Simple Summary
The European flounder (Platichthys flesus), which is closely related to the recently discovered Baltic flounder (Platichthys solemdali), is currently the third most commercially fished species in the Baltic Sea. The aim of this study was to obtain information on the current patterns of genetic diversity and the population structure of the European flounder and to verify whether the Baltic flounder is present in the southern Baltic Sea. Moreover, we aimed to verify whether the observed decline in the body condition indices of the species in the Baltic Sea might be associated with adaptive changes in its gene pool due to increased fishing pressure. The examined European flounder specimens displayed a high level of genetic diversity and represented a single genetic cluster. The applied molecular markers did not detect the presence of the Baltic flounder among the fish sampled from the studied area. Correlation analysis between genetic and morphological characteristics did not detect any signs of directional selection or density-dependent adaptive changes in the gene pool of the examined fish.
Abstract
The European flounder (Platichthys flesus), which is closely related to the recently discovered Baltic flounder (Platichthys solemdali), is currently the third most commercially fished species in the Baltic Sea. According to the available data from the Polish Fisheries Monitoring Center and fishermen’s observations, the body condition indices of the species in the Baltic Sea have declined in recent years. The aim of the present study was to obtain information on the current patterns of genetic variability and the population structure of the European flounder and to verify whether the Baltic flounder is present in the southern Baltic Sea. Moreover, we aimed to verify whether the observed decline in the body condition indices of the species in the Baltic Sea might be associated with adaptive alterations in its gene pool due to increased fishing pressure. For this purpose, 190 fish were collected from four locations along the central coastline of Poland, i.e., Mechelinki, Władysławowo, the Vistula Lagoon in 2018, and the Słupsk Bank in 2020. The fish were morphologically analyzed and then genetically screened by the application of nineteen microsatellite DNA and two diagnostic SNP markers. The examined European flounder specimens displayed a high level of genetic diversity (PIC = 0.832–0.903, I = 2.579–2.768). A lack of significant genetic differentiation (Fst = 0.004, p > 0.05) was observed in all the examined fish, indicating that the European flounder in the sampled area constitutes a single genetic cluster. A significant deficiency in heterozygotes (Fis = 0.093, p < 0.05) and overall deviations from Hardy–Weinberg expectations (H-WE) were only detected in fish sampled from the Słupsk Bank. The estimated effective population size (Ne) among the sampled fish groups varied from 712 (Słupsk Bank) to 10,115 (Władysławowo and Mechelinki). However, the recorded values of the Garza–Williamson indicator (M = 0.574–0.600) and the lack of significant (p > 0.05) differences in Heq > He under the SMM model did not support the species’ population size changes in the past. The applied SNP markers did not detect the presence of the Baltic flounder among the fish sampled from the studied area. The analysis of an association between biological traits and patterns of genetic diversity did not detect any signs of directional selection or density-dependent adaptive changes in the gene pool of the examined fish that might be caused by increased fishing pressure.
1. Introduction
The European flounder (Platichthys flesus) belongs to the Pleuronectidae family that is native to the coastline of the northeastern Atlantic Ocean and the Baltic Sea [1]. It is a euryhaline fish species that leads a demersal lifestyle, feeding on benthic invertebrates and small fish [2,3,4]. For a long time, two distinct ecological forms of the European flounder in the Baltic Sea have been considered. These two forms have shown different reproductive behaviors, i.e., pelagic and demersal [5]. Based on recent molecular evidence, the demersal form is now considered a new cryptic species: the Baltic flounder (Platichthys solemdali) [6].
The European flounder is an economically important, non-quota species fished in the Baltic Sea [7]. According to data from the International Council for the Exploration of the Sea (ICES), after the recent collapse of the cod (Gadus morhua) fishery, the European flounder has become the third most exploited species in the Baltic Sea [8]. Unfortunately, the available information shows about a 50% decrease in the total landings of the species in the Baltic Sea between 2016 and 2021 [8]. Other data show an almost 90% decline in the proportion of the European flounder in the total landings of flounder in the Gulf of Finland between 1980 and 2018 [9]. In addition, concerning reports from fishermen indicate that the body condition indices of the species have been decreasing, and caught fish are becoming smaller and thinner.
To date, at least three main hypotheses have been proposed to explain the observed decline in the population size/abundance and body condition indices of flounder in the Baltic Sea. The first hypothesis proposes that this decline is caused by the increasing deterioration of environmental conditions due to climate change and eutrophication, which shrink the species’ spawning grounds; cyanobacterial blooms in the nursery grounds; and higher rates of disease occurrence in the fish caused by invasive parasitic organisms [5,9]. The second hypothesis proposes that the decline is caused by growing competition for food and space from other native species that are better adapted to inhabit lower salinity environments, such as cyprinids and Baltic flounder [5,9]. The third hypothesis proposes that the decline is caused by adaptive genetic changes in the species’ gene pool toward limited growth rate and body size, and/or reduced population’s body condition indices due to fishery-mediated heavy selective pressure [10,11,12].
A research project recently launched by the National Marine Fisheries Research Institute (Poland) has indicated that the observed decrease in the flounders’ body condition indices in the Baltic Sea may be associated with environmental changes caused by climate change and anthropogenic pressure [13]. However, the research project did not include genetic examination of the species. This is significant because available data on the patterns of genetic diversity and the population structure of the European flounder were published more than a decade ago. Consequently, there is a lack of comprehensive molecular research examining the composition of the flounder species in the southern Baltic Sea. Moreover, the molecular research of Momigliano et al. [6,9] assessing the stock composition of flounders did not detect any signs of Baltic flounder presence in the southern Baltic Sea. In addition, the research of Momigliano et al. mainly focused on fish from the northern Baltic Sea and did not include areas located close to the Vistula River mouth, which potentially provides suitable environmental conditions for the Baltic flounder [6,9]. Because the research of Svedäng and Hornborg [12] demonstrated the strong impact of selective fishing on density-dependent adaptation in the body condition indices of Atlantic cod in the Baltic Sea, we might expect a similar effect to occur in the European flounder in recent years. If this selective effect was strong enough, it may be detected through the analysis of population genetic and morphological data. Moreover, in a severely overexploited population in which the number of individuals is drastically reduced, there is a higher likelihood of harmful alleles becoming fixed in the population. Harmful alleles are genetic variants that decrease an organism’s fitness; their fixation in a population can lower its viability, increasing the risk of extinction [5].
Therefore, the purpose of the present research was to obtain information on the current genetic status of the European flounder from the southern Baltic Sea and compare it with previous reports [14,15]. The study also sought to verify whether there are any signs of Baltic flounder presence in the southern Baltic Sea that could indicate species composition changes in relation to previously reported data [9]. Moreover, the study aimed to verify whether the observed decline in the species’ body condition indices might be associated with adaptive responses in its gene pool associated with the increased impact of fishing pressure in the Baltic Sea.
2. Materials and Methods
2.1. Fish Sampling and Morphological Data Collection
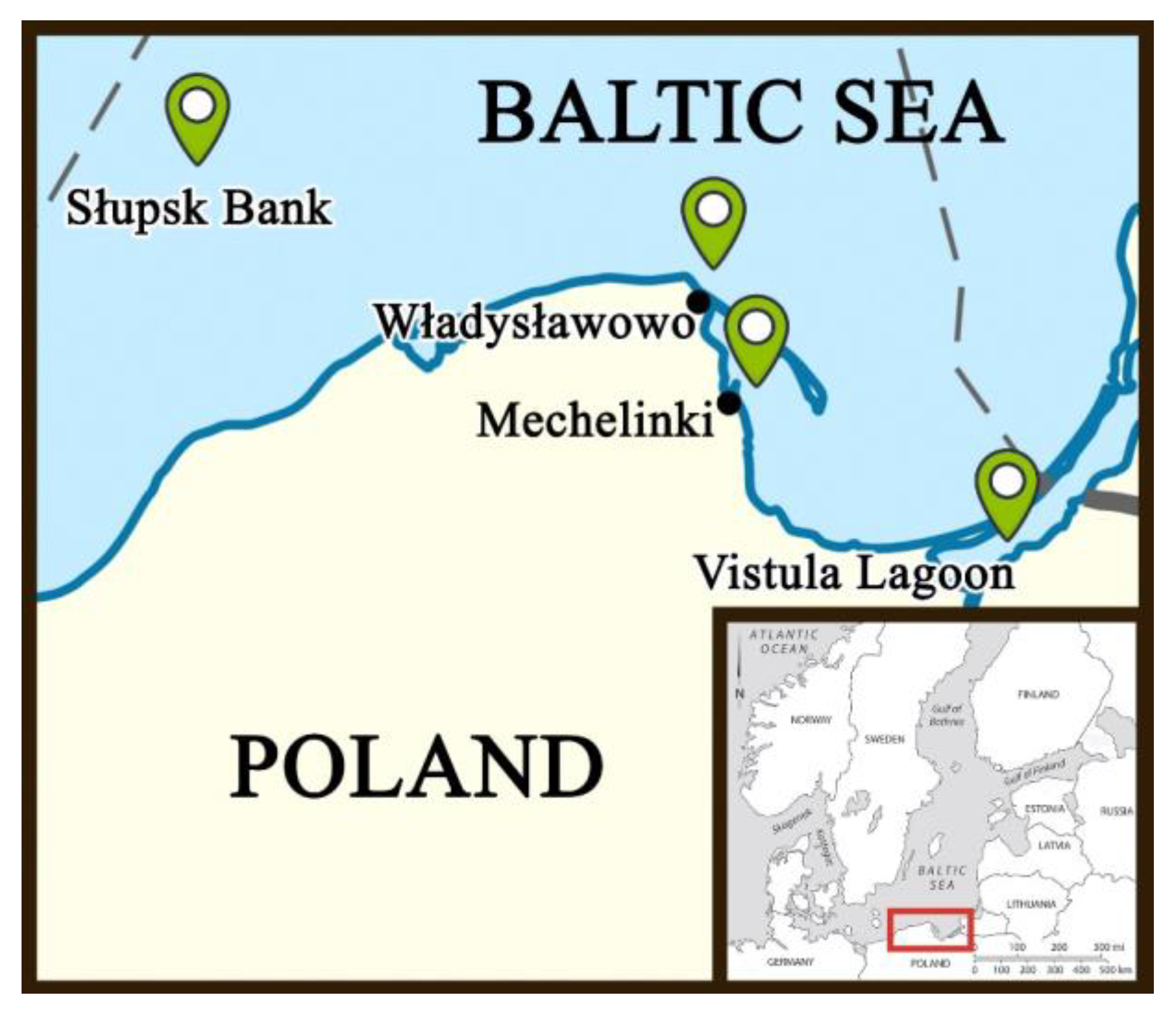
In total, 190 fish were sampled from four sampling sites along the central coastline of Poland, i.e., Mechelinki (54°37′07.0″ N 18°33′01.4″ E) (N = 50), Władysławowo (54°48′17.6″ N 18°24′33.1″ E) (N = 50), the Słupsk Bank (55°02′22.6″ N 16°22′03.0″ E) (N = 50), and the Vistula Lagoon (54°21′00.0″ N 19°33′00.0″ E) (N = 40) (Figure 1). Samples from Mechelinki, Władysławowo, and the Vistula Lagoon were collected in 2018 with the cooperation of local fishermen using square mesh panels. Samples from the Słupsk Bank were gathered in 2020 using the research vessel, Oceanograf, (University of Gdańsk). The fish sampled from Mechelinki, Władysławowo, and the Słupsk Bank represented mature fish that were at least 3+ years old. The fish sampled from the Vistula Lagoon were all young and immature, aged 1+ or 2+ years, indicating that the latter sampling site may be the nursery ground of the species.
Figure 1.
Map of the fish sampling sites along the central Polish coastline, namely, Mechelinki (54°37′07.0″ N 18°33′01.4″ E) (N = 50), Władysławowo (54°48′17.6″ N 18°24′33.1″ E) (N = 50), the Słupsk Bank (55°02′22.6″ N 16°22′03.0″ E) (N = 50), and the Vistula Lagoon (54°21′00.0″ N 19°33′00.0″ E) (N = 40).
The collected fish were measured to determine total body length (±0.1 cm) and mass (±0.01 g). The fish were then dissected to determine their sex and liver mass (± 0.01 g) (Supplementary Table S1). For the molecular analysis, fin clips were collected from each fish and stored in individual Eppendorf tubes filled with 96% ethanol.
2.2. DNA Isolation, Microsatellite DNA Markers Amplification and Genotyping
Genomic DNA were extracted and purified using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. The quality of the isolated DNA was checked using electrophoresis in 1.5% agarose gel.
Nineteen microsatellites that displayed satisfactory amplification performance were used for the genetic population study on the European flounder. Eleven of these had been previously used in studies on the species in the Baltic Sea: StPf1001, StPf1002, StPf1003, StPf1004, StPf1005, StPf1006, StPf1015, StPf1016, StPf1022, PL142, PL167 [14,15]. The remaining eight markers, FLAG5-83, FLAG4-71, FLAG2-76, FLAG4-65, FLAG8-37, Nplaf-33, FLAC4-67, FLAC4-69, originated from other studies on flounder along the Atlantic coast [16,17] (Table 1). Nested PCRs were performed in multiplexes according to the protocol described by Schuelke [18] and Nakano et al. [19].

Table 1.
Multilocus genetic diversity parameters of European flounder (Platichthys flesus) individuals examined in the current study. Ao: observed number of alleles, Ae: number of effective alleles, Ho: observed heterozygosity, He: expected heterozygosity, I: Shannon’s index, PIC: polymorphism information content, and Fis: fixation index. Values of the H–WE test and values of the Fis indicator are significant at a p < 0.05 and b p < 0.01. Significant values of the H–WE test and values of the Fis indicator after Bonferroni correction are bolded.
PCRs were prepared in 12.5 µL volume mixtures that contained 1X GoTaq® Hot Start Green Master Mix (Promega Corporation, San Luis Obispo, CA, USA) and varying primer concentrations, according to the recommendations described by Schuelke [18] (Supplementary Table S2). Around 10 ng of template DNA was used for each amplification. Nuclease-free water was added to the reaction mixture to obtain the desired final volume. Amplification was performed using a Thermal Cycler SimpliAMP (Applied Biosystems, Foster City, CA, USA) under the following conditions: 95 °C for 4 min, followed by 34 cycles of 95 °C for 30 s, 59 °C for 90 s, and 72 °C for 90 s, then eight cycles of 95 °C for 30 s, 55 °C for 90 s, and 72 °C for 90 s, and a final elongation step at 65 °C for 30 min. To reduce the probability of genotyping errors and the possible effect of a null allele, each homozygote was re-amplified in simplex reactions.
Genotyping was conducted using the Applied Biosystems 3130 Genetic Analyzer against the GeneScan 600 LIZ size standard (Applied Biosystems, Foster City, CA, USA). Electropherograms were analyzed using Peak Scanner Software ver. 1.0 (Applied Biosystems).
2.3. Amplification and Genotyping of SNP Markers
To analyze stock composition, we used two diagnostic SNP markers (SNPs 886_19 and 3599_4) that have been previously identified as being under divergent selection in the Baltic and European flounders [6,9]. The selected SNPs have been demonstrated to be almost fixed in both species, enabling the unambiguous determination of the species [20]. Both the SNP loci were amplified separately using primers and conditions described by Momigliano et al. [6]. To determine each SNP’s variants, the amplified DNA fragments were sequenced in both directions according to Sanger’s method using the Applied Biosystems 3130 Genetic Analyzer. Ten randomly selected fish from each sampling location were genotyped using the applied SNP markers.
2.4. Data Analysis
The obtained microsatellite raw data were checked for the presence of microsatellite null alleles, inconsistent values, scoring errors, and large allele dropouts using Micro-Checker software ver. 2.2.3 [21]. The observed microsatellite allele frequency and number (Ao), Shannon’s index (I), the polymorphism information content (PIC value), and the inbreeding coefficient (Fis) were calculated using GenAlEx (version 6.5) and PowerMarker (version 3.25) [22,23]. The Ewens–Watterson–Slatkin exact neutrality test, Beaumont and Nichols’ detection test of loci under selection, the Garza–Williamson index (M-ratio), the observed heterozygosity (Ho), and the expected heterozygosity (He), as well as the Hardy–Weinberg equilibrium (H–WE) test for each locus were performed using Arlequin software (version 3.5) [24]. To assess the global H–WE, Fisher’s and Smouse’s multilocus analysis methods were employed, using the Genepop (version 2.9.3.2) and Popgene (version 1.3.2) [25,26] computer programs. Significance levels for H–WE and the Fis indicator were adjusted using the sequential Bonferroni correction [27].
The genetic structure and differentiation among the fish from each sampling site were assessed using the estimation of genetic differentiation index (Fst) and the analysis of molecular variance (AMOVA) using Arlequin software (ver. 3.5). The genetic heterogeneity among tested fish groups was examined using ONCOR (ver. 2.0) and GeneClass (ver. 2.0) software [28,29]. The leave-one-out method and individual assignment tests were applied. Additionally, a set of analyses was carried out by applying algorithms that search for putative genetic clusters (K) without a priori information on the origin of the examined fish. For this purpose, we used two computer programs that apply different clustering principles, i.e., Structure (ver. 2.3.4) and Flock (ver. 3.1.) [30,31]. The first program applies a Bayesian clustering analysis method, detecting the most probable number of K within the sampled fish group. Ten runs were completed for each tested number of K (K = 1–8), setting the admixture model with 250,000 burn-in periods and one million Markov chain Monte Carlo (MCMC) replicates. The ΔK method of Evanno et al. [32] was used to estimate the most probable number of genetic clusters using Structure Harvester online software [33]. Flock software uses a frequentist and partly deterministic method that relies on iterative re-allocation. K is identified using Plateau analyses that are based on the repetition of identical cluster solutions. Additionally, the representation of the genetic structure independent of H–WE optimization––i.e., allele sharing distances (DAS), principal component analysis (PCA), and principal coordinate analysis (PCoA)––was also carried out using Populations (ver. 1.2.32) [34], R package Adegenet (ver. 2.1.5) [35], and GenAlEx software, respectively.
To delineate the historical demography of the studied fish group, effective population size (Ne) was estimated using the NeEstimator computer program (ver. 2.01) [36]. The single-sample method based on random linkage disequilibrium was applied. To obtain the best ratio between precision and bias, the criterion for excluding rare alleles of Pcrit = 0.02 and 0.05 was chosen, in line with Waples and Do [37]. Additionally, to detection of past population size decline in the studied fish groups was carried out using Bottleneck software (ver. 1.9) [38]. For this purpose, the equilibrium was tested using an infinite alleles model (IAM), stepwise mutation model (SMM), and two-phase mutation model (TPM). This method assumes that the excess of expected heterozygosity (He) over expected heterozygosity under a mutation–drift equilibrium (Heq) in the population indicates the population size reduction and associated bottlenecks.
The obtained DNA sequences of genomic regions bearing analyzed SNP markers were viewed using BioEdit software (ver. 7.2.5) [39] to recover the respective genotypes for each analyzed fish. The retrieved SNP genotype data were then manually compared with genotype profiles established for European and Baltic flounders by Momigliano et al. [6,14].
An analysis was conducted to identify an association between biological traits (i.e., sex, total body length, body mass, liver mass) and the pattern of genetic diversity (i.e., allele frequency, individual pairwise genetic distance, PIC, I, Ho, Fis) to assess the possible effect of selective pressure on the genetic structure of the studied fish. For this purpose, the recorded meristic parameters of the studied fish were transformed into relative values, i.e., Fulton’s condition factor (K) and the hepatosomatic index (HSI) [40,41]. Next, the sampled fish were divided into series of separate groups according to sex, the values of Fulton’s condition factor (K; (total weight × total lenght−3) × 100), and the hepatosomatic index (HSI; (liver weight × total weight−1) × 100) (Supplementary Table S3). All the abovementioned genetic parameters were then recalculated for each group. A non-parametric Kruskal–Wallis post hoc test analysis and a set of multidimensional analyses, such as principal components analysis (PCA/PCoA), correspondence analysis (DA), and discriminant function analysis (DFA), were implemented using Statistica (ver. 12.0) (StatSoft Company, Kraków, Poland) and GenAlEx software. These analyses were conducted to check for any significant (p < 0.05) associations in the estimated values of genetic parameters with any of the established fish groups as a sign of possible density-dependent genetic adaptation toward body condition indices. For PCoA analysis, individual pairwise genetic matrices for all analyzed fish were calculated, then each fish in the matrix was labeled according to the series group in their division. Lastly, a 2-dimensional plot was constructed. Because fish from the Vistula Lagoon displayed significant differences in terms of cohort composition, the analysis of correspondence between biological traits was carried out for all fish except for those sampled from the Vistula Lagoon.
3. Results
The Micro-Checker software did not detect any data consistency failures associated with stutter miscalls, scoring errors, or allelic dropout in the collected microsatellite DNA data. The estimated frequencies of possible null alleles across all the examined loci were below 5%, excluding the influence of possible amplification issues on the estimated population genetic parameters [42,43]. The overall number of alleles at the individual locus ranged from 6 to 47 (average = 23.9) (Supplementary Table S4). The Ewens–Watterson–Slatkin test was applied, showing the selective neutrality of the applied microsatellite markers.
The mean number of alleles per locus among fish from each sampling site varied between Ao = 17.9 and 19.4. The average values of polymorphism information content (PIC) and Shannon’s index ranged from 0.832 to 0.903 and from 2.579 to 2.768, respectively. The multilocus analysis revealed significant (p < 0.05) overall values of the fixation index (Fis = 0.074–0.093) in fish sampled from Mechelinki and the Słupsk Bank. However, after Bonferroni correction, significant values were observed only in fish from the latter sampling site. The recorded values of the fixation index (0.031 and 0.036) in fish sampled from Władysławowo and the Vistula Lagoon were not significant (p > 0.05). The mean values of observed heterozygosity (Ho) and expected heterozygosity (He) varied from 0.751 to 0.815 and from 0.841 to 0.847, respectively. Significant differences between values of observed heterozygosity (Ho) and expected (He) heterozygosity were observed across four (Władysławowo and the Vistula Lagoon) and five loci (Mechelinki and the Słupsk Bank). After Bonferroni correction, significant differences were observed only in one locus (Słupsk Bank). The H–WE probability global tests detected significant (p < 0.05) deviations from the H–WE expectations in fish sampled from Mechelinki and the Słupsk Bank. These deviations remained significant after Bonferroni correction only for fish from the latter group.
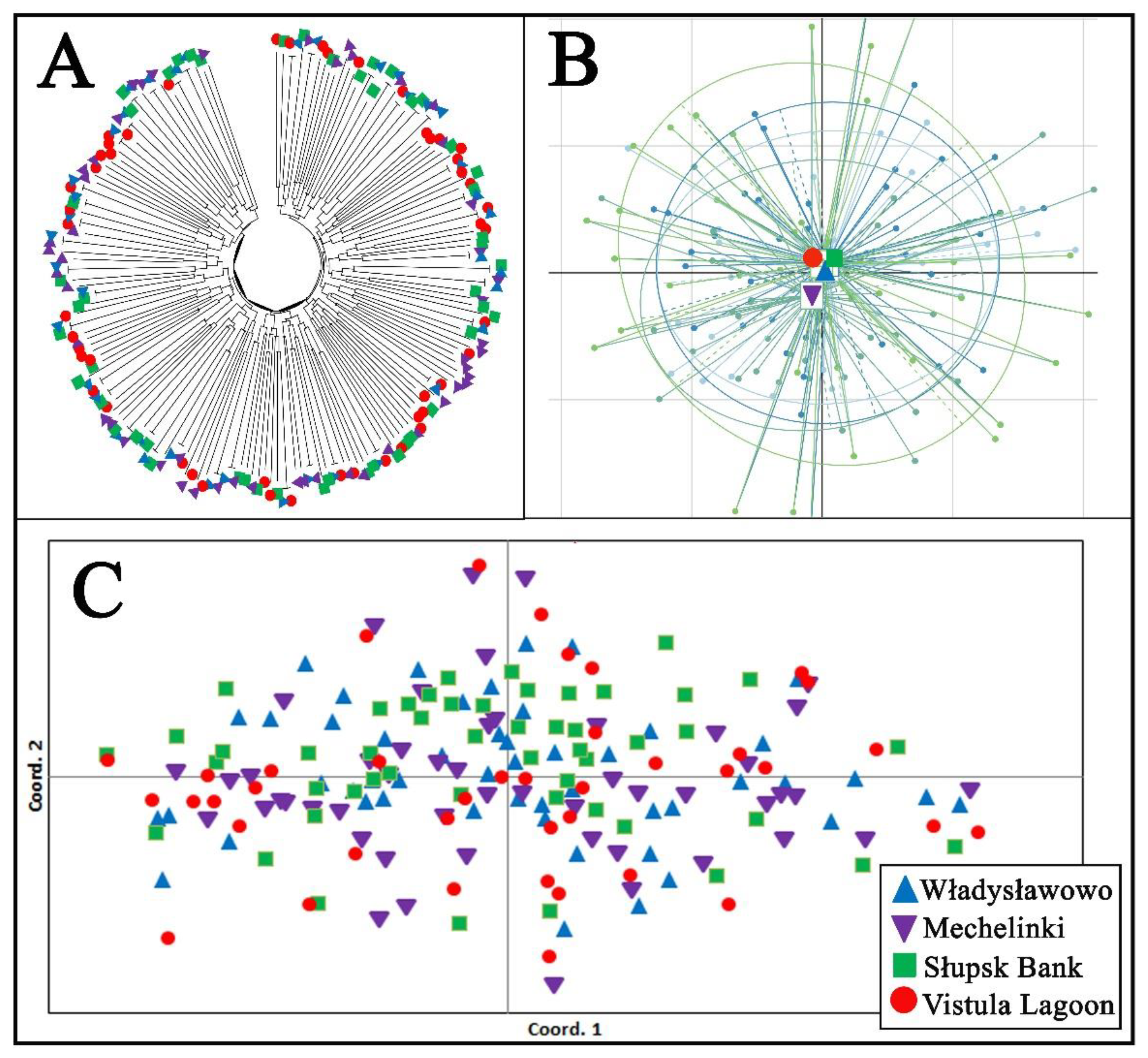
The AMOVA analysis revealed that most of the genetic variability occurred within individuals (93.6%), while only 0.04% occurred between fish groups originating from each sampling site. Similarly, the pairwise Fst genetic differentiation between the examined groups of fish from each sampling site varied from 0.001 to 0.003, while the global test of differentiation revealed an insignificant value (Fst = 0.004, p > 0.05). The individual assignments test showed a low level of fish classification correctness for each sampling location, ranging from 26% (Władysławowo) to 40% (Słupsk Bank) (quality index = 31.79%). The resolved individual’s tree based on DAS genetic distances, PCoA, and PCA analyses revealed that all the examined fish formed one common genetic cluster without any signs of significant genetic differentiation (Figure 2). Similarly, the results for the individual multilocus genotype based on Bayesian analyses and Flock software did not identify any signs of genetic clustering in the examined fish. The results of the diagnostic SNP marker analysis did not detect any signs of Baltic flounder presence among the studied fish (Supplementary Table S5).
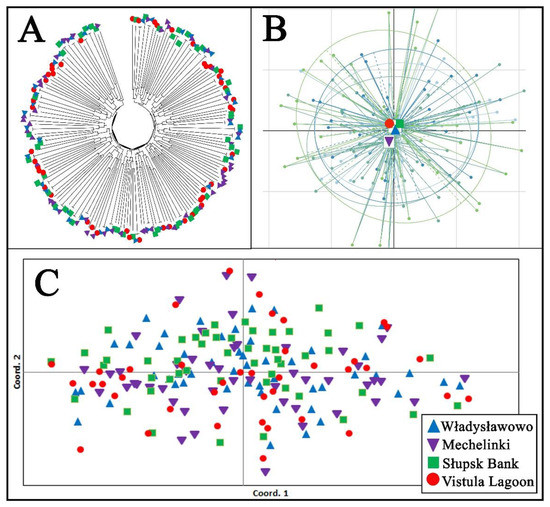
Figure 2.
Genetic relationship among examined fish, estimated by (A) the unrooted neighbor-joining tree of individuals based on allele sharing distances (DAS), (B) scatter plot of the principal component analysis (PCA), and (C) principal coordinate analysis (PCoA), based on individual pairwise genetic distances.
The estimated effective population size (Ne) for all examined fish equaled Ne = 9101 (95% CI = 3425-infinite, Pcrit = 0.05). In the fish sampled from Mechelinki, Władysławowo, and the Vistula Lagoon in 2018, the estimated value of Ne equaled Ne = 10115 (95% CI = 2113-infinite, Pcrit = 0.05). In the fish sampled from the Słupsk Bank in 2020, the estimated value of the indicator was considerably lower, i.e., Ne = 712 (95% CI = 207-infinite, Pcrit = 0.05). Significant (p > 0.05) He > Heq differences were recorded only under the infinite alleles model (IAM) and the two-phase mutation model (TPM), where nine loci (FLAG2-76, FLAG4-65, Stpf1002, Stpf1006, Stpf1015, Stpf1016, PL142, PL167, and FLAC4-69) exhibited significant heterozygosity excess (Supplementary Table S6). The two-tail Wilcoxon test detected a significant (p < 0.05) overall He > Heq excess only under the IAM mutation model. The mean values of the M-ratio in all examined fish groups were similar, ranging from 0.574 to 0.647. The lowest values of this indicator (M < 0.5) were recorded for FLAG5-83, FLAG4-65, FLAG8-37, Nplaf-33, Stpf1001, Stpf1002, Stpf1015, Stpf1016, and FLAC4-69 (Table 2).

Table 2.
Results of the Bottleneck analysis. Comparison of expected heterozygosity (He) vs. heterozygosity (Heq) expected under the infinite alleles model (IAM). Stepwise mutation model (SMM) and two-phase model of mutation (TPM) in examined European flounder (Platichthys flesus) from the Baltic Sea. M: Garza–Williamson Index and significant values of the Wilcoxon test for He > Heq are bolded (p < 0.05).
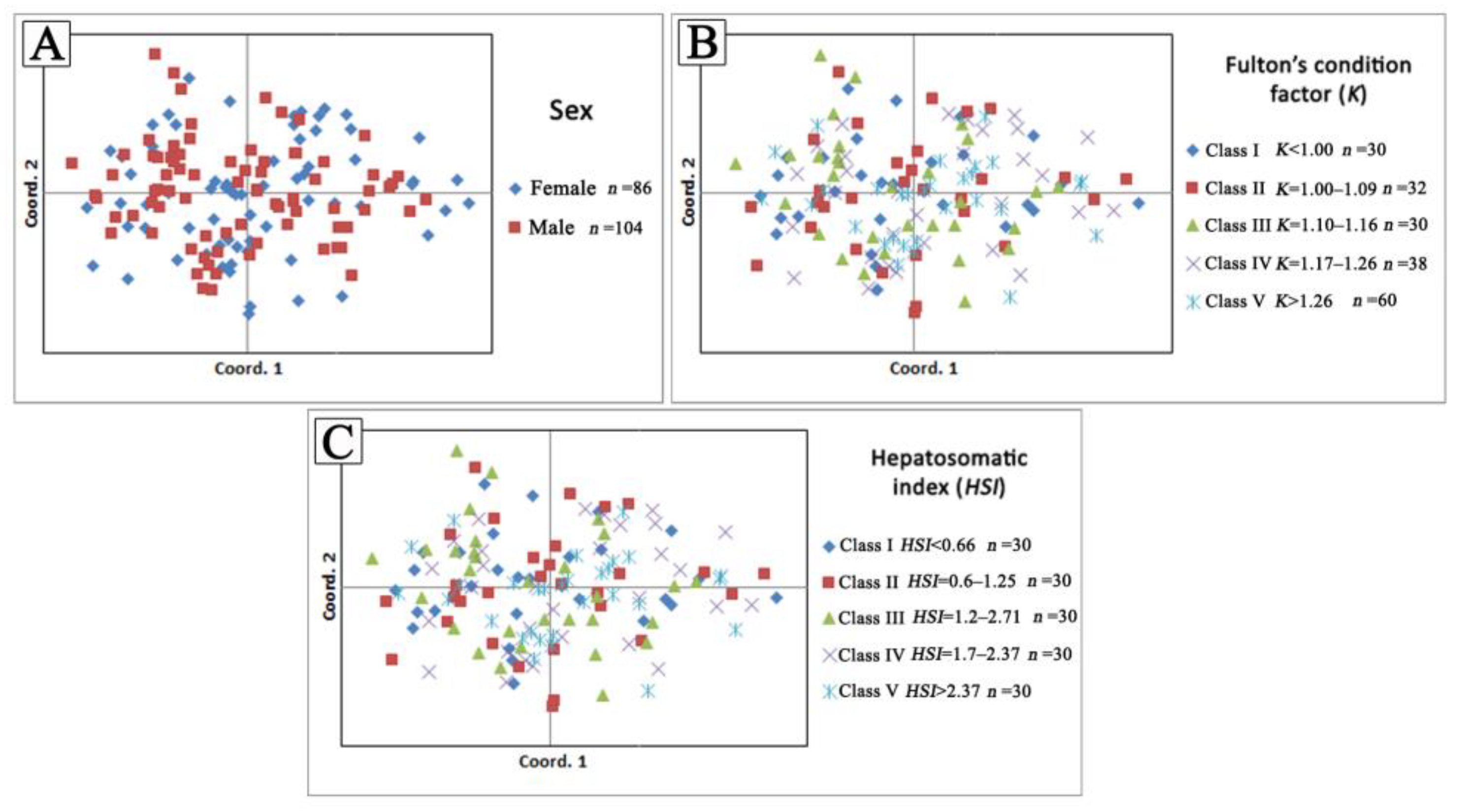
The Beaumont and Nichols’ method for the selection of signatures implemented using Arlequin software [24] did not detect any microsatellite loci, with outlier Fst values at the 95% confidence level in the examined fish groups. The performed statistical tests did not detect any significant differences (p > 0.05) in estimated genetic parameters among the predefined series of fish groups according to sex, the values of Fulton’s condition factor, and hepatosomatic index body condition indices in all comparisons. The performed multidimensional analyses showed no association between genetic parameters and biological characteristics in the examined fish (Figure 3).
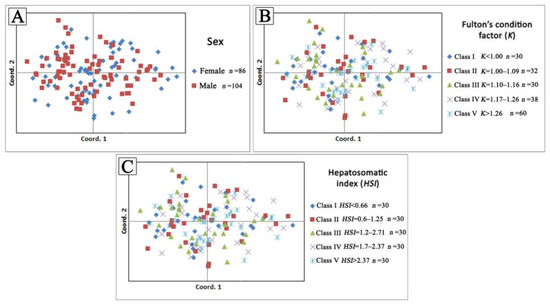
Figure 3.
Principal coordinate analysis (PCoA) based on individual pairwise genetic distances between each predefined fish group based on (A) sex, (B) Fulton’s condition factor (K), and (C) Hepatosomatic index (HSI) values.
4. Discussion
Marine ecosystems are dynamic complexes of biological, environmental, and anthropogenic factors such as life history, demography, inter-generic interactions, hydrology, climate, pollution, and fisheries, all of which shape the genetic diversity and structure of marine fish populations [44,45]. Selective pressures associated with environmental and anthropogenic factors may lead to the loss of genetic diversity in fish. This decreases their adaptation abilities to changing ecological conditions, making them more vulnerable to environmental stressors, disease, and other threats [46,47,48]. Moreover, in severely overexploited populations, the fixation of harmful alleles through genetic drift may decrease population viability, increasing the risk of extinction [44,46]. Thus, spatiotemporal genetic-based monitoring of marine resources is an important tool in sustainable management, providing information on the species/population’s past and current life history, degree of relationship and patterns of genetic diversity, and population structure [5,49,50,51].
The results of our study show that despite a significant increase in fishing pressure on the European flounder in the Baltic Sea over the last 20 years, the recorded values of genetic variability indicators (Ao = 17.9–19.4, PIC = 0.832–0.903, I = 2.579–2.768) in the flounder from all sampling sites were high, and comparable with data obtained over a decade ago from fish sampled in other parts of the Baltic Sea [15,16]. This temporal stability in the levels of estimated genetic variability parameters in the examined species might be caused by the large gene flow between flounder from the Baltic and the North Sea, as previously hypothesized by other researchers [15,16,52]. A similar mechanism of genetic diversity stabilization over time has also been reported in other heavily exploited marine fish species [53,54]. The estimated levels of genetic variability in the fish examined in our study were high and comparable with those reported in many other flounder species [55,56,57,58]. This suggests that the currently observed decline in the body condition indices of the European flounder in the Baltic Sea might not be directly associated with the reduction of the species’ genetic variability but rather, with the qualitative changes in the gene pool caused by directional selection pressure associated with environmental changes and intensive fishery exploitation [12]. However, the applied method for loci selection signatures and the correlation analysis between genetic and biological traits did not reveal signs of directional selection or density-dependent adaptative changes in the gene pool of the European flounder specimens examined in this study. The lack of a significant correlation between the patterns of genetic diversity and the reduced body condition indices of fish was also reported by Poćwierz-Kotus et al. [59] in Atlantic cod from the southern Baltic Sea.
The available studies indicate that the genetic structure of flounder populations along the European coastline from the Bay of Biscay to northern Norway is mainly shaped by the latitudinal temperature gradient; however, factors such as migratory behavior, the strict association of juveniles with estuaries or freshwater habitats, and the adaptive response to environmental pollution may also play an important role in shaping the genetic structure of the species across its distribution range [10,52,60,61,62]. To date, genetic population studies on the European flounder have reported low (Fst = 0.005–0.090) but significant and stable levels of genetic differentiation, supporting the existence of at least eight genetic clusters of the species in Europe, i.e., (1) Iberian Peninsula, (2) Bay of Biscay, (3) English Channel, (4) North Sea and Irish Sea, (5) Baltic Sea, (6) Faroe Islands, (7) central coast of Norway, and (8) northern coast of Norway [15,16,52,63,64]. The genetic analyses performed in our study did not detect any signs of significant genetic structure in any the studied fish from the sampled areas. Similar levels of genetic homogeneity (Fst = 0.005–0.015) have been reported among European flounder populations in the North Sea and Irish Sea [15,16]. The results of the diagnostic SNP marker analysis did not find any signs of Baltic flounder presence among the studied fish. This may suggest that the observed decline in the body condition indices of the European flounder in the Baltic Sea is not associated with the presence and competition for food and space with the Baltic flounder in the studied area. Nevertheless, the lack of any signs of Baltic flounder in the Vistula Lagoon, which theoretically provides suitable conditions for this species, is intriguing and suggests the presence of an unknown environmental barrier that prevents Baltic flounder from entering the southern Baltic Sea. Existing biological information, together with the age analysis of the fish sampled in this study, suggest that the Vistula Lagoon serves as an important nursery ground for juvenile European flounder [65].
In contrast to freshwater fish species, the estimated effective population size (Ne) of marine fish populations is frequently several times lower than census data. This makes them particularly vulnerable to the loss of genetic diversity due to population size bottlenecks caused by the impact of increased fishing pressure or changes in the environment [46,47,48,66,67,68,69]. Thus, estimating and tracking effective population size is a powerful tool in the conservation genetics of marine fish insofar as it can be used to determine fish population/stock susceptibility to the loss of genetic integrity. The estimated effective population size (Ne) of fish from Władysławowo, Mechelinki, and the Vistula Lagoon sampled in 2018 was high, and comparable with the previously estimated values of Ne for flounder populations in the North and Irish Seas [15]. Moreover, the estimated values of Ne for fish sampled from the Słupsk Bank in 2020 displayed visibly lower values of this indicator, i.e., Ne = 712. This finding might indicate a change in the population size of European flounder in the southern Baltic Sea. However, this conclusion is very speculative and requires further investigation. Interestingly, a similar number of differences in Ne values was reported in overexploited plaice populations from Iceland (Ne = c.a. 2000) and landlocked populations of Atlantic cod from Mogilnoe Lake (Ne = c.a. 200), where the decrease in this parameter was assumed to be accelerated by the fish non-random mating and the associated cryptic mating structure [48,70].
The uncoupling between observed heterozygosity (Ho) and expected heterozygosity (He) may indicate the presence of a recent demographic disturbance [68,69,70,71]. Numerous studies demonstrate that significant heterozygosity deficiency in marine fish may be associated with population size changes, inbreeding, the Wahlund effect, homoplasy, or selection [72,73,74]. In this study, the fish group from the Słupsk Bank was not in the Hardy–Weinberg equilibrium, which is similar to results reported a decade ago for European flounder sampled from other locations in the Baltic Sea [15,16,52]. The recorded values of the fixation index were the highest in fish sampled from the Słupsk Bank in 2020 (Fis = 0.093, p < 0.05), suggesting a recent reduction in population size in the sampled area. However, the recorded values of the Garza–Williamson indicator (M = 0.574–0.600) and the lack of significant differences of Heq > He under the SMM model do not support this conclusion [75]. Moreover, null alleles are known to be a frequent cause-factor that may be responsible for observed heterozygosity deficits. However, in the present study, all the algorithms used in the Micro-Checker software to estimate null alleles excluded this eventuality, as the recorded levels of excess homozygotes greatly exceed the estimated frequencies of possible null alleles. The lack of significant genetic differentiation among the examined fish groups seems to exclude the possibility of the Wahlund effect, which is congruent with the conclusions of Hemmer-Hansen et al. [15]. Thus, a more comprehensive study that includes series of retrospective cohort-labeled samples is required to confirm the species population size dynamics observed in the present study and to track the population over time.
5. Conclusions
In the present study, the recorded levels of genetic diversity among the studied European flounder from the southern Baltic Sea were high and temporally stable in relation to the data published about a decade ago. Moreover, the recorded levels of homozygosity were similar to previously reported genetic data. The results indicate that the European flounder from the southern Baltic Sea represent a single and homogenous genetic cluster. Significant differences in the effective population sizes (Ne) between fish sampled in 2018 and those sampled in 2020 might indicate recent changes in the population size of the European flounder in the southern Baltic Sea. However, this conclusion is speculative and requires further studies to be verified. The results do not support the hypothesis of adaptive genetic changes in the flounder’s gene pool toward a limited growth rate, body size, and/or reduced population body condition indices caused by fishery-mediated heavy selective pressure on the species’ populations in the southern Baltic Sea. Our study did not detect the presence of the Baltic flounder in the southern Baltic Sea. Broader geographic and time scale research based on more sensitive genetic analysis methods, such as whole genome sequencing or gene expression, is required to provide more detailed data on the European flounder’s status in the Baltic Sea.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ani13091448/s1. Supplementary Table S1: Summary of biological and morphological data used in the present study; Supplementary Table S2: Characterization of ten microsatellite loci used in the present study; Supplementary Table S3: Diagram of fish division on series of separate groups according to sex, age, calculated values of Fulton’s condition factor (K), and hepatosomatic index (HSI) to analyze the association between biological traits and patterns of genetic variability; Supplementary Table S4: Genetic diversity parameters of the European flounder (Platichthys flesus) individuals in the current study; Supplementary Table S5: Genotyping results of ten randomly selected European flounder from each sampled area located in the southern Baltic Sea using two diagnostic SNP markers based on data published by Momigliano et al. [9]; and Supplementary Table S6: Results of the Bottleneck analysis.
Author Contributions
Conceptualization, M.K., M.J.-L., A.G., Z.M., K.N.-A., J.S.-R. and K.O.; methodology, M.K.; software, M.K.; investigation, M.K.; writing—original draft preparation, M.K.; writing—review and editing, M.J.-L., A.G., Z.M., K.N.-A., J.S.-R. and K.O.; visualization, M.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by statutory funding provided by the Ministry of Science and Higher Education in Poland (Grant number [DS 531-O110-D423-23]).
Institutional Review Board Statement
The current study was carried out in strict accordance with the recommendations of the Polish ACT of 15 January 2015 on Animal Experiments (Dz. U. of. 2015 No 33, item 266) and special permission is not required by local law to carry out the experiments. However, this research has got the ap-proval from the Local Ethics Committee for Animal Experimentation in Bydgoszcz (Poland) (No 14/2022, issued on 24.04.2022) as a part of permission for wider scope of research.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data presented in the manuscript are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.
References
- Zander, C.D.; Whitehead, P.J.P.; Bauchot, M.L.; Hureau, J.C.; Nielsen, J.; Tortonese, E. Fishes of the North-Eastern Atlantic and the Mediterranean; United Nations Educational Scientific and Cultural Organization: Paris, France, 1986; Volume 3. [Google Scholar]
- Haase, K.; Orio, A.; Pawlak, J.; Pachur, M.; Casini, M. Diet of dominant demersal fish species in the Baltic Sea: Is flounder stealing benthic food from cod? Mar. Ecol. Prog. Ser. 2020, 645, 159–170. [Google Scholar] [CrossRef]
- Florin, A.B. Flatfishes in the Baltic Sea. A review of biology and fishery with a focus on Swedish conditions. Finfo 2005, 14, 1–45. [Google Scholar]
- Nissling, A.; Nyberg, S.; Petereit, C. Egg buoyancy of flounder, Platichthys flesus, in the Baltic Sea—Adaptation to salinity and implications for egg survival. Fish. Res. 2017, 191, 179–189. [Google Scholar] [CrossRef]
- Jokinen, H.; Momigliano, P.; Merilä, J. From ecology to genetics and back: The tale of two flounder species in the Baltic Sea. ICES J. Mar. Sci. 2019, 76, 2267–2275. [Google Scholar] [CrossRef]
- Momigliano, P.; Denys, G.P.J.; Jokinen, H.; Merilä, J. Platichthys solemdali sp. nov. (Actinopterygii, Pleuronectiformes): A New Flounder Species From the Baltic Sea. Front. Mar. Sci. 2018, 5, 225. [Google Scholar] [CrossRef]
- FAO. Food and Agriculture Organization of the United Nations. 2022. Available online: http://www.fao.org/fishery/about/en (accessed on 1 August 2022).
- ICES Baltic Fisheries Assessment Working Group (WGBFAS). ICES Scientific Reports; ICES Baltic Fisheries Assessment Working Group (WGBFAS): Storr-Paulsen, Denmark, 2022; Volume 4, 659p. [Google Scholar] [CrossRef]
- Momigliano, P.; Jokinen, H.; Calboli, F.; Aro, E.; Merilä, J. Cryptic temporal changes in stock composition explain the decline of a flounder (Platichthys spp.) assemblage. Evol. Appl. 2018, 12, 549–559. [Google Scholar] [CrossRef]
- Hemmer-Hansen, J.; Nielsen, E.E.; Frydenberg, J.; Loeschcke, V. Adaptive divergence in a high gene flow environment: Hsc70 variation in the European flounder (Platichthys flesus L.). Heredity 2007, 99, 592–600. [Google Scholar] [CrossRef]
- Larsen, P.F.; Nielsen, E.E.; Williams, T.; Hemmer-Hansen, J.; Chipman, J.K.; Kruhøffer, M.; Grønkjær, P.; George, S.G.; Dyrskjøt, L.; Loeschcke, V. Adaptive differences in gene expression in European flounder (Platichthys flesus). Mol. Ecol. 2007, 16, 4674–4683. [Google Scholar] [CrossRef]
- Svedäng, H.; Hornborg, S. Selective fishing induces density-dependent growth. Nat. Commun. 2014, 5, 4152. [Google Scholar] [CrossRef]
- NMFRI. A Study Programme on the Marine Environment of the Puck Bay, with Particular Emphasis on Factors Relevant to Fisheries in 2019–2021; NMFRI: Gdynia, Poland, 2020. [Google Scholar]
- Hemmer-Hansen, J.; Nielsen, E.E.; Grønkjaer, P.; Loeschcke, V. Evolutionary mechanisms shaping the genetic population structure of marine fishes; lessons from the European flounder (Platichthys flesus L.). Mol. Ecol. 2007, 16, 3104–3118. [Google Scholar] [CrossRef]
- Florin, A.-B.; Höglund, J. Population structure of flounder (Platichthys flesus) in the Baltic Sea: Differences among demersal and pelagic spawners. Heredity 2008, 101, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Tysklind, N.; Neuparth, T.; Ashcroft, G.R.; Taylor, M.I.; Lyons, B.P.; McCarthy, I.D.; Carvalho, G.R. Isolation and characterization of 28 new microsatellite markers for European flounder (Platichthys flesus L.). Mol. Ecol. Resour. 2009, 9, 1065–1068. [Google Scholar] [CrossRef] [PubMed]
- Pédron, N.; Morvezen, R.; Le Moan, A.; Guinand, B.; Zambonino-Infante, J.-L.; Laroche, J.; Charrier, G. New set of candidate gene SNPs and microsatellites to disentangle selective and neutral processes shaping population responses of European flounder (Platichthys flesus) to anthropogenic stress and contrasted environments. Conserv. Genet. Resour. 2015, 7, 823–826. [Google Scholar] [CrossRef]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Nakano, S.-I.; Fujimoto, M.; Hara, H.; Sugimoto, N. Nucleic acid duplex stability: Influence of base composition on cation effects. Nucleic Acids Res. 1999, 27, 2957–2965. [Google Scholar] [CrossRef] [PubMed]
- Momigliano, P.; Jokinen, H.; Fraimout, A.; Florin, A.-B.; Norkko, A.; Merilä, J. Extraordinarily rapid speciation in a marine fish. Proc. Natl. Acad. Sci. USA 2017, 114, 6074–6079. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Peakall, R.O.D.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Yeh, F.C.; Boylet, J.B. Population genetic analysis of codominant and dominant markers and quantitative traits. Belg. J. Bot. 1997, 129, 157. [Google Scholar]
- Rousset, F. GenePop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Rice, W.R. Analyzing tables of statistical tests. Evolution 1989, 43, 223. [Google Scholar] [CrossRef] [PubMed]
- Piry, S.; Alapetite, A.; Cornuet, J.-M.; Paetkau, D.; Baudouin, L.; Estoup, A. GeneClass2: A Software for Genetic Assignment and First-Generation Migrant Detection. J. Hered. 2004, 95, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T.; Manlove, K.R.; Taper, M.L. ONCOR: A Computer Program for Genetic Stock Identification; Department of Ecology, Montana State University: Bozeman, MT, USA, 2007; Available online: https://www.montana.edu/kalinowski/software/oncor.html (accessed on 15 June 2022).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Duchesne, P.; Turgeon, J. FLOCK provides reliable solutions to the “number of populations” problem. J. Hered. 2012, 103, 734–743. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Langella, O. Populations 1.2.28. Logiciel de Génétique des Populations. Laboratoire Populations, Génétique et Évolution, CNRS UPR 9034, Gif-sur-Yvette. Available online: http://bioinformatics.org/~tryphon/populations/#ancre_bibliographie (accessed on 15 May 2022).
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimatorv2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour. 2013, 14, 209–214. [Google Scholar] [CrossRef]
- Waples, R.S.; Do, C. Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: A largely untapped resource for applied conservation and evolution. Evol. Appl. 2009, 3, 244–262. [Google Scholar] [CrossRef] [PubMed]
- Piry, S.; Luikart, G.; Cornuet, J.-M. Bottleneck: A computer program for detecting recent reductions in effective population size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Costopoulos, C.; Fonds, M. Proximate body composition and energy content of plaice (Pleuronectes platessa) in relation to the condition factor. Neth. J. Sea Res. 1989, 24, 45–55. [Google Scholar] [CrossRef]
- Giguère, A.; Campbell, P.G.; Hare, L.; McDonald, D.G.; Rasmussen, J.B. Influence of lake chemistry and fish age on cadmium, copper, and zinc concentrations in various organs of indigenous yellow perch (Perca flavescens). Can. J. Fish. Aquat. Sci. 2004, 61, 1702–1716. [Google Scholar] [CrossRef]
- Chapuis, M.-P.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2006, 24, 621–631. [Google Scholar] [CrossRef]
- Huang, K.; Ritland, K.; Dunn, D.W.; Qi, X.; Guo, S.; Li, B. Estimating relatedness in the presence of null alleles. Genetics 2015, 202, 247–260. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; David, A.; Briscoe, D.A. Introduction to Conservation Genetics, 2nd ed.; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Bonanomi, S.; Pellissier, L.; Therkildsen, N.O.; Hedeholm, R.B.; Retzel, A.; Meldrup, D.; Olsen, S.M.; Nielsen, A.; Pampoulie, C.; Hemmer-Hansen, J.; et al. Archived DNA reveals fisheries and climate induced collapse of a major fishery. Sci. Rep. 2015, 5, 15395. [Google Scholar] [CrossRef]
- Hauser, L.; Adcock, G.J.; Smith, P.J.; Ramírez, J.H.B.; Carvalho, G.R. Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper (Pagrus auratus). Proc. Natl. Acad. Sci. USA 2002, 99, 11742–11747. [Google Scholar] [CrossRef]
- Hutchinson, W.F.; Van Oosterhout, C.; Rogers, S.I.; Carvalho, G.R. Temporal analysis of archived samples indicates marked genetic changes in declining North Sea cod (Gadus morhua). Proc. R. Soc. B Boil. Sci. 2003, 270, 2125–2132. [Google Scholar] [CrossRef]
- Hoarau, G.; Boon, E.; Jongma, D.N.; Ferber, S.; Palsson, J.; Van der Veer, H.W.; Rijnsdorp, A.D.; Stam, W.T.; Olsen, J.L. Low effective population size and evidence for inbreeding in an overexploited flatfish, plaice (Pleuronectes platessa L.). Proc. R. Soc. B Boil. Sci. 2005, 272, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Sterner, T. Unobserved diversity, depletion and irreversibility The importance of subpopulations for management of cod stocks. Ecol. Econ. 2007, 61, 566–574. [Google Scholar] [CrossRef]
- Hutchinson, W.F. The dangers of ignoring stock complexity in fishery management: The case of the North Sea cod. Biol. Lett. 2008, 4, 693–695. [Google Scholar] [CrossRef] [PubMed]
- Lindegren, M.; Waldo, S.; Nilsson, P.A.; Svedäng, H.; Persson, A. Towards sustainable fisheries of the Öresund cod (Gadus morhua) through sub-stock-specific assessment and management recommendations. ICES J. Mar. Sci. 2013, 70, 1140–1150. [Google Scholar] [CrossRef]
- Calvès, I.; Lavergne, E.; Meistertzheim, A.; Charrier, G.; Cabral, H.; Guinand, B.; Quiniou, L.; Laroche, J. Genetic structure of European flounder Platichthys flesus: Effects of both the southern limit of the species’ range and chemical stress. Mar. Ecol. Prog. Ser. 2013, 472, 257–273. [Google Scholar] [CrossRef]
- Larsson, L.C.; Laikre, L.; André, C.; Dahlgren, T.G.; Ryman, N. Temporally stable genetic structure of heavily exploited Atlantic herring (Clupea harengus) in Swedish waters. Heredity 2009, 104, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Cuveliers, E.L.; Volckaert, F.A.M.; Rijnsdorp, A.D.; Larmuseau, M.H.D.; Maes, G.E. Temporal genetic stability and high effective population size despite fisheries-induced life-history trait evolution in the North Sea sole. Mol. Ecol. 2011, 20, 3555–3568. [Google Scholar] [CrossRef] [PubMed]
- Sekino, M.; Hara, M.; Taniguchi, N. Loss of microsatellite and mitochondrial DNA variation in hatchery strains of Japanese flounder Paralichthys olivaceus. Aquaculture 2002, 213, 101–122. [Google Scholar] [CrossRef]
- Xu, D.; Li, S.; Lou, B.; Zhang, Y.; Zhan, W.; Shi, H. Genetic diversity in two Japanese flounder populations from China seas inferred using microsatellite markers and COI sequences. Chin. J. Oceanol. Limnol. 2012, 30, 604–610. [Google Scholar] [CrossRef]
- An, H.S.; Nam, M.M.; Myeong, J.I.; An, C.M. Genetic diversity and differentiation of the Korean starry flounder (Platichthys stellatus) between and within cultured stocks and wild populations inferred from microsatellite DNA analysis. Mol. Biol. Rep. 2014, 41, 7281–7292. [Google Scholar] [CrossRef]
- Pan, T.; Zhang, Y.; Gao, T.; Li, F. Genetic diversity of Pleuronectes yokohamae population revealed by fluorescence microsatellite labeled. Biochem. Syst. Ecol. 2014, 55, 118–124. [Google Scholar] [CrossRef]
- Poćwierz-Kotus, A.; Kijewska, A.; Petereit, C.; Bernaś, R.; Więcaszek, B.; Arnyasi, M.; Lien, S.; Kent, M.; Wenne, R. Genetic differentiation of brackish water populations of cod Gadus morhua in the southern Baltic, inferred from genotyping using SNP-arrays. Mar. Genom. 2015, 19, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Olsen, E.M.; Knutsen, H.; Gjøsaeter, J.; Jorde, P.E.; Knutsen, J.A.; Stenseth, N.C. Small-scale biocomplexity in coastal Atlantic cod supporting a Darwinian perspective on fisheries management. Evol. Appl. 2008, 1, 524–533. [Google Scholar] [CrossRef]
- Hermant, M.; Lobry, J.; Bonhommeau, S.; Poulard, J.-C.; Le Pape, O. Impact of warming on abundance and occurrence of flatfish populations in the Bay of Biscay (France). J. Sea Res. 2010, 64, 45–53. [Google Scholar] [CrossRef]
- Marchand, J.; Evrard, E.; Guinand, B.; Cachot, J.; Quiniou, L.; Laroche, J. Genetic polymorphism and its potential relation to environmental stress in five populations of the European flounder Platichthys flesus, along the French Atlantic coast. Mar. Environ. Res. 2010, 70, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Borsa, P.; Blanquer, A.; Berrebi, P. Genetic structure of the flounders Platichthys flesus and P. stellatus at different geographic scales. Mar. Biol. 1997, 129, 233–246. [Google Scholar] [CrossRef]
- Pédron, N.; Laroche, J.; Aboim, M.A.; Tanner, S.E.; Reis-Santos, P.; Vasconcelos, R.P. Genetic variation across the geographical range of the European flounder, with a focus on southern peripheral populations. Remerciements 2016, 51. Available online: https://theses.hal.science/tel-02928183 (accessed on 28 February 2023).
- Psuty, I.; Wilkońska, H. The stability of fish assemblages under unstable conditions: A ten-year series from the Polish part of the Vistula Lagoon. Fish Aquat. Sci. 2009, 17, 65–76. [Google Scholar] [CrossRef]
- Turner, T.F.; Wares, J.P.; Gold, J.R. Genetic effective size is three orders of magnitude smaller than adult census size in an abundant, estuarine-dependent marine fish (Sciaenops ocellatus). Genetics 2002, 162, 1329–1339. [Google Scholar] [CrossRef]
- Hauser, L.; Carvalho, G.R. Paradigm shifts in marine fisheries genetics: Ugly hypotheses slain by beautiful facts. Fish 2008, 9, 333–362. [Google Scholar] [CrossRef]
- Ruggeri, P.; Splendiani, A.; Bonanomi, S.; Arneri, E.; Cingolani, N.; Santojanni, A.; Belardinelli, A.; Giovannotti, M.; Caputo, V. Temporal genetic variation as revealed by a microsatellite analysis of European sardine (Sardina pilchardus) archived samples. Can. J. Fish. Aquat. Sci. 2012, 69, 1698–1709. [Google Scholar] [CrossRef]
- Ruggeri, P.; Splendiani, A.; Di Muri, C.; Fioravanti, T.; Santojanni, A.; Leonori, I.; De Felice, A.; Biagiotti, I.; Carpi, P.; Arneri, E.; et al. Coupling demographic and genetic variability from archived collections of European anchovy (Engraulis encrasicolus). PLoS ONE 2016, 11, e0151507. [Google Scholar] [CrossRef]
- Zhivotovsky, L.A.; Teterina, A.A.; Mukhina, N.V.; Stroganov, A.N.; Rubtsova, G.A.; Afanasiev, K.I. Effects of genetic drift in a small population of Atlantic cod (Gadus morhua kildinensis Derjugin) landlocked in a meromictic lake: Genetic variation and conservation measures. Conserv. Genet. 2015, 17, 229–238. [Google Scholar] [CrossRef]
- Leberg, P.L. Effects of population bottlenecks on genetic diversity as measured by allozyme electrophoresis. Evolution 1992, 46, 477–494. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.; Wright, J.M. Microsatellite DNA in fishes. Rev. Fish Biol. Fish. 1997, 7, 331–363. [Google Scholar] [CrossRef]
- Karlsson, S.; Mork, J. Deviation from Hardy–Weinberg equilibrium, and temporal instability in allele frequencies at microsatellite loci in a local population of Atlantic cod. ICES J. Mar. Sci. 2005, 62, 1588–1596. [Google Scholar] [CrossRef]
- Atarhouch, T.; Rüber, L.; Gonzalez, E.G.; Albert, E.M.; Rami, M.; Dakkak, A.; Zardoya, R. Signature of an early genetic bottleneck in a population of Moroccan sardines (Sardina pilchardus). Mol. Phylogenet. Evol. 2006, 39, 373–383. [Google Scholar] [CrossRef]
- Tzika, A.C.; Rosa, S.F.P.; Fabiani, A.; Snell, H.L.; Snell, H.M.; Marquez, C.; Tapia, W.; Rassmann, K.; Gentile, G.; Milinkovitch, M.C. Population genetics of Galápagos land iguana (genus Conolophus) remnant populations. Mol. Ecol. 2008, 17, 4943–4952. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).