Retrospective Multi-Locus Sequence Analysis of African Swine Fever Viruses by “PACT” Confirms Co-Circulation of Multiple Outbreak Strains in Uganda
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Acquisition
2.2. Antibody Detection ELISA for Recognition of ASF Antibodies
2.3. OIE(WOAH)-Recommended Diagnostic PCR for ASF
2.3.1. Extraction and Genomic Amplification of the Viral DNA
2.3.2. PCR Purification and Sanger Sequencing
2.4. Sequencing and Phylogeny
3. Results
3.1. Subsection
3.2. Molecular Detection of African Swine Fever Virus
3.2.1. Conventional PCR Using p72 Gene for ASF Diagnosis
3.2.2. Multilocus Typing of the Ten Positive DNA Extracts Using a Four Gene Region (PACT) Approach
3.3. Molecular Diagnosis of African Swine Fever Virus Phylogenetic Analyses
3.3.1. p72 Gene Phylogeny
3.3.2. CVR- 9RL ORF Tetramer Alignment of the Positive 5 Amplicons
3.3.3. p54 Gene Phylogeny
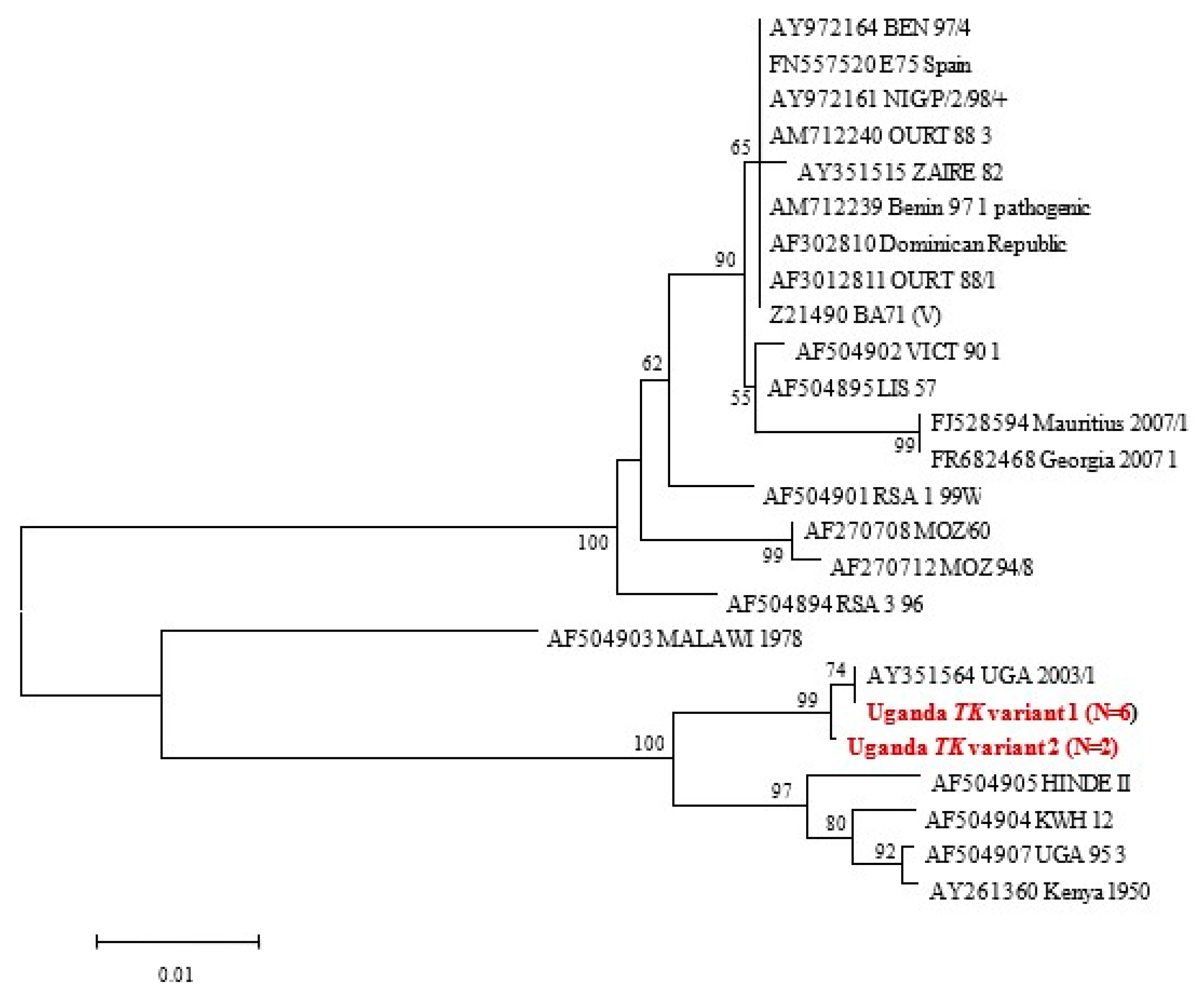
3.3.4. TK Gene Phylogeny
3.3.5. Combined PACT Results
4. Discussion
4.1. Determination of ASF Outbreaks Using Serology
4.2. Determination of ASF Outbreaks Using Molecular Tools
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Acronyms
References
- MacLachlan, J.N.; Dubovi, J.E. Asfaviridae and Iridoviridae, Ch. 8. In Fenner’s Veterinary Virology, 4th ed.; Academic Press: London, UK, 2011; pp. 167–169. [Google Scholar] [CrossRef]
- Bastos, A.D.S.; Fasina, F.O.; King, D.P. African swine fever virus. In Manual of Security Sensitive Microbes and Toxins; Liu, D., Ed.; CRC Press: Sydney, NSW, Australia, 2014; pp. 579–587, E-Book; Available online: http://www.crcpress.com/product/isbn/9781466553965 (accessed on 25 August 2023).
- Montgomery, R.E. On a farm of swine fever occurring in British East Africa (Kenya Colony). J. Comparative Pathol. 1921, 34, 159–191. [Google Scholar] [CrossRef]
- Dixon, L.K.; Escribano, J.M.; Martins, C.; Rock, D.L.; Salas, M.L.; Wilkinson, P.J. Asfarviridae. In Virus Taxonomy, VIIIth Report of the ICTV; Fauquet, C.M., Mayo, M.A., Maniloff, J., Desselberger, U., Ball, L.A., Eds.; Elsevier: Amsterdam, The Netherlands; Academic Press: London, UK, 2005; pp. 135–143. [Google Scholar]
- Bastos, A.D.S.; Penrith, M.L.; Crucière, C.; Edrich, J.L.; Hutchings, G.; Roger, F.; Couacy-Hymann, E.; Thomson, G.R. Genotyping field strains of African swine fever virus by partial p72 gene characterisation. Archives Virol. 2003, 148, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Boshoff, C.I.; Bastos, A.D.S.; Gerber, L.J.; Vosloo, W. Genetic Characterisation of African swine fever viruses from outbreaks in Southern Africa (1973–1999). Vet. Microbiol. 2007, 121, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Lubisi, B.A.; Bastos, A.D.S.; Dwarka, R.M.; Vosloo, W. Molecular epidemiology of African swine fever in East Africa. Arch. Virol. 2005, 150, 2439–2452. [Google Scholar] [CrossRef] [PubMed]
- Achenbach, J.E.; Gallardo, C.; Nieto-Pelegrin, E.; Rivera-Arroyo, B.; Degefa-Negi, T.; Arias, M.; Jenberie, S.; Mulisa, D.D.; Gizaw, D.; Gelaye, E.; et al. Identification of a new genotype of African swine fever virus in domestic pigs from Ethiopia. Transbound. Emerg. Dis. 2017, 64, 1393–1404. [Google Scholar] [CrossRef] [PubMed]
- Penrith, M.-L.; Thomson, G.R.; Bastos, A.D.S. African swine fever. In Infectious Diseases of Livestock with Special Reference to Southern Africa, 2nd ed.; Coetzer, J.A.W., Tustin, R.C., Eds.; Oxford University Press: Cape Town, South Africa, 2004; pp. 1087–1119. [Google Scholar]
- Oleaga-Perez, A.; Perez-Sanchez, R.; Encinas-Grandes, A. Distribution and biology of Ornithodoros erracticus in parts of Spain affected by African swine fever. Vet. Rec. 1990, 126, 32–37. [Google Scholar] [PubMed]
- Boinas, F.S.; Wilson, A.J.; Hutchings, G.H.; Martins, C.; Dixon, L.J. The Persistence of African swine fever virus in field-infected Ornithodoros erraticus during the ASF Endemic Period in Portugal. PLoS ONE 2011, 6, e20383. [Google Scholar] [CrossRef]
- Penrith, M. African swine fever. Onderstepoort J. Vet. Res. 2009, 76, 91–95. [Google Scholar] [CrossRef]
- Jori, F.; Bastos, A.D.S. Role of wild suids in the epidemiology of African swine fever. EcoHealth 2009, 6, 296–310. [Google Scholar] [CrossRef]
- Sánchez-Vizcaíno, J.M.; Martínez-López, B.; Martínez-Avilés, M.; Martins, C.; Boinas, F.; Vialc, L.; Michaud, V.; Jori, F.; Etter, E.; Albina, E.; et al. Scientific review on African swine fever. EFSA Support Publ. 2009, 6, 5E. [Google Scholar] [CrossRef]
- Haresnape, J.M.; Wilkinson, P.J. A study of African swine fever infected ticks (Ornithodoros moubata) collected from three villages in the ASF enzootic area of Malawi following an outbreak of the disease in domestic pigs. Epidemiol. Infect. 1989, 102, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Atuhaire, K.D.; Ochwo, S.; Afayoa, M.; Mwiine, F.N.; Ikwap, K.; Eugene, A.; Ademun-Okurut, R.A.; Okuni, J.B.; Nanteza, A.; Ayebazibwe, C.; et al. Epidemiological overview of African swine fever in Uganda (2001–2012). J. Vet. Med. 2013, 2013, 949638. [Google Scholar] [CrossRef]
- Björnheden, L. A Study of Domestic Pigs, Wild Suids and Ticks as Reservoirs for African Swine Fever Virus in Uganda; Swedish University of Agricultural Sciences (SLU), Dept. of Biomedical Sciences and Veterinary Public Health: Uppsala, Sweden, 2011; Available online: http://stud.epsilon.slu.se/2355/ (accessed on 15 May 2013).
- Atuhaire, D.K.; Afayoa, M.; Ochwo, S.; Mwesigwa, S.; Okuni, J.B.; Olaho-Mukani, W.; Ojok, L. Molecular characterization and phylogenetic study of African swine fever virus isolates from recent outbreaks in Uganda (2010–2013). Virol. J. 2013, 10, 247. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Ademun, A.R.; Nieto, R.; Nantima, N.; Arias, M.; Martín, E.; Pelayo, V.; Bishop, R.P. Genotyping of African swine fever virus (ASFV) isolates associated with disease outbreaks in Uganda in 2007. Afr. J. Biotech 2011, 10, 3488–3497. [Google Scholar]
- Perez-Filguera, D.M.; Gonzalez-Camacho, F.; Gallardo, C.; Resino-Talavan, P.; Blanco, E.; Gomez-Casado, E.; Alonso, C.; Escribano, J.M. Optimization and Validation of Recombinant Serological Tests for African Swine Fever Diagnosis Based on Detection of the p30 Protein Produced in Trichoplusia ni Larvae. J. Clin. Microbiol. 2006, 44, 3114–3121. [Google Scholar] [CrossRef] [PubMed]
- Fosgate, G.T. Practical Sample Size Calculations for Surveillance and Diagnostic Investigations. J. Vet. Diagn. Invest. 2009, 21, 3–14. [Google Scholar] [CrossRef]
- World Organisation for Animal Health. African swine fever (infection with African swine fever virus), Chapter 3.9.1. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 12th ed.; World Organisation for Animal Health: Paris, France, 2023; Available online: https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/3.09.01_ASF.pdf (accessed on 27 November 2023).
- Onzere, C.K.; Bastos, A.D.; Okoth, E.A.; Lichoti, J.K.; Bochere, E.N.; Owido, M.G.; Ndambuki, G.; Bronsvoort, M.; Bishop, R.P. Multi-locus sequence typing of African swine fever viruses from endemic regions of Kenya and Eastern Uganda (2011–2013) reveals rapid B602L central variable region evolution. Virus Genes 2018, 54, 111–123. [Google Scholar] [CrossRef]
- Gallardo, C.; Mwaengo, D.M.; Macharia, J.M.; Arias, M.; Taracha, E.A.; Soler, A.; Okoth, E.; Martín, E.; Kasiti, J.; Bishop, R.P. Enhanced discrimination of African swine fever virus isolates through nucleotide sequencing of the p54, p72, and pB602L (CVR) genes. Virus Genes 2009, 38, 85–95. [Google Scholar] [CrossRef]
- Bastos, A.D.; Penrith, M.L.; Macome, F.; Pinto, F.; Thomson, G.R. Co-circulation of two genetically distinct viruses in an outbreak of African swine fever in Mozambique: No evidence for individual co-infection. Vet. Microbiol. 2004, 103, 169–182. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Geneious, Biomatters, Auckland, New Zealand. Geneious Version 10.0. Available online: http://www.geneious.com (accessed on 13 February 2014).
- Muwonge, A.; Munang’andu, H.M.; Kankya, C.; Biffa, D.; Oura, C.; Skjerve, E.; Oloya, J. African swine fever among slaughter pigs in Mubende district, Uganda. Trop. Anim. Health Prod. 2012, 44, 1593–1598. [Google Scholar] [CrossRef] [PubMed]
- Fasina, F.O.; Shamaki, D.; Makinde, A.A.; Lombin, L.H.; Lazarus, D.D.; Rufai, S.A.; Adamu, S.S.; Agom, D.; Pelayo, V.; Soler, A.; et al. Surveillance for African swine fever in Nigeria, 2006–2009. Transbound Emerg. Dis. 2010, 57, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Etter, E.M.C.; Seck, I.; Grosbois, V.; Jori, F.; Blanco, E.; Vial, L.; Akakpo, A.J.; Bada-Alhambedji, R.; Kone, P.; Roger, F.L. Seroprevalence of African swine fever in Senegal, 2006. Emerg. Infect. Dis. 2011, 17, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, G.H.; Ferris, N.P. Indirect sandwich ELISA for antigen detection of African swine fever virus: Comparison of polyclonal and monoclonal antibodies. J. Virol. Methods 2005, 131, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Kabuuka, T.; Kasaija, P.D.; Mulindwa, H.; Shittu, A.; Bastos, A.D.S.; Fasina, F.O. Drivers and risk factors for circulating African swine fever virus in Uganda, 2012–2013. Res. Vet. Sci. 2014, 97, 218–225. [Google Scholar] [CrossRef]
- Muhangi, D.; Masembe, C.; Emanuelson, U.; Boqvist, S.; Mayega, L.; Ademun, R.O.; Bishop, R.P.; Ocaido, M.; Berg, M.; Ståhl, K. A longitudinal survey of African swine fever in Uganda reveals high apparent disease incidence rates in domestic pigs, but absence of detectable persistent virus infections in blood and serum. BMC Vet. Res. 2015, 11, 106. [Google Scholar] [CrossRef]
- Masembe, C.; Sreenu, V.B.; Da Silva Filipe, A.; Wilkie, G.S.; Ogweng, P.; Mayega, F.J.; Muwanika, V.B.; Biek, R.; Palmarini, M.; Davison, A.J. Genome sequences of five African swine fever virus genotype IX isolates from domestic pigs in Uganda. Microbiol. Resour. Announc. 2018, 7, e01018-18. [Google Scholar] [CrossRef]
- Okwasiimire, R.; Flint, J.F.; Kayaga, E.B.; Lakin, S.; Pierce, J.; Barrette, R.W.; Faburay, B.; Ndoboli, D.; Ekakoro, J.E.; Wampande, E.M.; et al. Whole genome sequencing shows that African swine Fever virus genotype IX is still circulating in domestic pigs in all regions of Uganda. Pathogens 2023, 12, 912. [Google Scholar] [CrossRef]



| District | Samples Positive (%) | Minimum Optical Density | Maximum Optical Density | Samples Negative (%) | Minimum Optical Density | Maximum Optical Density |
|---|---|---|---|---|---|---|
| Pallisa | 0 (0) | - | - | 28 (100) | 1.876 | 2.509 |
| Lira | 0 (0) | - | - | 28 (100) | 1.901 | 2.349 |
| Abim | 6 (24) | 0.058 | 0.739 | 19 (76) | 1.749 | 2.254 |
| Nebbi | 0 (0) | - | - | 28 (100) | 1.844 | 2.489 |
| Kabarole | 0 (0) | - | - | 28 (100) | 1.815 | 2.519 |
| Kibaale | 0 (0) | - | - | 28 (100) | 1.9 | 2.736 |
| Mukono | 0 (0) | - | - | 28 (100) | 1.905 | 2.55 |
| Total | 6 (3.1) | 187 (96.9) |
| District of Origin | Laboratory Sample Name | Number Assigned for Multilocus Typing | Tissue Type | WOAH (OIE) Diagnostic PCR | Four Primer Sets | |||
|---|---|---|---|---|---|---|---|---|
| [A] PPA89 + PPA722 | [C] CVR-FLF + CVR-FLR (Tetramer Number) | [P] P72-U + p72-D | [T] * TK1 + TK-rev | |||||
| Pallisa | 10 | 1 | Kidney | + | A1 | C1 (Tet-23) | P1 | T1 |
| Nebbi | 36 | 2 | Spleen | + | A2 | C2 (Tet-23) | P2 | T2 |
| Kibaale | 46 | 3 | Liver | + | - | - | - | T3 * |
| Kibaale | 48 | 4 | Mesenteric lymph node | + | - | - | - | T4 (NS) |
| Kibaale | 52 | 5 | Spleen | + | - | C5 (Tet-29) | P5 | T5 * |
| Mukono | 58 | 6 | Liver | + | A6 | C6 (Tet-24) | P6 | T6 |
| Kibaale | 59 | 7 | Liver | + | A7 | C7 (Tet-24) | P7 | T7 |
| Nebbi | 2a | 2a | Spleen | - | - | - | - | T2A (NS) |
| Abim | 15 | 15 | Mesenteric lymph node | - | - | - | - | T15 |
| Lira | 17 | 17 | Kidney | - | - | - | - | T17 |
| PCR-positivity | 11.86% | 6.78% | 8.47% | 8.47% | 16.95% | |||
| Relative sensitivity (compared to TK) | 70% | 40% | 50% | 50% | - | |||
| Degree of agreement with diagnostic primers (%) based on Kappa statistics | 70 | 80 | 80 | 100 | ||||
| Kappa statistics | 0.44 (moderate) | 0.60 (moderate) | 0.60 (moderate) | 0.00 (slight) | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kabuuka, T.; Mulindwa, H.; Bastos, A.D.S.; van Heerden, J.; Heath, L.; Fasina, F.O. Retrospective Multi-Locus Sequence Analysis of African Swine Fever Viruses by “PACT” Confirms Co-Circulation of Multiple Outbreak Strains in Uganda. Animals 2024, 14, 71. https://doi.org/10.3390/ani14010071
Kabuuka T, Mulindwa H, Bastos ADS, van Heerden J, Heath L, Fasina FO. Retrospective Multi-Locus Sequence Analysis of African Swine Fever Viruses by “PACT” Confirms Co-Circulation of Multiple Outbreak Strains in Uganda. Animals. 2024; 14(1):71. https://doi.org/10.3390/ani14010071
Chicago/Turabian StyleKabuuka, Tonny, Henry Mulindwa, Armanda D. S. Bastos, Juanita van Heerden, Livio Heath, and Folorunso O. Fasina. 2024. "Retrospective Multi-Locus Sequence Analysis of African Swine Fever Viruses by “PACT” Confirms Co-Circulation of Multiple Outbreak Strains in Uganda" Animals 14, no. 1: 71. https://doi.org/10.3390/ani14010071
APA StyleKabuuka, T., Mulindwa, H., Bastos, A. D. S., van Heerden, J., Heath, L., & Fasina, F. O. (2024). Retrospective Multi-Locus Sequence Analysis of African Swine Fever Viruses by “PACT” Confirms Co-Circulation of Multiple Outbreak Strains in Uganda. Animals, 14(1), 71. https://doi.org/10.3390/ani14010071








