Kompetitive Allele Specific PCR Genotyping of 89 SNPs in Romanian Spotted and Romanian Brown Cattle Breeds and Their Association with Clinical Mastitis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Animals and General Management
2.3. Data Collection
2.4. Selection of SNPs
2.5. DNA Extraction and Genotyping
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seegers, H.; Fourichon, C.; Beaudeau, F. Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet. Res. 2003, 34, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Halasa, T.; Huijps, K.; Østerås, O.; Hogeveen, H. Economic effects of bovine mastitis and mastitis management: A review. Vet. Q. 2007, 29, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Molano, E.; Bay, V.; Smith, R.F.; Oikonomou, G.; Banos, G. Quantitative Trait Loci Mapping for Lameness Associated Phenotypes in Holstein-Friesian Dairy Cattle. Front. Genet. 2019, 10, 926. [Google Scholar] [CrossRef]
- Oltenacu, P.A.; Broom, D.M. The impact of genetic selection for increased milk yield on the welfare of dairy cows. Anim. Welf. 2010, 19, 39–49. [Google Scholar] [CrossRef]
- Kadri, N.K.; Guldbrandtsen, B.; Lund, M.S.; Sahana, G. Genetic dissection of milk yield traits and mastitis resistance quantitative trait loci on chromosome 20 in dairy cattle. J. Dairy Sci. 2015, 98, 9015–9025. [Google Scholar] [CrossRef] [PubMed]
- Moretti, R.; Soglia, D.; Chessa, S.; Sartore, S.; Finocchiaro, R.; Rasero, R.; Sacchi, P. Identification of SNPs Associated with Somatic Cell Score in Candidate Genes in Italian Holstein Friesian Bulls. Animals. 2021, 11, 366. [Google Scholar] [CrossRef]
- Pawlik, A.; Sender, G.; Sobczyńska, M.; Korwin-Kossakowska, A.; Lassa, H.; Oprządek, J. Lactoferrin gene variants, their expression in the udder and mastitis susceptibility in dairy cattle. Anim. Prod. Sci. 2014, 55, 999–1004. [Google Scholar] [CrossRef]
- Sharma, P.; Parmar, S.N.S.; Thakur, M.S.; Nauriyal, D.S.; Ranjan, R. Association of bovine lactoferrin gene with mastitis in frieswal cattle. Iran. J. Appl. Anim. Sci. 2015, 5, 859–863. [Google Scholar]
- Wang, X.P.; Xu, S.Z.; Gao, X.; Li, J.Y.; Ren, H.Y.; Luoren, Z.M. Cloning and SNP screening of the TLR4 gene and the association between its polymorphism and somatic cell score in dairy cattle. S. Afr. J. Anim. Sci. 2008, 38, 101–109. [Google Scholar]
- Mesquita, A.Q.; Rezende, C.S.E.; Mesquita, A.J.; Jardim, E.A.; Kipnis, A.P. Association of TLR4 polymorphisms with subclinical mastitis in Brazilian holsteins. Braz. J. Microbiol. 2012, 43, 692–697. [Google Scholar] [CrossRef]
- Gupta, P.H.; Patel, N.A.; Rank, D.N.; Joshi, C.G. Genetic polymorphism of toll-like receptors 4 gene by polymerase chain reaction-restriction fragment length polymorphisms, polymerase chain reaction-single-strand conformational polymorphism to correlate with mastitic cows. Vet. World. 2015, 8, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Youngerman, S.M.; Saxton, A.M.; Oliver, S.P.; Pighetti, G.M. Association of CXCR2 polymorphisms with subclinical and clinical mastitis in dairy cattle. J. Dairy Sci. 2004, 87, 2442–2448. [Google Scholar] [CrossRef] [PubMed]
- Magotra, A.; Gupta, I.D.; Ahmad, T.; Alex, R. Polymorphism in DNA repair gene BRCA1 associated with clinical mastitis and production traits in indigenous dairy cattle. Res. Vet. Sci. 2020, 133, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Kell, D.B.; Heyden, E.L.; Pretorius, E. The Biology of Lactoferrin, an Iron-Binding Protein That Can Help Defend Against Viruses and Bacteria. Front. Immunol. 2020, 11, 1221. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Song, H.; Zhu, X.; Xing, S.; Zhang, M.; Zhang, H.; Wang, X.; Yang, Z.; Ding, X.; Karrow, N.A.; et al. Toll-like receptor 4 gene polymorphisms influence milk production traits in Chinese Holstein cows. J. Dairy Res. 2018, 85, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Russo, R.C.; Garcia, C.C.; Teixeira, M.M.; Amaral, F.A. The CXCL8/IL-8 chemokine family and its receptors in inflammatory diseases. Expert Rev. Clin. Immunol. 2014, 10, 593–619. [Google Scholar] [CrossRef]
- Bohórquez, M.D.; Ordoñez, D.; Suárez, C.F.; Vicente, B.; Vieira, C.; López-Abán, J.; Muro, A.; Ordóñez, I.; Patarroyo, M.A. Major Histocompatibility Complex Class II (DRB3) Genetic Diversity in Spanish Morucha and Colombian Normande Cattle Compared to Taurine and Zebu Populations. Front. Genet. 2020, 10, 1293. [Google Scholar] [CrossRef]
- Pelmus, R.S.; Rotar, M.C.; Lazar, C.; Uta, R.A. Estimation the genetic parameters for milk yield in Romanian Spotted, Simmental type cattle breed. Arch. Zootec. 2021, 24, 105–121. [Google Scholar] [CrossRef]
- Gradinaru, A.C.; Petrescu-Mag, I.V.; Oroian, F.C.; Balint, C.; Oltean, I. Milk Protein Polymorphism Characterization: A Modern Tool for Sustainable Conservation of Endangered Romanian Cattle Breeds in the Context of Traditional Breeding. Sustainability 2018, 10, 534. [Google Scholar] [CrossRef]
- Erina, S.; Cziszter, L.T.; Acatincai, S. Tehnologii Zootehnice-Bovine; Ed. Eurobit: Timisoara, Romania, 2018; p. 98. [Google Scholar]
- Cziszter, L.T.; Ilie, D.E.; Neamt, R.I.; Neciu, F.C.; Saplacan, S.I.; Gavojdian, D. Comparative study on production, reproduction and functional traits between Fleckvieh and Braunvieh cattle. Asian-australas. J. Anim. Sci. 2017, 30, 666. [Google Scholar] [CrossRef]
- Maciuc, V.; Leonte, C.; Radu-Rusu, R. Manual de Bune Practici in Cresterea Bovinelor; Ed. Alfa: Iasi, Romania, 2015; pp. 31–32. [Google Scholar]
- Wang, X.; Ma, P.; Liu, J.; Zhang, Q.; Zhang, Y.; Ding, X.; Jiang, L.; Wang, Y.; Zhang, Y.; Sun, D.; et al. Genome-wide association study in Chinese Holstein cows reveal two candidate genes for somatic cell score as an indicator for mastitis susceptibility. BMC Genet. 2015, 16, 111. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Song, Y.; Riaz, H.; Yang, L. Effect of BoLA-DRB3 exon2 polymorphisms on lameness of Chinese Holstein cows. Mol. Biol. Rep. 2013, 40, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Ogorevc, J.; Kunej, T.; Razpet, A.; Dovc, P. Database of cattle candidate genes and genetic markers for milk production and mastitis. Anim. Genet. 2009, 40, 832–851. [Google Scholar] [CrossRef]
- Yeh, F.C.; Yang, R.C.; Boyle, T. Microsoft Window Based Freeware for Population Genetic Analysis. Popgene Ver. 1. 31; University of Alberta: Edmonton, AB, Canada, 1999. [Google Scholar]
- Hill, T.; Lewicki, P. Statistics: Methods and Applications, 1st ed.; StatSoft, Inc.: Tulsa, OK, USA, 2006. [Google Scholar]
- Shete, S.; Tiwari, H.; Elston, R.C. On estimating the heterozygosity and polymorphism information content value. Theor. Popul. Biol. 2000, 57, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Rupp, R.; Boichard, D. Genetics of resistance to mastitis in dairy cattle. Vet. Res. 2003, 34, 671–688. [Google Scholar] [CrossRef] [PubMed]
- Koeck, A.; Heringstad, B.; Egger-Danner, C.; Fuerst, C.; Winter, P.; Fuerst-Waltl, B. Genetic analysis of clinical mastitis and somatic cell count traits in Austrian Fleckvieh cows. J. Dairy Sci. 2010, 93, 5987–5995. [Google Scholar] [CrossRef]
- Graves, H.; Rayburn, A.L.; Gonzalez-Hernandez, J.L.; Nah, G.; Kim, D.S.; Lee, D.K. Validating DNA Polymorphisms Using KASP Assay in Prairie Cordgrass (Spartina pectinata Link) Populations in the U.S. Front. Plant Sci. 2016, 6, 1271. [Google Scholar] [CrossRef]
- Semagn, K.; Babu, R.; Hearne, S.; Olsen, M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol. Breed. 2014, 33, 1–14. [Google Scholar] [CrossRef]
- Kusza, S.; Cziszter, L.T.; Ilie, D.E.; Sauer, M.; Padeanu, I.; Gavojdian, D. Kompetitive Allele Specific PCR (KASP™) genotyping of 48 polymorphisms at different caprine loci in French Alpine and Saanen goat breeds and their association with milk composition. PeerJ 2018, 6, e4416. [Google Scholar] [CrossRef]
- Nascimento, C.S.D.; Machado, M.A.; Martinez, M.L.; Silva, M.V.G.; Guimarães, M.F.M.; Campos, A.L.; Azevedo, A.L.S.; Teodoro, R.L.; Verneque, R.D.S.; Guimarães, S.E.F.; et al. Association of the bovine major histocompatibility complex (BoLA) BoLA-DRB3 gene with fat and protein production and somatic cell score in Brazilian Gyr dairy cattle (Bos indicus). Genet. Mol. Biol. 2006, 29, 641–647. [Google Scholar] [CrossRef]
- Kulberg, S.; Heringstad, B.; Guttersrud, O.A.; Olsaker, I. Study on the association of BoLA-DRB3.2 alleles with clinical mastitis in Norwegian Red cows. J. Anim. Breed. Genet. 2007, 124, 201–207. [Google Scholar] [CrossRef]
- Martinez, M.L.; Machado, M.A.; Nascimento, C.S.; Silva, M.V.; Teodoro, R.L.; Furlong, J.; Prata, M.C.; Campos, A.L.; Guimarães, M.F.; Azevedo, A.L.; et al. Association of BoLA-DRB3.2 alleles with tick (Boophilus microplus) resistance in cattle. Genet. Mol. Res. 2006, 5, 513–524. [Google Scholar] [PubMed]
- Nikbakht Brujeni, G.; Ghorbanpour, R.; Esmailnejad, A. Association of BoLA-DRB3. 2 alleles with BLV infection profiles (persistent lymphocytosis/lymphosarcoma) and lymphocyte subsets in Iranian Holstein cattle. Biochem. Genet. 2016, 54, 194–207. [Google Scholar] [CrossRef]
- Sharif, S.; Mallard, B.A.; Sargeant, J.M. Presence of glutamine at position 74 of pocket 4 in the BoLA-DR antigen binding groove is associated with occurrence of clinical mastitis caused by Staphylococcus species. Vet. Immunol. Immunopathol. 2000, 76, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Furuta, H.; Kondo, Y.; Mukoyama, H. Association of BoLA-DRB3 alleles with mastitis resistance and susceptibility in Japanese Holstein cows. Anim. Sci. J. 2012, 83, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Duangjinda, M.; Buayai, D.; Pattarajinda, V.; Phasuk, Y.; Katawatin, S.; Vongpralub, T.; Chaiyotvittayakul, A. Detection of bovine leukocyte antigen DRB3 alleles as candidate markers for clinical mastitis resistance in Holstein x Zebu. J. Anim. Sci. 2009, 87, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Carignano, H.A.; Beribe, M.J.; Caffaro, M.E.; Amadio, A.; Nani, J.P.; Gutierrez, G.; Alvarez, I.; Trono, K.; Miretti, M.M.; Poli, M.A. BOLA-DRB3 gene polymorphisms influence bovine leukaemia virus infection levels in Holstein and Holstein × Jersey crossbreed dairy cattle. Anim. Genet. 2017, 48, 420–430. [Google Scholar] [CrossRef] [PubMed]
- Suprovych, T.M.; Suprovych, M.P.; Kolinchuk, R.V.; Karchevska, T.M.; Chornyi, I.O.; Kolodiy, V.A. Association of BoLA-DRB3. 2 alleles with fusobacteriosis in cows. Regul. Mech. Biosyst. 2020, 11, 249–254. [Google Scholar] [CrossRef]
| SNP ID | Gene Symbol | Chr | Position 1 | Alleles | Remarks | Substitution/Position in Coding Region/Change in Amino Acid |
|---|---|---|---|---|---|---|
| rs478194554 | CXCR2 | 2 | 106,191,506 | A/T | missense | c.73A>T, p.Asn25Tyr |
| rs480434223 | CXCR2 | 2 | 106,191,772 | G/T | missense | c.339G>T, p.Lys113Asn |
| rs468701549 | CXCR2 | 2 | 106,191,779 | G/A | missense | c.346G>A, p.Val116Ile |
| rs209319366 | CXCR2 | 2 | 106,192,040 | A/C | missense | c.607A>C, p.Met203Leu |
| rs464576793 | CXCR2 | 2 | 106,192,155 | T/G | missense | c.722T>G, p.Leu241Arg |
| rs433132004 | CXCR2 | 2 | 106,192,157 | T/C | missense | c.724T>C, p.Phe242Leu |
| rs467207447 | CXCR2 | 2 | 106,192,187 | G/T | missense | c.754G>T, p.Ala252Ser |
| rs455755453 | CXCR2 | 2 | 106,192,194 | G/T | missense | c.761G>T, p.Arg254Leu |
| rs474074698 | CXCR2 | 2 | 106,192,265 | A/C | missense | c.832A>C, p.Thr278Pro |
| rs446793067 | CXCR2 | 2 | 106,192,320 | G/C | missense | c.887G>C, p.Gly296Ala |
| rs381099940 | CXCR2 | 2 | 106,192,422 | A/G | missense | c.989A>G, p.Lys330Arg |
| rs384398669 | CXCR2 | 2 | 106,192,437 | A/G | missense | c.1004A>G, p.His335Arg |
| rs134178216 | IL8 (CXCL8) | 6 | 88,810,896 | T/G | missense | c.20T>G, p.Val7Gly |
| rs468955241 | IL8 (CXCL8) | 6 | 88,812,530 | A/C | missense | c.81A>C, p.Arg27Ser |
| rs436292881 | IL8 (CXCL8) | 6 | 88,812,541 | A/T | missense | c.92A>T, Glu31Val |
| rs448378725 | IL8 (CXCL8) | 6 | 88,812,575 | A/C | synonymous | c.126A>C, p.Thr42= |
| rs466813222 | IL8 (CXCL8) | 6 | 88,812,578 | T/C | synonymous | c.129T>C, p.Pro43= |
| rs433737959 | IL8 (CXCL8) | 6 | 88,812,616 | A/T | missense | c.167A>T, p.Glu56Val |
| rs453466519 | IL8 (CXCL8) | 6 | 88,812,623 | G/T | synonymous | c.174G>T, p.Gly58= |
| rs452742998 | IL8 (CXCL8) | 6 | 88,812,944 | C/A | missense | c.222C>A, p.Asn74Lys |
| rs445886432 | IL8 (CXCL8) | 6 | 88,812,994 | T/G | missense | c.272T>G, p.Val91Gly |
| rs8193041 | TLR4 | 8 | 107,058,305 | C/T | missense | c.10C>T, p.Arg4Cys |
| rs460053411 | TLR4 | 8 | 107,058,332 | C/T | missense | c.37C>T, p.Pro13Ser |
| rs8193048 | TLR4 | 8 | 107,062,990 | G/A | missense | c.148G>A, p.Asp50Asn |
| rs8193049 | TLR4 | 8 | 107,066,042 | A/C | missense | c.452A>C, p.Asn151Thr |
| rs8193050 | TLR4 | 8 | 107,066,304 | C/G | missense | c.714C>G, p.Asn238Lys |
| rs8193053 | TLR4 | 8 | 107,066,630 | C/A | missense | c.1040C>A, p.Ala347Glu |
| rs8193055 | TLR4 | 8 | 107,066,732 | A/G | missense | c.1142A>G, p.Lys381Arg |
| rs135659119 | TLR4 | 8 | 107,067,191 | C/G | missense | c.1601C>G, p.Ser534Ter |
| rs8193066 | TLR4 | 8 | 107,067,538 | G/A | missense | c.1948G>A, p.Val650Ile |
| rs437880924 | BRCA1 | 19 | 43,070,163 | A/G | missense | c.5543T>C, p.Val1848Ala |
| rs457333759 | BRCA1 | 19 | 43,070,167 | G/T | missense | c.5539C>A, p.Leu1847Met |
| rs470945089 | BRCA1 | 19 | 43,070,173 | T/G | missense | c.5533A>C, p.Thr1845Pro |
| rs439749719 | BRCA1 | 19 | 43,070,175 | T/G | missense | c.5531A>C, p.Asp1844Ala |
| rs453404290 | BRCA1 | 19 | 43,070,180 | C/G | missense | c.5526G>C, p.Glu1842Asp |
| rs473473146 | BRCA1 | 19 | 43,070,181 | T/C | missense | c.5525A>G, p.Glu1842Gly |
| rs442243059 | BRCA1 | 19 | 43,070,189 | C/A | missense | c.5517G>T, p.Gln1839His |
| rs462317177 | BRCA1 | 19 | 43,070,190 | T/G | missense | c.5516A>C, p.Gln1839Pro |
| rs482618972 | BRCA1 | 19 | 43,070,194 | A/C | missense | c.5512T>G, p.Tyr1838Asp |
| rs457592746 | BRCA1 | 19 | 43,070,199 | G/T | missense | c.5507C>A, p.Ala1836Asp |
| rs477576896 | BRCA1 | 19 | 43,070,207 | G/C | missense | c.5499C>G, p.Asp1833Glu |
| rs466537670 | BRCA1 | 19 | 43,070,214 | A/C | missense | c.5492T>G, p.Val1831Gly |
| rs480362322 | BRCA1 | 19 | 43,070,229 | A/C | missense | c.5477T>G, p.Val1826Gly |
| rs208338549 | BRCA1 | 19 | 43,070,233 | C/T | missense | c.5473G>A, p.Val1825Met |
| rs469103334 | BRCA1 | 19 | 43,070,236 | G/C | missense | c.5470C>G, p.Pro1824Ala |
| rs437904779 | BRCA1 | 19 | 43,070,244 | C/G | missense | c.5462G>C, p.Cys1821Ser |
| rs451586626 | BRCA1 | 19 | 43,070,247 | A/C | missense | c.5459T>G, p.Met1820Arg |
| rs456463862 | BRCA1 | 19 | 43,070,254 | C/T | missense | c.5452G>A, p.Gly1818Arg |
| rs384176726 | LTF | 22 | 52,953,375 | A/G | missense | c.5A>G, p.Lys2Arg |
| rs461137889 | LTF | 22 | 52,953,380 | T/C | missense | c.10T>C, p.Phe4Leu |
| rs483118032 | LTF | 22 | 52,953,384 | T/G | missense | c.14T>G, p.Val5Gly |
| rs450733501 | LTF | 22 | 52,953,387 | C/T | missense | c.17C>T, p.Pro6Leu |
| rs478177296 | LTF | 22 | 52,953,392 | C/G | missense | c.22C>G, p.Leu8Val |
| rs445052454 | LTF | 22 | 52,953,398 | T/G | missense | c.28T>G, p.Ser10Ala |
| rs478065524 | LTF | 22 | 52,957,166 | G/A | missense | c.55G>A, p.Ala19Thr |
| rs460348489 | LTF | 22 | 52,957,271 | A/C | missense | c.160A>C, p.Thr54Pro |
| rs453982323 | LTF | 22 | 52,960,346 | G/T | missense | c.260G>T, p.Gly87Val |
| rs472508993 | LTF | 22 | 52,960,352 | A/G | missense | c.266A>G, p.Asp89Gly |
| rs461145277 | LTF | 22 | 52,960,367 | G/T | missense | c.281G>T, p.Arg94Leu |
| rs482729559 | LTF | 22 | 52,960,370 | C/G | missense | c.284C>G, p.Pro95Arg |
| rs476824727 | LTF | 22 | 52,960,393 | A/G | missense | c.307A>G, p.Thr103Ala |
| rs447239896 | LTF | 22 | 52,960,402 | T/G | missense | c.316T>G, p.Ser106Ala |
| rs433547801 | BOLA-DRB3 | 23 | 25,730,098 | A/T | missense | c.104A>T, p.His35Leu |
| rs208667846 | BOLA-DRB3 | 23 | 25,730,106 | G/C | missense | c.112G>C, p.Glu38Gln |
| rs110364925 | BOLA-DRB3 | 23 | 25,730,113 | A/C | missense | c.119A>C, p.Tyr40Ser |
| rs137786474 | BOLA-DRB3 | 23 | 25,730,116 | A/C | missense | c.122A>C, p.Lys41Thr |
| rs210718478 | BOLA-DRB3 | 23 | 25,730,118 | A/G | missense | c.124A>G, p.Arg42Gly |
| rs461191302 | BOLA-DRB3 | 23 | 25,730,119 | G/A | missense | c.125G>A, p.Arg42Lys |
| rs379932839 | BOLA-DRB3 | 23 | 25,730,151 | G/T | missense | c.157G>T, p.Val53Leu |
| rs42313447 | BOLA-DRB3 | 23 | 25,730,159 | C/G | missense | c.165C>G, p.Phe55Leu |
| rs448858468 | BOLA-DRB3 | 23 | 25,730,163 | G/C | missense | c.169G>C, p.Asp57His |
| rs467367584 | BOLA-DRB3 | 23 | 25,730,165 | C/G | missense | c.171C>G, p.Asp57Glu |
| rs209865172 | BOLA-DRB3 | 23 | 25,730,169 | T/C | missense | c.175T>C, p.Tyr59His |
| rs465057416 | BOLA-DRB3 | 23 | 25,730,173 | T/A | missense | c.179T>A, p.Phe60Tyr |
| rs467605669 | BOLA-DRB3 | 23 | 25,730,176 | A/C | missense | c.182A>C, p.His61Pro |
| rs109928428 | BOLA-DRB3 | 23 | 25,730,191 | A/C/ | missense | c.197A>C, p.Tyr66Ser |
| rs436108988 | BOLA-DRB3 | 23 | 25,730,194 | T/C | missense | c.200T>C, p.Val67Ala |
| rs476406737 | BOLA-DRB3 | 23 | 25,730,194 | A/G | missense | c.200T>C, p.Val67Ala |
| rs42309897 | BOLA-DRB3 | 23 | 25,730,215 | G/A | missense | c.221G>A, p.Gly74Asp |
| rs136458736 | BOLA-DRB3 | 23 | 25,730,239 | T/A | missense | c.245T>A, p.Leu82Gln |
| rs133097997 | BOLA-DRB3 | 23 | 25,730,248 | G/C | missense | c.254G>C, p.Arg85Pro |
| rs434083272 | BOLA-DRB3 | 23 | 25,732,613 | G/A | missense | c.448G>A, p.Gly150Ser |
| rs209853925 | BOLA-DRB3 | 23 | 25,732,628 | C/A | missense | c.463C>A, p.His155Asn |
| rs454104745 | BOLA-DRB3 | 23 | 25,732,631 | A/G | missense | c.466A>G, p.Ile156Val |
| rs437148562 | BOLA-DRB3 | 23 | 25,732,650 | G/A | missense | c.485G>A, p.Arg162Gln |
| rs42312242 | BOLA-DRB3 | 23 | 25,732,780 | G/T | missense | c.615G>T, p.Glu205Asp |
| rs209467115 | BOLA-DRB3 | 23 | 25,732,791 | G/A | missense | c.626G>A, p.Arg209Gln |
| rs208816121 | BOLA-DRB3 | 23 | 25,733,626 | C/T | missense | c.718C>T, p.Leu240Phe |
| rs110124025 | BOLA-DRB3 | 23 | 25,734,153 | A/G | missense | c.767A>G, p.His256Arg |
| Status | Number | Success Ratio | SNP/Gene |
|---|---|---|---|
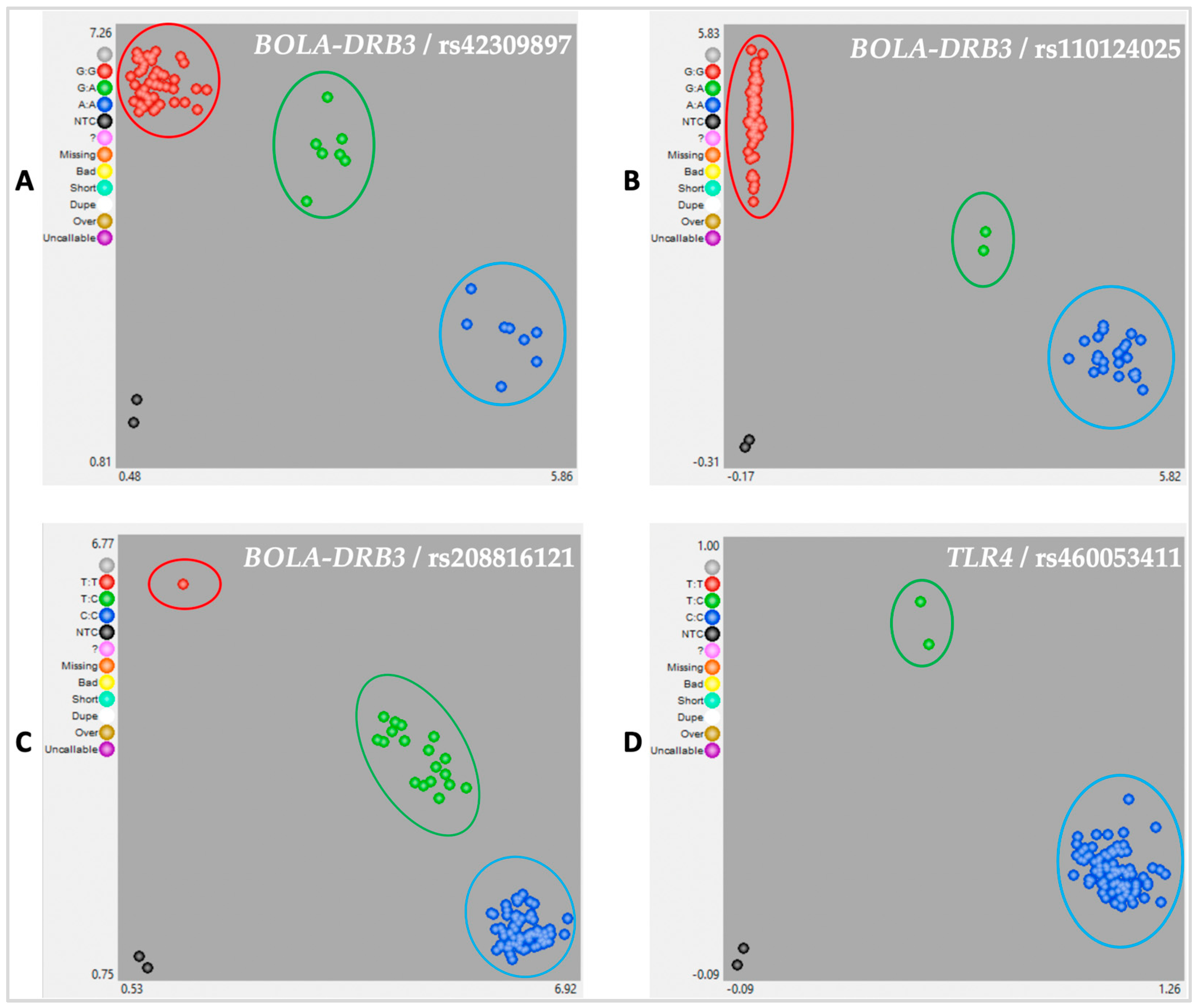
| Polymorphic | 4 | 4.50% | rs460053411/TLR4, rs42309897/BOLA-DRB3, rs208816121/BOLA-DRB3, rs110124025/BOLA-DRB3 |
| Monomorphic | 31 | 34.83% | rs480434223/CXCR2, rs209319366/CXCR2, rs464576793/CXCR2, rs467207447/CXCR2, rs446793067/CXCR2, rs134178216/CXCL8, rs468955241/CXCL8, rs448378725/CXCL8, rs466813222/CXCL8, rs453466519/CXCL8, rs452742998/CXCL8, rs445886432/CXCL8, rs8193041/TLR4, rs8193048/TLR4, rs8193049/TLR4, rs8193050/TLR4, rs8193053/TLR4, rs8193055/TLR4, rs135659119/TLR4, rs8193066/TLR4, rs437880924/BRCA1, rs470945089/BRCA1, rs480362322/BRCA1, rs469103334/BRCA1, rs460348489/LTF, rs461145277/LTF, rs476824727/LTF, rs447239896/LTF, rs208667846/BOLA-DRB3, rs476406737/BOLA-DRB3, rs454104745/BOLA-DRB3 |
| Failed | 54 | 60.67% | rs478194554/CXCR2, rs468701549/CXCR2, rs433132004/CXCR2, rs455755453/CXCR2, rs474074698/CXCR2, rs381099940/CXCR2, rs384398669/CXCR2, rs436292881/CXCL8, rs433737959/CXCL8, rs457333759/BRCA1, rs439749719/BRCA1, rs453404290/BRCA1, rs473473146/BRCA1, rs442243059/BRCA1, rs462317177/BRCA1, rs482618972/BRCA1, rs457592746/BRCA1, rs477576896/BRCA1, rs466537670/BRCA1, rs208338549/BRCA1, rs437904779/BRCA1, rs451586626/BRCA1, rs456463862/BRCA1, rs384176726/LTF, rs461137889/LTF, rs483118032/LTF, rs450733501/LTF, rs478177296/LTF, rs445052454/LTF, rs478065524/LTF, rs453982323/LTF, rs472508993/LTF, rs482729559/LTF, rs433547801/BOLA-DRB3, rs110364925/BOLA-DRB3, rs137786474/BOLA-DRB3, rs210718478/BOLA-DRB3, rs461191302/BOLA-DRB3, rs379932839/BOLA-DRB3, rs42313447/BOLA-DRB3, rs448858468/BOLA-DRB3, rs467367584/BOLA-DRB3, rs209865172/BOLA-DRB3, rs465057416/BOLA-DRB3, rs467605669/BOLA-DRB3, rs109928428/BOLA-DRB3, rs436108988/BOLA-DRB3, rs136458736/BOLA-DRB3, rs133097997/BOLA-DRB3, rs434083272/BOLA-DRB3, rs209853925/BOLA-DRB3, rs437148562/BOLA-DRB3, rs42312242/BOLA-DRB3, rs209467115/BOLA-DRB3 |
| Gene/SNP ID | Breed | Genotype Frequency | Allele Frequency | PIC | He | Ho | |||
|---|---|---|---|---|---|---|---|---|---|
| BOLA-DRB3 rs42309897 | GG | GA | AA | G | A | ||||
| RS | 0.826 (n = 123) | 0.074 (n = 11) | 0.101 (n = 15) | 0.86 | 0.14 | 0.400 | 0.238 | 0.073 | |
| RB | 0.611 (n = 22) | 0.083 (n = 3) | 0.306 (n = 11) | 0.65 | 0.35 | 0.645 | 0.459 | 0.083 | |
| BOLA-DRB3 rs208816121 | CC | TC | TT | C | T | ||||
| RS * | 0.884 (n = 221) | 0.108 (n = 27) | 0.008 (n = 2) | 0.94 | 0.06 | 0.232 | 0.116 | 0.108 | |
| RB | 0.375 (n = 18) | 0.604 (n = 29) | 0.021 (n = 1) | 0.68 | 0.32 | 0.629 | 0.441 | 0.604 | |
| BOLA-DRB3 rs110124025 | GG | GA | AA | G | A | ||||
| RS | 0.740 (n = 185) | 0.008 (n = 2) | 0.252 (n = 63) | 0.74 | 0.26 | 0.568 | 0.381 | 0.008 | |
| RB | 0.771 (n = 37) | 0.042 (n = 2) | 0.188 (n = 9) | 0.79 | 0.21 | 0.511 | 0.333 | 0.041 | |
| TLR4 rs460053411 | CC | TC | TT | C | T | ||||
| RS * | 0.984 (n = 246) | 0.016 (n = 4) | 0.000 (n = 0) | 0.99 | 0.01 | 0.046 | 0.015 | 0.016 | |
| RB * | 0.979 (n = 47) | 0.021 (n = 1) | 0.000 (n = 0) | 0.99 | 0.01 | 0.059 | 0.020 | 0.020 | |
| Breed | SNP | Genotype | Mastitis Frequency (%) | Chi-Square d.f. p Value | Number of Affected Quarters (%) | Chi-Square d.f. p Value | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 0 | 1 | 2 | 4 | |||||
| RS | rs42309897 | GG | 85.1 | 13.2 | 1.7 | 2.836 | 85.1 | 9.6 | 1.8 | 3.5 | 7.774 |
| AA | 93.3 | 6.7 | 0.0 | 4 | 93.3 | 0.0 | 6.7 | 0.0 | 4 | ||
| GA | 72.7 | 27.3 | 0.0 | 0.5855 | 72.7 | 27.3 | 0.0 | 0.0 | 0.2551 | ||
| rs208816121 | CC | 85.4 | 13.1 | 1.5 | 0.500 | 85.4 | 9.2 | 1.9 | 3.5 | 1.383 | |
| TC | 88.9 | 11.1 | 0.0 | 2 | 88.9 | 7.4 | 3.7 | 0.0 | 2 | ||
| TT | - | - | - | 0.7787 | - | - | - | - | 0.7094 | ||
| rs110124025 | GG | 88.6 | 10.3 | 1.1 | 12.611 | 88.6 | 8.1 | 1.1 | 2.2 | 12.791 | |
| AA | 69.0 | 29.3 | 1.7 | 2 | 69.0 | 20.8 | 5.1 | 5.1 | 2 | ||
| GA | - | - | - | 0.0018 | - | - | - | - | 0.0051 | ||
| RB | rs42309897 | GG | 85.0 | 5.0 | 10.0 | 3.856 | 85.0 | 5.0 | 5.0 | 5.0 | 5.415 |
| AA | 81.8 | 18.2 | 0.0 | 4 | 81.8 | 9.1 | 9.1 | 0.0 | 4 | ||
| GA | 66.7 | 33.3 | 0.0 | 0.4258 | 66.7 | 0.0 | 0.0 | 33.3 | 0.4918 | ||
| rs208816121 | CC | 82.2 | 11.8 | 5.9 | 0.152 | 82.3 | 5.9 | 5.9 | 5.9 | 0.923 | |
| TC | 85.7 | 10.7 | 3.6 | 2 | 85.7 | 7.1 | 3.6 | 3.6 | 2 | ||
| TT | - | - | - | 0.9269 | - | - | - | - | 0.9612 | ||
| rs110124025 | GG | 88.9 | 8.3 | 2.8 | 6.654 | 88.9 | 5.5 | 2.8 | 2.8 | 7.775 | |
| AA | 50.0 | 37.5 | 12.5 | 2 | 50.0 | 12.5 | 25 | 12.5 | 2 | ||
| GA | - | - | - | 0.0359 | - | - | - | - | 0.0500 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ilie, D.E.; Gavojdian, D.; Kusza, S.; Neamț, R.I.; Mizeranschi, A.E.; Mihali, C.V.; Cziszter, L.T. Kompetitive Allele Specific PCR Genotyping of 89 SNPs in Romanian Spotted and Romanian Brown Cattle Breeds and Their Association with Clinical Mastitis. Animals 2023, 13, 1484. https://doi.org/10.3390/ani13091484
Ilie DE, Gavojdian D, Kusza S, Neamț RI, Mizeranschi AE, Mihali CV, Cziszter LT. Kompetitive Allele Specific PCR Genotyping of 89 SNPs in Romanian Spotted and Romanian Brown Cattle Breeds and Their Association with Clinical Mastitis. Animals. 2023; 13(9):1484. https://doi.org/10.3390/ani13091484
Chicago/Turabian StyleIlie, Daniela Elena, Dinu Gavojdian, Szilvia Kusza, Radu Ionel Neamț, Alexandru Eugeniu Mizeranschi, Ciprian Valentin Mihali, and Ludovic Toma Cziszter. 2023. "Kompetitive Allele Specific PCR Genotyping of 89 SNPs in Romanian Spotted and Romanian Brown Cattle Breeds and Their Association with Clinical Mastitis" Animals 13, no. 9: 1484. https://doi.org/10.3390/ani13091484
APA StyleIlie, D. E., Gavojdian, D., Kusza, S., Neamț, R. I., Mizeranschi, A. E., Mihali, C. V., & Cziszter, L. T. (2023). Kompetitive Allele Specific PCR Genotyping of 89 SNPs in Romanian Spotted and Romanian Brown Cattle Breeds and Their Association with Clinical Mastitis. Animals, 13(9), 1484. https://doi.org/10.3390/ani13091484





