The Use of a Brief Synchronization Treatment after Weaning, Combined with Superovulation, Has Moderate Effects on the Gene Expression of Surviving Pig Blastocysts
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Ethics
2.2. Experimental Design
2.3. Treatments, Detection of Estrus, and Insemination
2.4. Collection of Embryos
2.5. Embryo Quality Evaluation
2.6. Total RNA Extraction
2.7. Microarray Processing
2.8. Analysis of the Microarray Data
2.9. Real-Time, Reverse Transcription Quantitative PCR (RT-qPCR) Assay
3. Results
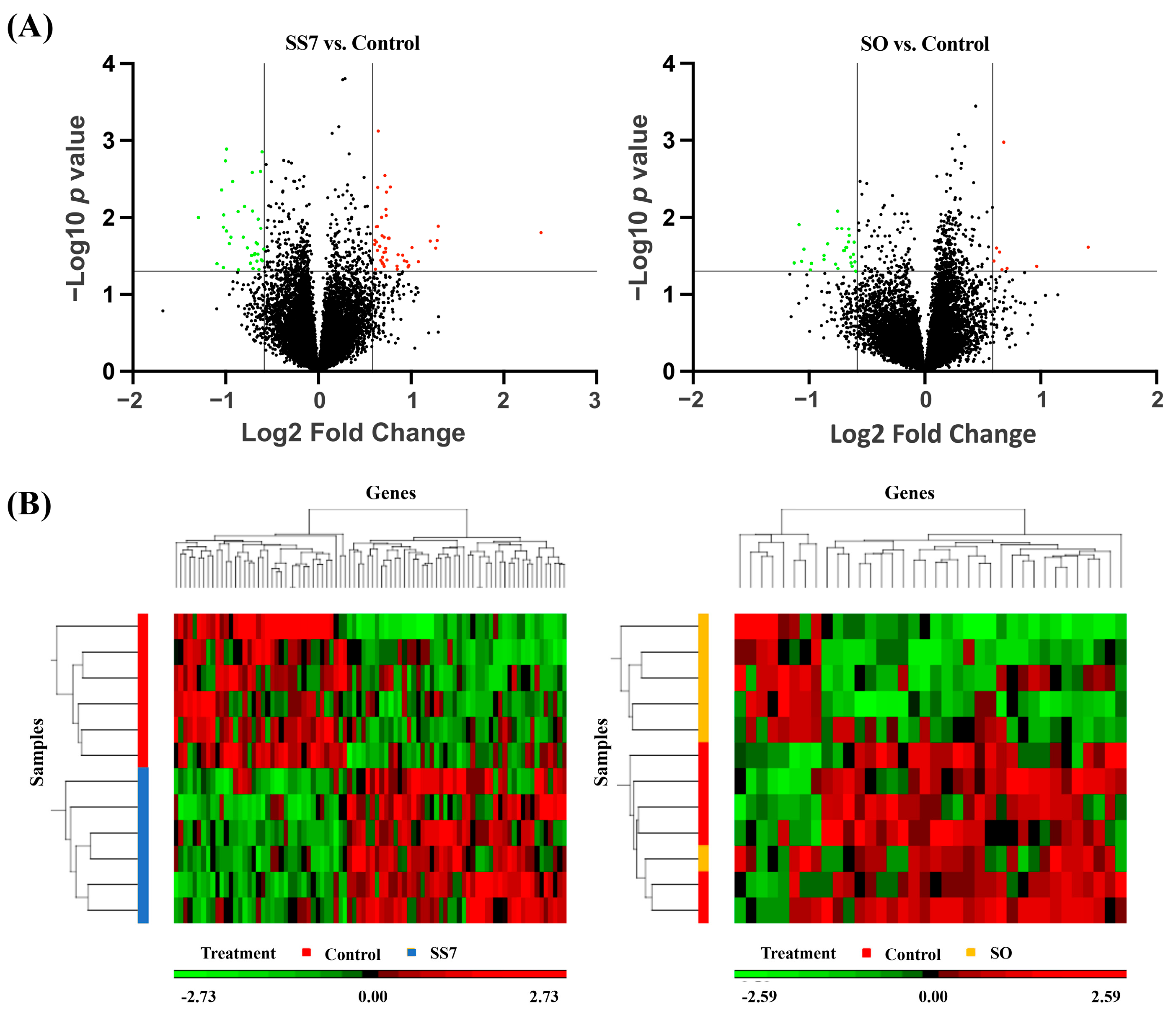
3.1. Gene Expression Profiles of Embryos
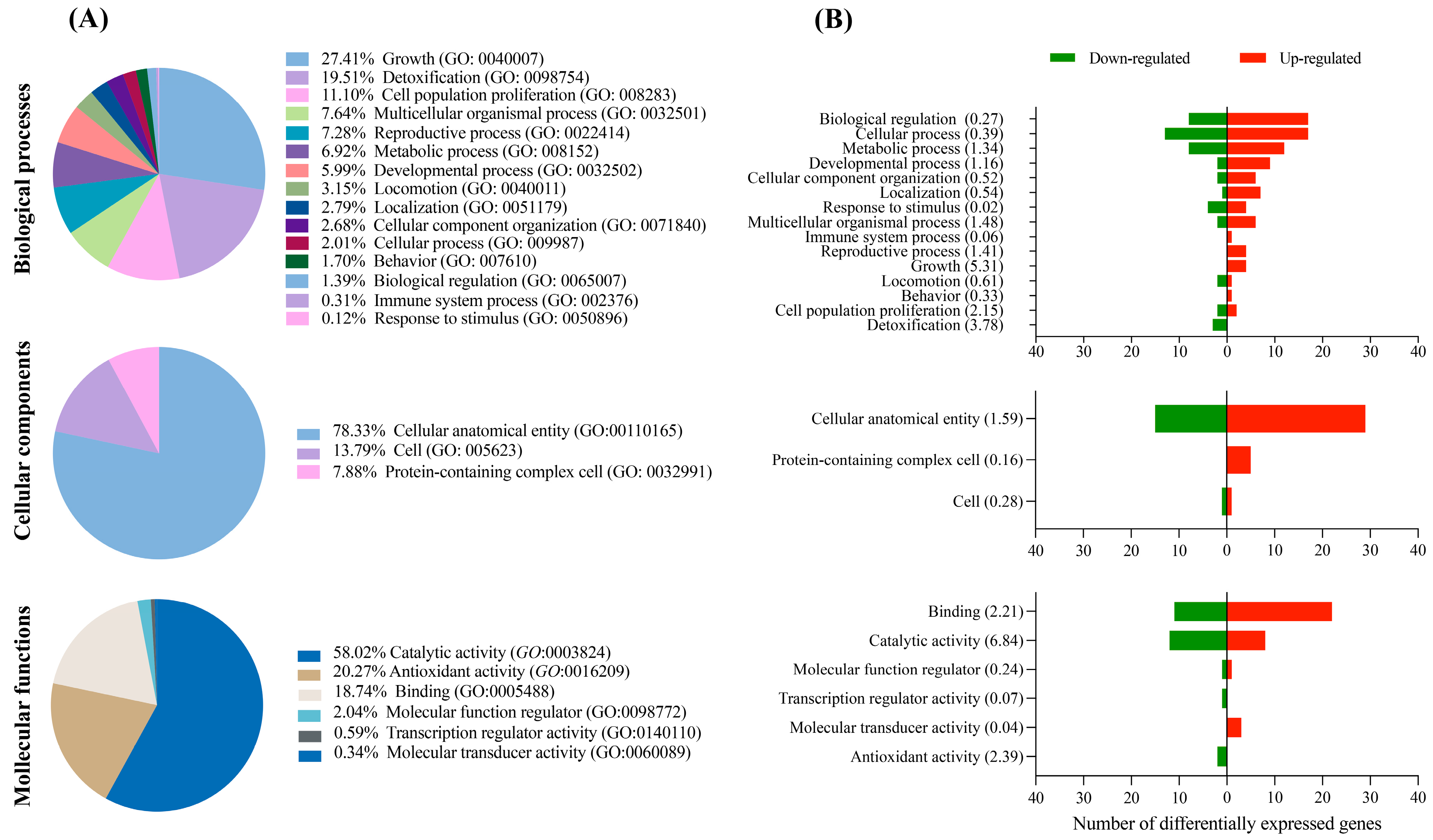
3.2. GO and Pathway Enrichment of DEGs
3.3. Microarray Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martinez, E.A.; Martinez, C.A.; Nohalez, A.; Sanchez-Osorio, J.; Vazquez, J.M.; Roca, J.; Parrilla, I.; Gil, M.A.; Cuello, C. Nonsurgical deep uterine transfer of vitrified, in vivo-derived, porcine embryos is as effective as the default surgical approach. Sci. Rep. 2015, 5, 10587. [Google Scholar] [CrossRef]
- Martinez, E.A.; Nohalez, A.; Martinez, C.A.; Parrilla, I.; Vila, J.; Colina, I.; Diaz, M.; Reixach, J.; Vazquez, J.L.; Roca, J.; et al. The Recipients’ Parity Does Not Influence Their Reproductive Performance Following Non-Surgical Deep Uterine Porcine Embryo Transfer. Reprod. Domest. Anim. 2016, 51, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Nohalez, A.; Martinez, C.A.; Reixach, J.; Diaz, M.; Vila, J.; Colina, I.; Parrilla, I.; Vazquez, J.L.; Roca, J.; Gil, M.A.; et al. Factors of importance when selecting sows as embryo donors. Animal 2017, 11, 1330–1335. [Google Scholar] [CrossRef]
- Martinez, C.A.; Nohalez, A.; Parrilla, I.; Lucas, X.; Sanchez-Osorio, J.; Roca, J.; Cuello, C.; Rodriguez-Martinez, H.; Martinez, E.A.; Gil, M.A. Simple storage (CO2-free) of porcine morulae for up to three days maintains the in vitro viability and developmental competence. Theriogenology 2018, 108, 229–238. [Google Scholar] [CrossRef]
- Cuello, C.; Martinez, C.A.; Nohalez, A.; Parrilla, I.; Roca, J.; Gil, M.A.; Martinez, E.A. Effective vitrification and warming of porcine embryos using a pH-stable, chemically defined medium. Sci. Rep. 2016, 6, 33915. [Google Scholar] [CrossRef]
- Martinez, C.A.; Cambra, J.M.; Nohalez, A.; Parrilla, I.; Roca, J.; Vazquez, J.L.; Rodriguez-Martinez, H.; Gil, M.A.; Martinez, E.A.; Cuello, C. Prevention of hatching of porcine morulae and blastocysts by liquid storage at 20 degrees C. Sci. Rep. 2019, 9, 6219. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.A.; Angel, M.A.; Cuello, C.; Sanchez-Osorio, J.; Gomis, J.; Parrilla, I.; Vila, J.; Colina, I.; Diaz, M.; Reixach, J.; et al. Successful non-surgical deep uterine transfer of porcine morulae after 24 hour culture in a chemically defined medium. PLoS ONE 2014, 9, e104696. [Google Scholar] [CrossRef]
- Martinez, E.A.; Cuello, C.; Parrilla, I.; Martinez, C.A.; Nohalez, A.; Vazquez, J.L.; Vazquez, J.M.; Roca, J.; Gil, M.A. Recent advances toward the practical application of embryo transfer in pigs. Theriogenology 2016, 85, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Angel, M.A.; Gil, M.A.; Cuello, C.; Sanchez-Osorio, J.; Gomis, J.; Parrilla, I.; Vila, J.; Colina, I.; Diaz, M.; Reixach, J.; et al. The effects of superovulation of donor sows on ovarian response and embryo development after nonsurgical deep-uterine embryo transfer. Theriogenology 2014, 81, 832–839. [Google Scholar] [CrossRef]
- Hazeleger, W.; Bouwman, E.G.; Noordhuizen, J.P.; Kemp, B. Effect of superovulation induction on embryonic development on day 5 and subsequent development and survival after nonsurgical embryo transfer in pigs. Theriogenology 2000, 53, 1063–1070. [Google Scholar] [CrossRef]
- Patterson, J.; Wellen, A.; Hahn, M.; Pasternak, A.; Lowe, J.; DeHaas, S.; Kraus, D.; Williams, N.; Foxcroft, G. Responses to delayed estrus after weaning in sows using oral progestagen treatment. J. Anim. Sci. 2008, 86, 1996–2004. [Google Scholar] [CrossRef]
- van Leeuwen, J.J.J.; Williams, S.I.; Kemp, B.; Soede, N.M. Post-weaning Altrenogest treatment in primiparous sows; the effect of duration and dosage on follicular development and consequences for early pregnancy. Anim. Reprod. Sci. 2010, 119, 258–264. [Google Scholar] [CrossRef]
- Gonzalez-Ramiro, H.; Cuello, C.; Cambra, J.M.; Gonzalez-Plaza, A.; Vazquez, J.M.; Vazquez, J.L.; Rodriguez-Martinez, H.; Gil, M.A.; Lucas-Sanchez, A.; Parrilla, I.; et al. A Short-Term Altrenogest Treatment Post-weaning Followed by Superovulation Reduces Pregnancy Rates and Embryo Production Efficiency in Multiparous Sows. Front. Vet. Sci. 2021, 8, 771573. [Google Scholar] [CrossRef]
- Gonzalez-Ramiro, H.; Parrilla, I.; Cambra, J.M.; Gonzalez-Plaza, A.; Gil, M.A.; Cuello, C.; Martinez, E.A.; Rodriguez-Martinez, H.; Martinez, C.A. Combined synchronization and superovulation treatments negatively impact embryo viability possibly by the downregulation of WNT/β-catenin and Notch signaling genes in the porcine endometrium. J. Anim. Sci. 2022, 100, skac315. [Google Scholar] [CrossRef] [PubMed]
- Theil, P.K.; Krogh, U.; Bruun, T.S.; Feyera, T. Feeding the modern sow to sustain high productivity. Mol. Reprod. Dev. 2022. [Google Scholar] [CrossRef]
- Lugar, D.W.; Harlow, K.E.; Hundley, J.; Goncalves, M.; Bergstrom, J.; Stewart, K.R. Effects of increased levels of supplemental vitamins during the summer in a commercial artificial insemination boar stud. Animal 2019, 13, 2556–2568. [Google Scholar] [CrossRef]
- Pursel, V.G.; Johnson, L.A. Freezing of boar spermatozoa: Fertilizing capacity with concentrated semen and a new thawing procedure. J. Anim. Sci. 1975, 40, 99–102. [Google Scholar] [CrossRef]
- Martinez, C.A.; Nohalez, A.; Parrilla, I.; Vazquez, J.L.; Roca, J.; Cuello, C.; Rodriguez-Martinez, H.; Martinez, E.A.; Gil, M.A. Surgical embryo collection but not nonsurgical embryo transfer compromises postintervention prolificacy in sows. Theriogenology 2017, 87, 316–320. [Google Scholar] [CrossRef]
- Funahashi, H.; Ekwall, H.; Rodriguez-Martinez, H. Zona reaction in porcine oocytes fertilized in vivo and in vitro as seen with scanning electron microscopy. Biol. Reprod. 2000, 63, 1437–1442. [Google Scholar] [CrossRef]
- Martinez, C.A.; Gil, M.A.; Parrilla, I.; Martinez, E.A.; Cuello, C. Protocol for Porcine Embryo Transfer. Part 1: Embryos Produced In Vivo. In Manual of the International Embryo Transfer Society; International Embryo Transfer Society 2441 Village Green Place: Champaign, IL, USA, 2020; pp. 3a1–3a9. [Google Scholar]
- Bolstad, B.M.; Irizarry, R.A.; Åstrand, M.; Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. The KEGG database. Novartis Found. Symp. 2002, 247, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Khatib, H. Comparison of transcriptomic landscapes of bovine embryos using RNA-Seq. BMC Genom. 2010, 11, 711. [Google Scholar] [CrossRef]
- Cuello, C.; Berthelot, F.; Delaleu, B.; Venturi, E.; Pastor, L.M.; Vazquez, J.M.; Roca, J.; Martinat-Botté, F.; Martinez, E.A. The effectiveness of the stereomicroscopic evaluation of embryo quality in vitrified-warmed porcine blastocysts: An ultrastructural and cell death study. Theriogenology 2007, 67, 970–982. [Google Scholar] [CrossRef]
- Fabian, D.; Gjørret, J.O.; Berthelot, F.; Martinat-Botté, F.; Maddox-Hyttel, P. Ultrastructure and cell death of in vivo derived and vitrified porcine blastocysts. Mol. Reprod. Dev. 2005, 70, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.A.; Martinez, C.A.; Cambra, J.M.; Maside, C.; Lucas, X.; Vazquez, J.L.; Vazquez, J.M.; Roca, J.; Rodriguez-Martinez, H.; Gil, M.A.; et al. Achievements and future perspectives of embryo transfer technology in pigs. Reprod. Domest. Anim. 2019, 54 (Suppl. 4), 4–13. [Google Scholar] [CrossRef]
- Cuello, C.; Martinez, C.A.; Cambra, J.M.; Parrilla, I.; Rodriguez-Martinez, H.; Gil, M.A.; Martinez, E.A. Effects of vitrification on the blastocyst gene expression profile in a porcine model. Int. J. Mol. Sci. 2021, 22, 1222. [Google Scholar] [CrossRef]
- Cuello, C.; Martinez, C.A.; Cambra, J.M.; González-Plaza, A.; Parrilla, I.; Rodriguez-Martinez, H.; Gil, M.A.; Martinez, E.A. Vitrification Effects on the Transcriptome of in vivo-Derived Porcine Morulae. Front. Vet. Sci. 2021, 8, 771996. [Google Scholar] [CrossRef]
- Wang, W.; Shi, L.; Qin, Y.; Li, F. Research and Application of Chondroitin Sulfate/Dermatan Sulfate-Degrading Enzymes. Front. Cell Dev. Biol. 2020, 8, 560442. [Google Scholar] [CrossRef]
- Mizumoto, S.; Yamada, S. An Overview of in vivo Functions of Chondroitin Sulfate and Dermatan Sulfate Revealed by Their Deficient Mice. Front. Cell Dev. Biol. 2021, 9, 764781. [Google Scholar] [CrossRef]
- Izumikawa, T.; Kanagawa, N.; Watamoto, Y.; Okada, M.; Saeki, M.; Sakano, M.; Sugahara, K.; Sugihara, K.; Asano, M.; Kitagawa, H. Impairment of embryonic cell division and glycosaminoglycan biosynthesis in glucuronyltransferase-I-deficient mice. J. Biol. Chem. 2010, 285, 12190–12196. [Google Scholar] [CrossRef]
- Endo, M.; Yosizawa, Z. Hormonal effect on glycoproteins and glycosaminoglycans in rabbit uteri. Arch. Biochem. Biophys. 1973, 156, 397–403. [Google Scholar] [CrossRef]
- Takata, K.; Terayama, H. Hormonal effect on glycosaminoglycans and glycoproteins in uteri of ovariectomized rats. Biochim. Biophys. Acta 1977, 500, 333–343. [Google Scholar] [CrossRef]
- Wilson, D.G.; Phamluong, K.; Lin, W.Y.; Barck, K.; Carano, R.A.D.; Diehl, L.; Peterson, A.S.; Martin, F.; Solloway, M.J. Chondroitin sulfate synthase 1 (Chsy1) is required for bone development and digit patterning. Dev. Biol. 2012, 363, 413–425. [Google Scholar] [CrossRef]
- Leese, H.J.; Sturmey, R.G.; Baumann, C.G.; McEvoy, T.G. Embryo viability and metabolism: Obeying the quiet rules. Hum. Reprod. 2007, 22, 3047–3050. [Google Scholar] [CrossRef]
- Baumann, C.G.; Morris, D.G.; Sreenan, J.M.; Leese, H.J. The quiet embryo hypothesis: Molecular characteristics favoring viability. Mol. Reprod. Dev. 2007, 74, 1345–1353. [Google Scholar] [CrossRef]
- Clagett-Dame, M.; Knutson, D. Vitamin A in reproduction and development. Nutrients 2011, 3, 385–428. [Google Scholar] [CrossRef]
- Niederreither, K.; Subbarayan, V.; Dollé, P.; Chambon, P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat. Genet. 1999, 21, 444–448. [Google Scholar] [CrossRef]
- See, A.W.-M.; Kaiser, M.E.; White, J.C.; Clagett-Dame, M. A nutritional model of late embryonic vitamin A deficiency produces defects in organogenesis at a high penetrance and reveals new roles for the vitamin in skeletal development. Dev. Biol. 2008, 316, 171–190. [Google Scholar] [CrossRef]
- Collins, M.D.; Mao, G.E. Teratology of retinoids. Annu. Rev. Pharmacol. Toxicol. 1999, 39, 399–430. [Google Scholar] [CrossRef]
- Sandell, L.L.; Sanderson, B.W.; Moiseyev, G.; Johnson, T.; Mushegian, A.; Young, K.; Rey, J.-P.; Ma, J.; Staehling-Hampton, K.; Trainor, P.A. RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes Dev. 2007, 21, 1113–1124. [Google Scholar] [CrossRef]
- Hanada, K. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim. Biophys. Acta 2003, 1632, 16–30. [Google Scholar] [CrossRef]
- Hojjati, M.R.; Li, Z.; Jiang, X.-C. Serine palmitoyl-CoA transferase (SPT) deficiency and sphingolipid levels in mice. Biochim. Biophys. Acta 2005, 1737, 44–51. [Google Scholar] [CrossRef]
- Ohanian, J.; Ohanian, V. Sphingolipids in mammalian cell signalling. Cell. Mol. Life Sci. 2001, 58, 2053–2068. [Google Scholar] [CrossRef]
- Chalfant, C.E.; Spiegel, S. Sphingosine 1-phosphate and ceramide 1-phosphate: Expanding roles in cell signaling. J. Cell Sci. 2005, 118, 4605–4612. [Google Scholar] [CrossRef]
- Ding, N.-Z.; Qi, Q.-R.; Gu, X.-W.; Zuo, R.-J.; Liu, J.; Yang, Z.-M. De novo synthesis of sphingolipids is essential for decidualization in mice. Theriogenology 2018, 106, 227–236. [Google Scholar] [CrossRef]
- Lu, H.; Gonzalez, F.J.; Klaassen, C. Alterations in hepatic mRNA expression of phase II enzymes and xenobiotic transporters after targeted disruption of hepatocyte nuclear factor 4 alpha. Toxicol. Sci. 2010, 118, 380–390. [Google Scholar] [CrossRef]
- Ramkumar, K.; Samanta, S.; Kyani, A.; Yang, S.; Tamura, S.; Ziemke, E.; Stuckey, J.A.; Li, S.; Chinnaswamy, K.; Otake, H.; et al. Mechanistic evaluation and transcriptional signature of a glutathione S-transferase omega 1 inhibitor. Nat. Commun. 2016, 7, 13084. [Google Scholar] [CrossRef]
- Mamo, S.; Bodo, S.; Kobolak, J.; Polgar, Z.; Tolgyesi, G.; Dinnyes, A. Gene expression profiles of vitrified in vivo derived 8-cell stage mouse embryos detected by high density oligonucleotide microarrays. Mol. Reprod. Dev. 2006, 73, 1380–1392. [Google Scholar] [CrossRef]
- Eroglu, B.; Szurek, E.A.; Schall, P.; Latham, K.E.; Eroglu, A. Probing lasting cryoinjuries to oocyte-embryo transcriptome. PLoS ONE 2020, 15, e0231108. [Google Scholar] [CrossRef]
- Cuvillier, O.; Pirianov, G.; Kleuser, B.; Vanek, P.G.; Coso, O.A.; Gutkind, S.; Spiegel, S. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature 1996, 381, 800–803. [Google Scholar] [CrossRef]
- Eliyahu, E.; Park, J.-H.; Shtraizent, N.; He, X.; Schuchman, E.H. Acid ceramidase is a novel factor required for early embryo survival. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2007, 21, 1403–1409. [Google Scholar] [CrossRef]


| Gene | Accession Number | Primers (5′-3′) | Size (pb) | Efficiency (%) | R2 |
|---|---|---|---|---|---|
| CHSY1 | NM_001244442.2 | F: GAGATGCGTGCGAAGGTT R: GTAGGGTGGGTTCTTGTTGG | 166 | 83.9 | 0.997 |
| CSGALNACT2 | XM_001926417 | F: CTGCCATTGTTTATGCCAAT R: ATTCCAAAGCCAAAATCTCG | 100 | 110.5 | 0.998 |
| CTPS1 | XM_003128105.4 | F: GGAACATTCTCTCCCTATGAGC R: GTCGGATGTCAAGGAAACG | 103 | 101.9 | 1.000 |
| RDH10 | XM_021089230.1 | F: ACATCAACACGCAGAGCAAC R: CTCTCTTTCCCACATCACAGG | 168 | 93.0 | 0.993 |
| SPTLC2 | XM_005656439.2 | F: ATCTGCGGACACATTCTCAC R: CCTGGTGTTTTCGGCTAACT | 152 | 102.6 | 0.999 |
| GSTK1 | NM_001315574.1 | F: GCAGGAGAAAGGGAACGAT R: TGCAGGTTGACATTCCAGAT | 183 | 116.5 | 0.996 |
| ATG4C | NM_001190284.1 | F: GTCGAAATGTTCAGGACTTCA R: GACAAACTCTTCTGTGCTAAATCTG | 206 | 99.1 | 0.995 |
| ACTB | XM_003357928 | F: CTCGATCATGAAGTGCGACGT R: TGATCTCCTTCTGCATCCTGTC | 114 | 101.8 | 0.998 |
| Group | Sows (n) | Viable Embryos (n, Mean ± SD) | Oocytes/Degenerated Embryos (n, Mean ± SD) | Embryo Stage * | Embryo Quality ** (n, %) |
|---|---|---|---|---|---|
| SS7 | 6 | 132 (22.0 ± 3.9) a | 55 (9.2 ± 4.3) a | 2.4 ± 0.4 | 115 (87.1) |
| SO | 6 | 174 (29.0 ± 1.8) b | 12 (2.0 ± 2.4) b | 2.5 ± 0.4 | 162 (93.1) |
| Control | 6 | 105 (17.5 ± 4.7) a | 9 (1.5 ± 1.4) b | 2.5 ± 0.2 | 99 (94.3) |
| Pathway ID | Pathway Name | Enrichment p-Value * | ||
|---|---|---|---|---|
| All | Up | Down | ||
| ssc00532 | Glycosaminoglycan biosynthesis—chondroitin sulfate/dermatan sulfate | 0.002 | 0.001 | - |
| ssc01100 | Metabolic pathways | 0.002 | 0.007 | 0.078 |
| ssc00980 | Metabolism of xenobiotics by cytochrome P450 | 0.007 | - | 0.003 |
| ssc00982 | Drug metabolism—cytochrome P450 | 0.007 | - | 0.003 |
| ssc00480 | Glutathione metabolism | 0.009 | - | 0.003 |
| ssc00600 | Sphingolipid metabolism | 0.009 | 0.053 | 0.088 |
| ssc04142 | Lysosome | 0.055 | - | 0.026 |
| ssc00900 | Terpenoid backbone biosynthesis | 0.066 | - | 0.042 |
| Pathway ID | Pathway Name | Enrichment p-Value * | ||
|---|---|---|---|---|
| All | Up | Down | ||
| ssc04966 | Collecting duct acid secretion | 0.028 | - | 0.024 |
| ssc04136 | Autophagy—other | 0.034 | - | 0.030 |
| ssc00510 | N-Glycan biosynthesis | 0.051 | - | 0.045 |
| ssc04650 | Natural killer cell-mediated cytotoxicity | 0.102 | 0.013 | - |
| Pathway ID | Pathway Name | Pathway Alteration | ES * | Altered Genes (%) | Gene List |
|---|---|---|---|---|---|
| ssc00532 | Glycosaminoglycan biosynthesis—chondroitin sulfate/dermatan sulfate | Up-regulation | 8.1 | 13.3 | CHSY1, CSGALNACT2 |
| ssc01100 | Metabolic pathways | Up-regulation | 4.9 | 0.6 | CHSY1, CSGALNACT2, CTPS1, RDH10, SPTLC2 |
| ssc00980 | Metabolism of xenobiotics by cytochrome P450 | Down-regulation | 5.9 | 7.7 | GSTK1, GSTO1 |
| ssc00982 | Drug metabolism—cytochrome P450 | Down-regulation | 5.8 | 7.4 | GSTK1, GSTO1 |
| ssc00480 | Glutathione metabolism | Down-regulation | 5.7 | 6.9 | GSTK1, GSTO1 |
| ssc04142 | Lysosome | Down-regulation | 3.6 | 2.4 | ASAH1, SUMF1 |
| ssc00900 | Terpenoid backbone biosynthesis | Down-regulation | 3.2 | 7.1 | FDPS |
| Pathway ID | Pathway Name | Pathway Alteration | ES * | Altered Genes (%) | Gene List |
|---|---|---|---|---|---|
| ssc04650 | Natural killer cell-mediated cytotoxicity | Up-regulation | 4.3 | 1.0 | HCST |
| ssc04966 | Collecting duct acid secretion | Down-regulation | 3.7 | 3.8 | ATP6V1B1 |
| ssc04136 | Autophagy—other | Down-regulation | 3.5 | 3.1 | ATG4C |
| ssc00510 | N-Glycan biosynthesis | Down-regulation | 3.1 | 2.0 | MAN1A2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez-Ramiro, H.; Gil, M.A.; Cuello, C.; Cambra, J.M.; Gonzalez-Plaza, A.; Vazquez, J.M.; Vazquez, J.L.; Rodriguez-Martinez, H.; Lucas-Sanchez, A.; Parrilla, I.; et al. The Use of a Brief Synchronization Treatment after Weaning, Combined with Superovulation, Has Moderate Effects on the Gene Expression of Surviving Pig Blastocysts. Animals 2023, 13, 1568. https://doi.org/10.3390/ani13091568
Gonzalez-Ramiro H, Gil MA, Cuello C, Cambra JM, Gonzalez-Plaza A, Vazquez JM, Vazquez JL, Rodriguez-Martinez H, Lucas-Sanchez A, Parrilla I, et al. The Use of a Brief Synchronization Treatment after Weaning, Combined with Superovulation, Has Moderate Effects on the Gene Expression of Surviving Pig Blastocysts. Animals. 2023; 13(9):1568. https://doi.org/10.3390/ani13091568
Chicago/Turabian StyleGonzalez-Ramiro, Henar, Maria A. Gil, Cristina Cuello, Josep M. Cambra, Alejandro Gonzalez-Plaza, Juan M. Vazquez, Jose L. Vazquez, Heriberto Rodriguez-Martinez, Alejandro Lucas-Sanchez, Inmaculada Parrilla, and et al. 2023. "The Use of a Brief Synchronization Treatment after Weaning, Combined with Superovulation, Has Moderate Effects on the Gene Expression of Surviving Pig Blastocysts" Animals 13, no. 9: 1568. https://doi.org/10.3390/ani13091568








