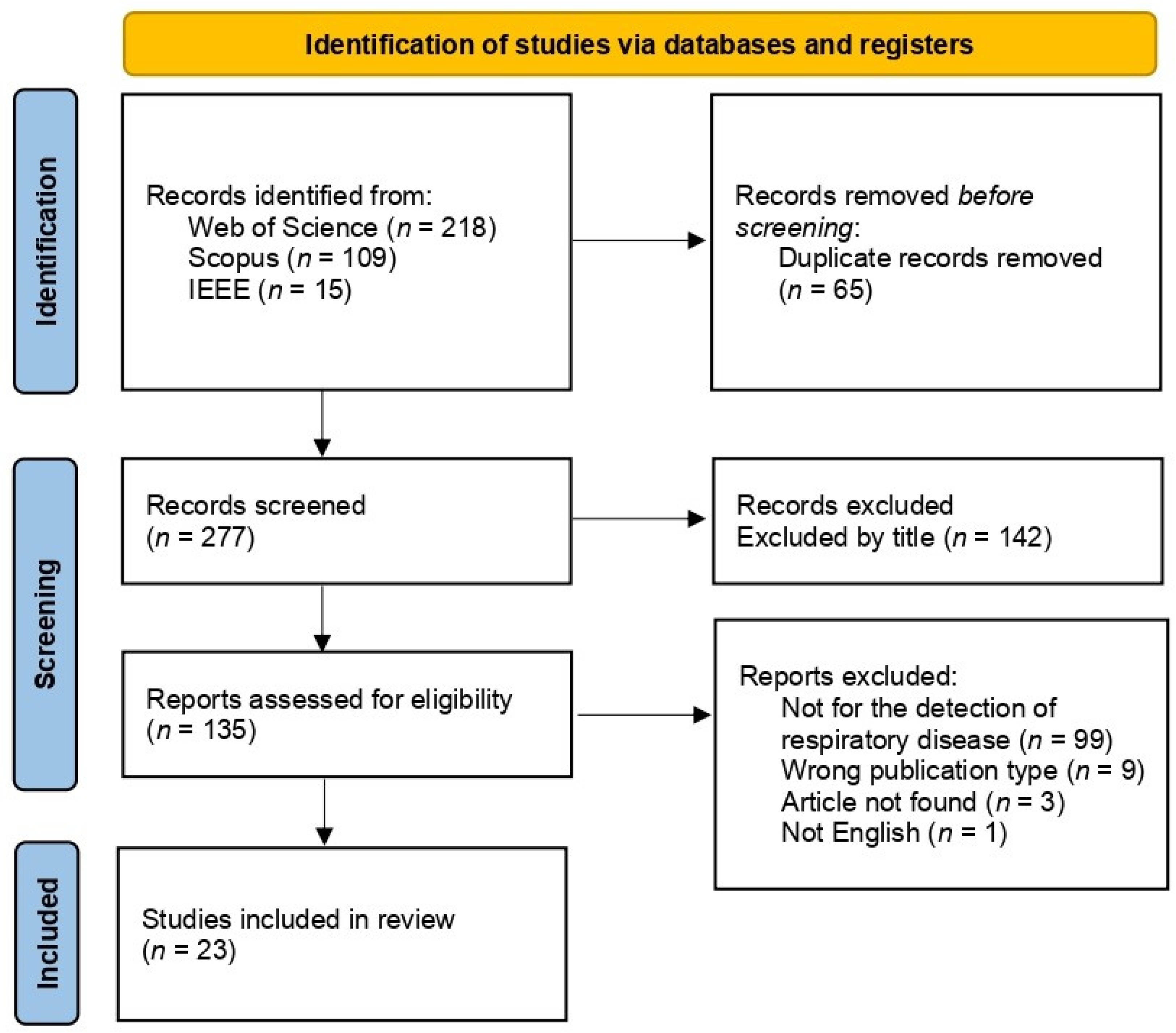
2.1. Literature Search
2.2. Data Gathered from Articles
2.3. Risk of Bias
2.4. Technology Reliability
3. Results
3.1. Studies’ Condition and Reference Tests
3.2. Risk of Bias
3.3. Respiratory Disease PLF Technologies for Poultry Production
3.4. Respiratory Disease PLF Technologies for Bovine Production
3.5. Respiratory Disease PLF Technologies for Swine Production
4. Discussion
4.1. Performance Measures
4.2. Reference Test
4.3. Risk of Bias
4.4. Can We Reliably Detect Livestock Respiratory Disease through Precision Farming?
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buczinski, S.; Achard, D.; Timsit, E. Effects of calfhood respiratory disease on health and performance of dairy cattle: A systematic review and meta-analysis. J. Dairy Sci. 2021, 104, 8214–8227. [Google Scholar] [CrossRef] [PubMed]
- Michiels, T.; Welby, S.; Vanrobaeys, M.; Quinet, C.; Rouffaer, L.; Lens, L.; Martel, A.; Butaye, P. Prevalence of Mycoplasma gallisepticum and Mycoplasma synoviae in commercial poultry, racing pigeons and wild birds in Belgium. Avian Pathol. 2016, 45, 244–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haji-Abdolvahab, H.; Ghalyanchilangeroudi, A.; Bahonar, A.; Ghafouri, S.A.; Marandi, M.V.; Mehrabadi, M.H.F.; Tehrani, F. Prevalence of avian influenza, Newcastle disease, and infectious bronchitis viruses in broiler flocks infected with multifactorial respiratory diseases in Iran, 2015–2016. Trop. Anim. Health Prod. 2018, 51, 689–695. [Google Scholar] [CrossRef]
- Roussan, D.; Haddad, R.; Khawaldeh, G. Molecular Survey of Avian Respiratory Pathogens in Commercial Broiler Chicken Flocks with Respiratory Diseases in Jordan. Poult. Sci. 2008, 87, 444–448. [Google Scholar] [CrossRef]
- Pessoa, J.; Rodrigues da Costa, M.; García Manzanilla, E.; Norton, T.; McAloon, C.; Boyle, L. Managing respiratory disease in finisher pigs: Combining quantitative assessments of clinical signs and the prevalence of lung lesions at slaughter. Prev. Vet. Med. 2021, 186, 105208. [Google Scholar] [CrossRef]
- Hassan, K.E.; Shany, S.A.; Ali, A.; Dahshan, A.-H.M.; El-Sawah, A.A.; El-Kady, M.F. Prevalence of avian respiratory viruses in broiler flocks in Egypt. Poult. Sci. 2016, 95, 1271–1280. [Google Scholar] [CrossRef]
- Dubrovsky, S.; Van Eenennaam, A.; Karle, B.; Rossitto, P.; Lehenbauer, T.; Aly, S. Bovine respiratory disease (BRD) cause-specific and overall mortality in preweaned calves on California dairies: The BRD 10K study. J. Dairy Sci. 2019, 102, 7320–7328. [Google Scholar] [CrossRef]
- Urie, N.J.; Lombard, J.E.; Shivley, C.B.; Kopral, C.A.; Adams, A.E.; Earleywine, T.J.; Olson, J.D.; Garry, F.B. Preweaned heifer management on US dairy operations: Part V. Factors associated with morbidity and mortality in preweaned dairy heifer calves. J. Dairy Sci. 2018, 101, 9229–9244. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.K.; Goyal, S.M.; Joo, H.S. Retrospective analysis of etiologic agents associated with respiratory diseases in pigs. Can. Vet. J. 2003, 44, 735–737. [Google Scholar] [PubMed]
- Nidzworski, D.; Wasilewska, E.; Smietanka, K.; Szewczyk, B.; Minta, Z. Detection and differentiation of Newcastle disease virus and influenza virus by using duplex real-time PCR. Acta Biochim. Pol. 2013, 60, 475–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, V.H.; Agnol, A.M.D.; Fritzen, J.T.; Lorenzetti, E.; Alfieri, A.A.; Alfieri, A.F. Microbial diversity involved in the etiology of a bovine respiratory disease outbreak in a dairy calf rearing unit. Comp. Immunol. Microbiol. Infect. Dis. 2020, 71, 101494. [Google Scholar] [CrossRef] [PubMed]
- Berckmans, D. General introduction to precision livestock farming. Anim. Front. 2017, 7, 6–11. [Google Scholar] [CrossRef] [Green Version]
- Vandermeulen, J.; Bahr, C.; Johnston, D.; Earley, B.; Tullo, E.; Fontana, I.; Guarino, M.; Exadaktylos, V.; Berckmans, D. Early recognition of bovine respiratory disease in calves using automated continuous monitoring of cough sounds. Comput. Electron. Agric. 2016, 129, 15–26. [Google Scholar] [CrossRef]
- Carpentier, L.; Vranken, E.; Berckmans, D.; Paeshuyse, J.; Norton, T. Development of sound-based poultry health monitoring tool for automated sneeze detection. Comput. Electron. Agric. 2019, 162, 573–581. [Google Scholar] [CrossRef]
- Hong, M.; Ahn, H.; Atif, O.; Lee, J.; Park, D.; Chung, Y. Field-Applicable Pig Anomaly Detection System Using Vocalization for Embedded Board Implementations. Appl. Sci. 2020, 10, 6991. [Google Scholar] [CrossRef]
- Bowen, J.; Haskell, M.; Miller, G.; Mason, C.; Bell, D.; Duthie, C.-A. Early prediction of respiratory disease in preweaning dairy calves using feeding and activity behaviors. J. Dairy Sci. 2021, 104, 12009–12018. [Google Scholar] [CrossRef]
- Hogeveen, H.; Kamphuis, C.; Steeneveld, W.; Mollenhorst, H. Sensors and Clinical Mastitis—The Quest for the Perfect Alert. Sensors 2010, 10, 7991–8009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominiak, K.; Kristensen, A. Prioritizing alarms from sensor-based detection models in livestock production—A review on model performance and alarm reducing methods. Comput. Electron. Agric. 2017, 133, 46–67. [Google Scholar] [CrossRef]
- Norton, T.; Berckmans, D. Developing precision livestock farming tools for precision dairy farming. Anim. Front. 2017, 7, 18–23. [Google Scholar] [CrossRef] [Green Version]
- Stachowicz, J.; Umstätter, C. Do we automatically detect health- or general welfare-related issues? A framework. Proc. R. Soc. B Boil. Sci. 2021, 288, 20210190. [Google Scholar] [CrossRef]
- Benjamin, M.; Yik, S. Precision Livestock Farming in Swine Welfare: A Review for Swine Practitioners. Animals 2019, 9, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez, Y.; Stygar, A.H.; Boumans, I.J.M.M.; Bokkers, E.A.M.; Pedersen, L.J.; Niemi, J.K.; Pastell, M.; Manteca, X.; Llonch, P. A Systematic Review on Validated Precision Livestock Farming Technologies for Pig Production and Its Potential to Assess Animal Welfare. Front. Vet. Sci. 2021, 8, 660565. [Google Scholar] [CrossRef]
- Stygar, A.H.; Gómez, Y.; Berteselli, G.V.; Dalla Costa, E.; Canali, E.; Niemi, J.K.; Llonch, P.; Pastell, M. A Systematic Review on Commercially Available and Validated Sensor Technologies for Welfare Assessment of Dairy Cattle. Front. Vet. Sci. 2021, 8, 634338. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Polanin, J.R.; Pigott, T.; Espelage, D.L.; Grotpeter, J. Best practice guidelines for abstract screening large-evidence systematic reviews and meta-analyses. Res. Synth. Methods 2019, 10, 330–342. [Google Scholar] [CrossRef] [Green Version]
- Page, M.J.; Moher, D.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. PRISMA 2020 explanation and elaboration: Updated guidance and exemplars for reporting systematic reviews. BMJ 2021, 372, n160. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, S.; Phyn, C.; Huzzey, J.; Mueller, K.; Turner, S.-A.; Donaghy, D.; Roche, J. Graduate Student Literature Review: Evaluating the appropriate use of wearable accelerometers in research to monitor lying behaviors of dairy cows. J. Dairy Sci. 2020, 103, 12140–12157. [Google Scholar] [CrossRef]
- ISO 20966:2007; Automatic Milking Installations—Requirements and Testing. International Organization for Standardization: Geneva, Switzerland, 2007. Available online: https://www.iso.org/standard/35593.html (accessed on 31 October 2022).
- Aerts, J.-M.; Jans, P.; Halloy, D.; Gustin, P.; Berckmans, D. Labeling of Cough Data from Pigs for on-Line Disease Monitoring by Sound Analysis. Trans. ASAE 2005, 48, 351–354. [Google Scholar] [CrossRef] [Green Version]
- Moshou, D.; Chedad, A.; Van Hirtum, A.; De Baerdemaeker, J.; Berckmans, D.; Ramon, H. An Intelligent Alarm for Early Detection of Swine Epidemics Based on Neural Networks. Trans. ASAE 2001, 44, 167–174. [Google Scholar] [CrossRef]
- Moshou, D.; Chedad, A.; Van Hirtum, A.; De Baerdemaeker, J.; Berckmans, D.; Ramon, H. Neural recognition system for swine cough. Math. Comput. Simul. 2001, 56, 475–487. [Google Scholar] [CrossRef]
- Van Hirtum, A.; Guarino, M.; Costa, A.; Jans, P.; Ghesquiere, K.; Aerts, J.M.; Navarotto, P.L.; Berckmans, D. Automatic Detection of Chronic Pig Coughing From Continuous Registration in Field Situations. In Proceedings of the Third International Workshop on Models and Analysis of Vocal Emissions for Biomedical Applications (MAVEBA 2003), Florence, Italy, 10–12 December 2003; pp. 251–254. [Google Scholar]
- Jans, P.; Guarino, M.; Costa, A.; Aerts, J.-M.; Berckmans, D. Field Test of Algorithm for Cough Detection in Pig Houses. In Proceedings of the ASAE Annual International Meeting, Ottawa, ON, Canada, 1–4 August 2004; pp. 4483–4494. [Google Scholar]
- Exadaktylos, V.; Silva, M.; Aerts, J.-M.; Taylor, C.; Berckmans, D. Real-time recognition of sick pig cough sounds. Comput. Electron. Agric. 2008, 63, 207–214. [Google Scholar] [CrossRef]
- Exadaktylos, V.; Silva, M.; Ferrari, S.; Guarino, M.; Taylor, C.J.; Aerts, J.-M.; Berckmans, D. Time-series analysis for online recognition and localization of sick pig (Sus scrofa) cough sounds. J. Acoust. Soc. Am. 2008, 124, 3803–3809. [Google Scholar] [CrossRef]
- Guarino, M.; Jans, P.; Costa, A.; Aerts, J.-M.; Berckmans, D. Field test of algorithm for automatic cough detection in pig houses. Comput. Electron. Agric. 2008, 62, 22–28. [Google Scholar] [CrossRef]
- Chung, Y.; Oh, S.; Lee, J.; Park, D.; Chang, H.-H.; Kim, S. Automatic Detection and Recognition of Pig Wasting Diseases Using Sound Data in Audio Surveillance Systems. Sensors 2013, 13, 12929–12942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Li, X.; Liu, W.; Gao, Y.; Lei, M.; Tan, H.; Yang, D. DNN-HMM based acoustic model for continuous pig cough sound recognition. Int. J. Agric. Biol. Eng. 2020, 13, 186–193. [Google Scholar] [CrossRef]
- Shen, W.; Tu, D.; Yin, Y.; Bao, J. A new fusion feature based on convolutional neural network for pig cough recognition in field situations. Inf. Process. Agric. 2020, 8, 573–580. [Google Scholar] [CrossRef]
- Yin, Y.; Tu, D.; Shen, W.; Bao, J. Recognition of sick pig cough sounds based on convolutional neural network in field situations. Inf. Process. Agric. 2020, 8, 369–379. [Google Scholar] [CrossRef]
- Shen, W.; Ji, N.; Yin, Y.; Dai, B.; Tu, D.; Sun, B.; Hou, H.; Kou, S.; Zhao, Y. Fusion of acoustic and deep features for pig cough sound recognition. Comput. Electron. Agric. 2022, 197, 106994. [Google Scholar] [CrossRef]
- Banakar, A.; Sadeghi, M.; Shushtari, A. An intelligent device for diagnosing avian diseases: Newcastle, infectious bronchitis, avian influenza. Comput. Electron. Agric. 2016, 127, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, M.; Carroll, B.T.; Anderson, D.V.; Daley, W.; Harbert, S.; Britton, D.F.; Jackwood, M.W. Identifying rale sounds in chickens using audio signals for early disease detection in poultry. In Proceedings of the 2016 IEEE Global Conference on Signal and Information Processing (GlobalSIP), Washington, DC, USA, 7–9 December 2016; pp. 55–59. [Google Scholar] [CrossRef]
- Cuan, K.; Zhang, T.; Huang, J.; Fang, C.; Guan, Y. Detection of avian influenza-infected chickens based on a chicken sound convolutional neural network. Comput. Electron. Agric. 2020, 178, 105688. [Google Scholar] [CrossRef]
- Liu, L.; Li, B.; Zhao, R.; Yao, W.; Shen, M.; Yang, J. A Novel Method for Broiler Abnormal Sound Detection Using WMFCC and HMM. J. Sensors 2020, 2020, 2985478. [Google Scholar] [CrossRef] [Green Version]
- Cuan, K.; Zhang, T.; Li, Z.; Huang, J.; Ding, Y.; Fang, C. Automatic Newcastle disease detection using sound technology and deep learning method. Comput. Electron. Agric. 2022, 194, 106740. [Google Scholar] [CrossRef]
- Schaefer, A.; Cook, N.; Bench, C.; Chabot, J.; Colyn, J.; Liu, T.; Okine, E.; Stewart, M.; Webster, J. The non-invasive and automated detection of bovine respiratory disease onset in receiver calves using infrared thermography. Res. Vet. Sci. 2012, 93, 928–935. [Google Scholar] [CrossRef]
- Carpentier, L.; Berckmans, D.; Youssef, A.; Berckmans, D.; van Waterschoot, T.; Johnston, D.; Ferguson, N.; Earley, B.; Fontana, I.; Tullo, E.; et al. Automatic cough detection for bovine respiratory disease in a calf house. Biosyst. Eng. 2018, 173, 45–56. [Google Scholar] [CrossRef]
- Taylor, J.D.; Fulton, R.W.; Lehenbauer, T.W.; Step, D.L.; Confer, A.W. The Epidemiology of Bovine Respiratory Disease: What Is the Evidence for Predisposing Factors? Can. Vet. J. 2010, 51, 1095. [Google Scholar]
- McGuirk, S.M.; Peek, S.F. Timely diagnosis of dairy calf respiratory disease using a standardized scoring system. Anim. Health Res. Rev. 2014, 15, 145–147. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, X.; He, Y.; Wang, K. Cough sound analysis to assess air quality in commercial weaner barns. Comput. Electron. Agric. 2019, 160, 8–13. [Google Scholar] [CrossRef]
- Van Hirtum, A.; Berckmans, D. Objective recognition of cough sound as biomarker for aerial pollutants. Indoor Air 2003, 14, 10–15. [Google Scholar] [CrossRef]
- Podolsky, S.H. The evolving response to antibiotic resistance (1945–2018). Palgrave Commun. 2018, 4, 124. [Google Scholar] [CrossRef] [Green Version]
- Lalkhen, A.; McCluskey, A. Clinical tests: Sensitivity and specificity. Contin. Educ. Anaesth. Crit. Care Pain 2008, 8, 221–223. [Google Scholar] [CrossRef] [Green Version]
- Callens, B.; Persoons, D.; Maes, D.; Laanen, M.; Postma, M.; Boyen, F.; Haesebrouck, F.; Butaye, P.; Catry, B.; Dewulf, J. Prophylactic and metaphylactic antimicrobial use in Belgian fattening pig herds. Prev. Vet. Med. 2012, 106, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Tuyttens, F.A.M.; Molento, C.F.M.; Benaissa, S. Twelve Threats of Precision Livestock Farming (PLF) for Animal Welfare. Front. Vet. Sci. 2022, 9, 889623. [Google Scholar] [CrossRef]
- McGuirk, S.M. Disease Management of Dairy Calves and Heifers. Vet. Clin. N. Am.—Food Anim. Pract. 2008, 24, 139–153. [Google Scholar] [CrossRef]
- Ollivett, T.L.; Buczinski, S. On-Farm Use of Ultrasonography for Bovine Respiratory Disease. Vet. Clin. N. Am.—Food Anim. Pract. 2016, 32, 19–35. [Google Scholar] [CrossRef]
- Decaris, N.; Buczinski, S.; Tárdon, D.I.C.; Camargo, L.; Schllemer, N.R.; Hagen, S.C.F.; Woolums, A.R.; Gomes, V. Diagnostic accuracy of Wisconsin and California scoring systems to detect bovine respiratory disease in preweaning dairy calves under subtropical environmental conditions. J. Dairy Sci. 2022, 105, 7750–7763. [Google Scholar] [CrossRef]
- Lowie, T.; Van Leenen, K.; Jourquin, S.; Pas, M.; Bokma, J.; Pardon, B. Differences in the association of cough and other clinical signs with ultrasonographic lung consolidation in dairy, veal, and beef calves. J. Dairy Sci. 2022, 105, 6111–6124. [Google Scholar] [CrossRef] [PubMed]
- Buczinski, S.; Ollivett, T.L.; Dendukuri, N. Bayesian estimation of the accuracy of the calf respiratory scoring chart and ultrasonography for the diagnosis of bovine respiratory disease in pre-weaned dairy calves. Prev. Vet. Med. 2015, 119, 227–231. [Google Scholar] [CrossRef] [PubMed]
| Species Terms | Technology Terms | Type of Conditions Terms |
|---|---|---|
| Dairy cow(s) | Precision Livestock Farming | Respiratory Disease(s) |
| Cow | Noninvasive Technology | Cough |
| Cattle | Smart sensor | Fever |
| Calf | Smart Farming | BRD |
| Calves | Automated technology | Vocalization |
| Pig | Online health monitoring | Infectious disease(s) |
| Sow | Computer vision | Sneeze |
| Swine | Cough recognition | Respiratory disease detection |
| Broiler | Sound analysis | |
| Laying hen | Sound classification | |
| Chicken | Convolutional neural network | |
| Poultry | ||
| Goat | ||
| Sheep | ||
| Ewe | ||
| Lamb |
| Species | Study | Sensor Type | Performance Measures | Study Conditions | Reference Test |
|---|---|---|---|---|---|
| Swine | [30] | Sound Based | Positive cough recognition | Laboratory | Remote audio labeling |
| [31] | Sound Based | Positive cough recognition | Laboratory | Remote audio labeling | |
| [32] | Sound Based | Accuracy | Field | Live audio labeling | |
| [33] | Sound Based | Accuracy | Field | Live audio labeling | |
| [34] | Sound Based | Correct identification ratio | Laboratory | Remote audio labeling | |
| [35] | Sound Based | Correct identification ratio | Field | Remote audio labeling | |
| [36] | Sound Based | Accuracy | Field | Live audio labeling | |
| [37] | Sound Based | Sensitivity, Precision, Accuracy, and cough detection rate | Field | Remote audio labeling and blood analysis | |
| [15] | Sound Based | Sensitivity, Precision, cough detection rate, and F1-score | Field | Video labeling and blood analysis | |
| [38] | Sound Based | Word error rate | Laboratory | Remote audio labeling | |
| [39] | Sound Based | Sensitivity, Specificity, Precision, Accuracy, and F1-score | Field | Remote audio labeling | |
| [40] | Sound Based | Sensitivity, Specificity, Precision, Accuracy, and F1-score | Field | Remote audio labeling | |
| [41] | Sound Based | Sensitivity, Precision, Accuracy, and F1-score | Field | Remote audio labeling | |
| Poultry | [42] | Sound Based | Sensitivity, Specificity, and Accuracy | Laboratory | PCR |
| [43] | Sound Based | Sensitivity, Precision, and Accuracy | Laboratory | Remote audio labeling | |
| [14] | Sound Based | Sensitivity, Specificity, and Precision | Laboratory | Remote audio labeling | |
| [44] | Sound Based | Accuracy | Laboratory | PCR | |
| [45] | Sound Based | Sensitivity, Precision, Accuracy, and F1-score | Field | Remote audio labeling | |
| [46] | Sound Based | Sensitivity, Precision, Accuracy, and F1-score | Laboratory | Video labeling and PCR | |
| Bovine | [47] | Image | Sensitivity, Specificity, PPV, NPV, and Cut off value | Field | Clinical assessment |
| [13] | Sound Based | Sensitivity, Specificity, and Precision | Field | Clinical assessment and blood analysis | |
| [48] | Sound Based | Sensitivity, Specificity, and Precision | Field | Blood analysis | |
| [16] | Accelerometer | Sensitivity, Specificity, Accuracy, and MCC | Field | Clinical assessment |
| Species | Study | Study Conditions | Housing | Hardware | How It Was Installed | Software | Population Description | Number of Animals | Raw Data | Risk of Bias |
|---|---|---|---|---|---|---|---|---|---|---|
| Swine | [30] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | low |
| [31] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | low | |
| [32] | ✔ | ✔ | ✖ | ✔ | ✔ | ✔ | ✖ | ✔ | high | |
| [33] | ✔ | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | high | |
| [34] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | low | |
| [35] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | low | |
| [36] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | low | |
| [37] | ✔ | ✔ | ✔ | ✔ | ✔ | ✖ | ✔ | ✖ | high | |
| [15] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | low | |
| [38] | ✔ | ✖ | ✔ | ✖ | ✔ | ✖ | ✔ | ✔ | high | |
| [39] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | low | |
| [40] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | low | |
| [41] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✖ | high | |
| Poultry | [42] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | low |
| [43] | ✔ | ✔ | ✖ | ✔ | ✔ | ✔ | ✔ | ✔ | high | |
| [14] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | low | |
| [44] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✖ | high | |
| [45] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | low | |
| [46] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | low | |
| Bovine | [47] | ✔ | ✔ | ✔ | ✔ | ✖ | ✔ | ✔ | ✔ | high |
| [13] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | low | |
| [48] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | low | |
| [16] | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ | low |
| Sensitivity (%) | Specificity (%) | Precision (%) | Accuracy (%) | F1-Score (%) | |
|---|---|---|---|---|---|
| [42] | 93.30 | 96.73 | N/A | 91.15 | N/A |
| [43] | 85.20 | N/A | 86.60 | 97.60 | N/A |
| [14] | 66.70 | N/A | 88.40 | N/A | N/A |
| [44] | N/A | N/A | N/A | 97.00 | N/A |
| [45] | 94.10 | N/A | 94.40 | 93.80 | 94.20 |
| [46] | 96.60 | N/A | 96.54 | 98.50 | 97.33 |
| Sensitivity (%) | Specificity (%) | Precision (%) | Accuracy (%) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|---|
| [47] | 100.00 | 97.40 | N/A | N/A | 86.30 | 100.00 |
| [13] | 50.30 | 99.20 | 87.50 | N/A | N/A | N/A |
| [48] | 41.40 | 99.90 | 94.20 | N/A | N/A | N/A |
| [16] | 54.00 | 95.00 | N/A | 75.00 | N/A | N/A |
| Sensitivity (%) | Specificity (%) | Precision (%) | Accuracy (%) | F1-Score (%) | Cough Detection Rate (%) | |
|---|---|---|---|---|---|---|
| [30] | N/A | N/A | N/A | N/A | N/A | 94.80 |
| [31] | N/A | N/A | N/A | N/A | N/A | 94.80 |
| [32] | N/A | N/A | N/A | N/A | N/A | 90.00 |
| [33] | N/A | N/A | N/A | 86.20 | N/A | N/A |
| [34] | N/A | N/A | N/A | N/A | N/A | 82.00 |
| [35] | N/A | N/A | N/A | N/A | N/A | 88.00 |
| [36] | N/A | N/A | N/A | 86.20 | N/A | 85.50 |
| [37] | 92.00 1 | N/A | 90.80 1 | 91.00 1 | N/A | 94.00 |
| [15] | 98.60 1 | N/A | 95.50 1 | N/A | 94.70 1 | 99.00 |
| [39] | 97.72 | 95.01 | 96.81 | 96.68 | 97.26 | 97.72 |
| [40] | 96.80 | 93.20 | 95.50 | 95.40 | 96.20 | 96.80 |
| [41] | 96.51 | N/A | 98.41 | 97.35 | 97.46 | 96.51 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garrido, L.F.C.; Sato, S.T.M.; Costa, L.B.; Daros, R.R. Can We Reliably Detect Respiratory Diseases through Precision Farming? A Systematic Review. Animals 2023, 13, 1273. https://doi.org/10.3390/ani13071273
Garrido LFC, Sato STM, Costa LB, Daros RR. Can We Reliably Detect Respiratory Diseases through Precision Farming? A Systematic Review. Animals. 2023; 13(7):1273. https://doi.org/10.3390/ani13071273
Chicago/Turabian StyleGarrido, Luís F. C., Sabrina T. M. Sato, Leandro B. Costa, and Ruan R. Daros. 2023. "Can We Reliably Detect Respiratory Diseases through Precision Farming? A Systematic Review" Animals 13, no. 7: 1273. https://doi.org/10.3390/ani13071273
APA StyleGarrido, L. F. C., Sato, S. T. M., Costa, L. B., & Daros, R. R. (2023). Can We Reliably Detect Respiratory Diseases through Precision Farming? A Systematic Review. Animals, 13(7), 1273. https://doi.org/10.3390/ani13071273








