Identification of Differentially Expressed Genes and microRNAs in the Gray and White Feather Follicles of Shitou Geese
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
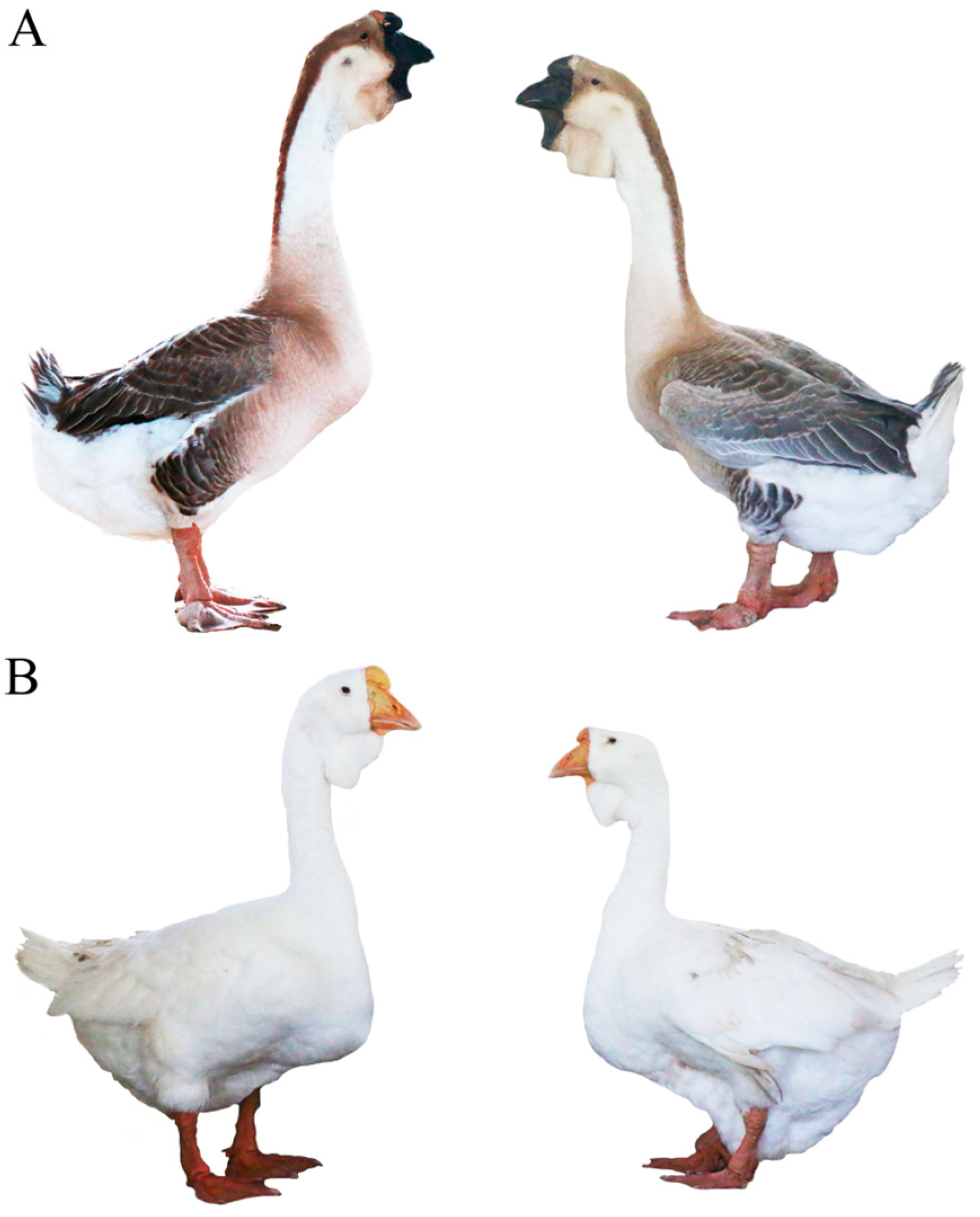
2.1. Sample Collection
2.2. Total RNA Extraction, cDNA Library Preparation, and Transcriptome Sequencing
2.3. Small RNA Library Construction and Deep Sequencing
2.4. Bioinformatic Analyses of Transcriptomes
2.5. Processing and Analysis of Small RNA Sequences
2.6. Construction of a miRNA-mRNA Interaction Network
2.7. Quantitative Real-Time PCR Validation
3. Results
3.1. Summary of mRNA Sequencing Data
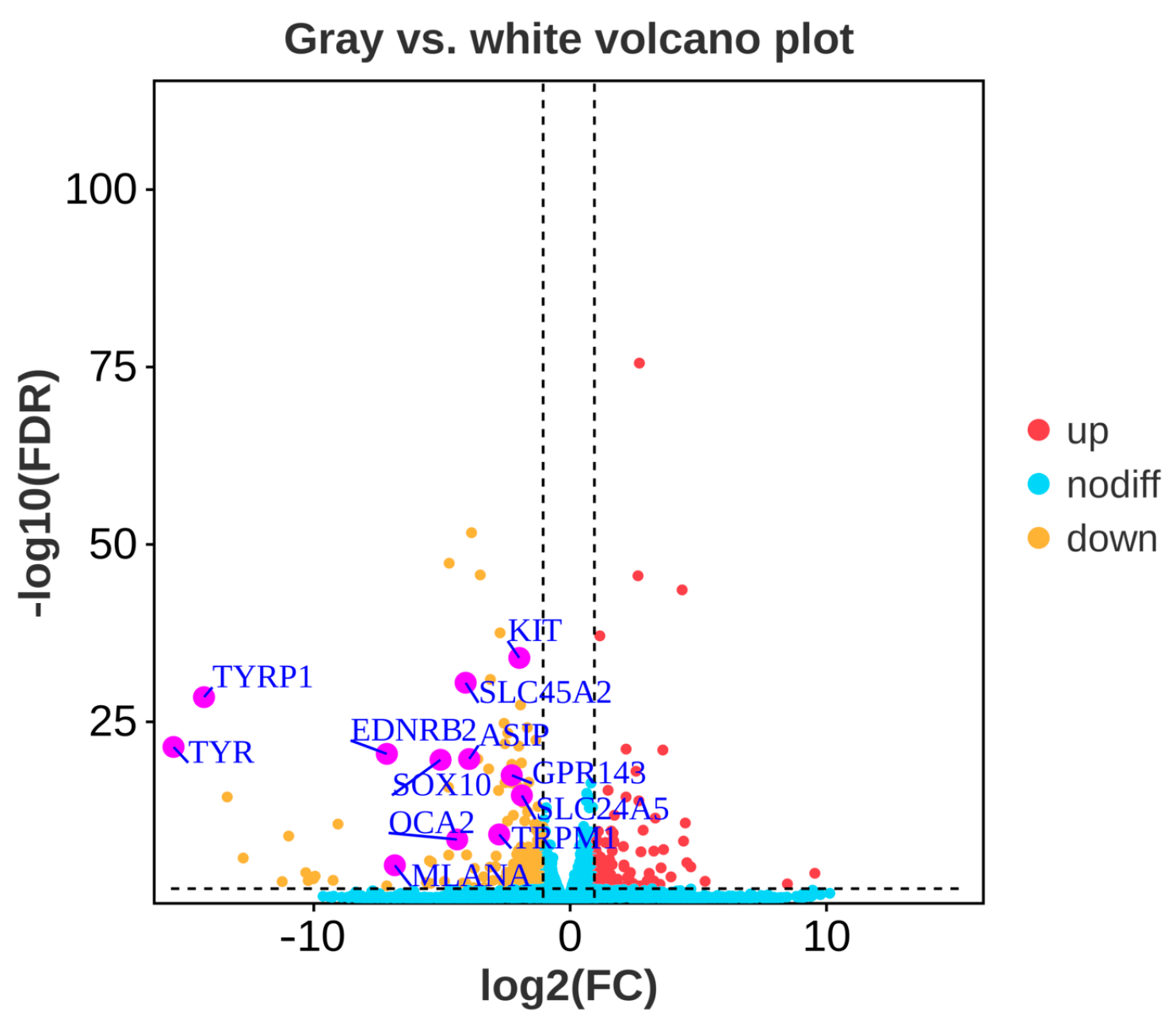
3.2. Differentially Expressed Genes between Gray and White Feather Follicles
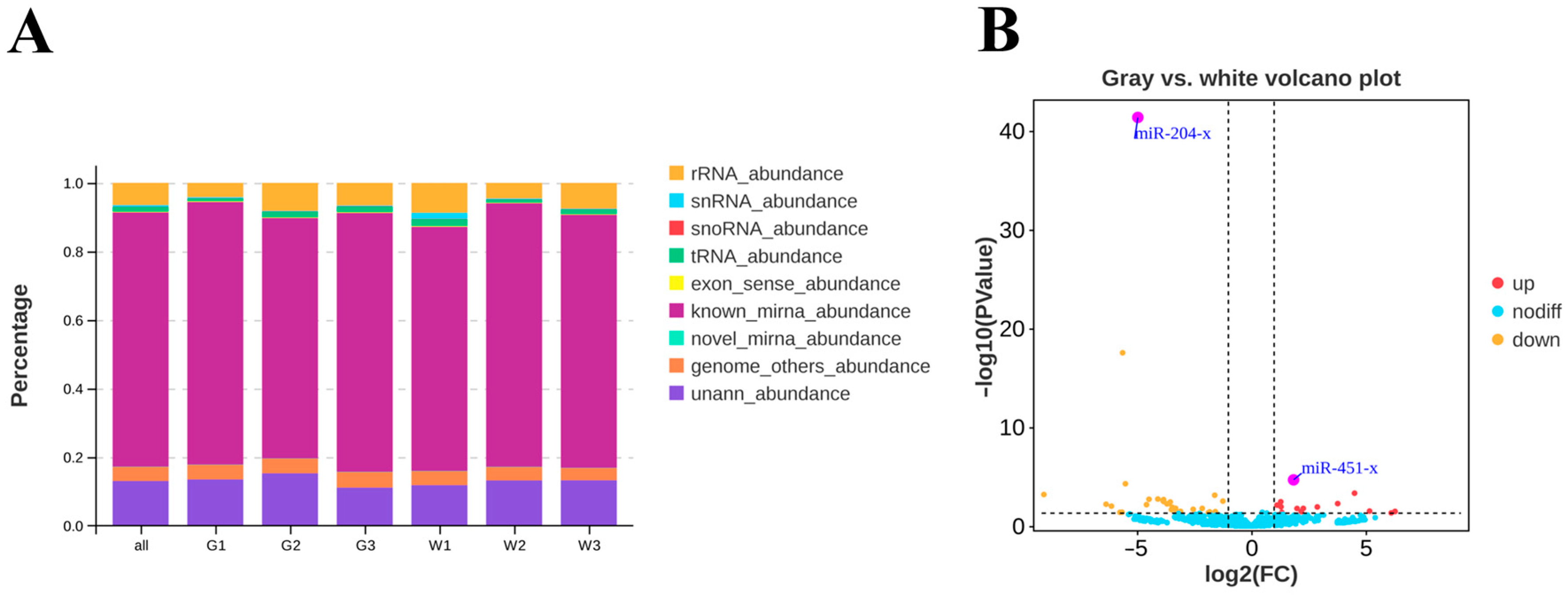
3.3. Differentially Expressed miRNAs and Their Target Genes in the Gray and White Feather Follicles
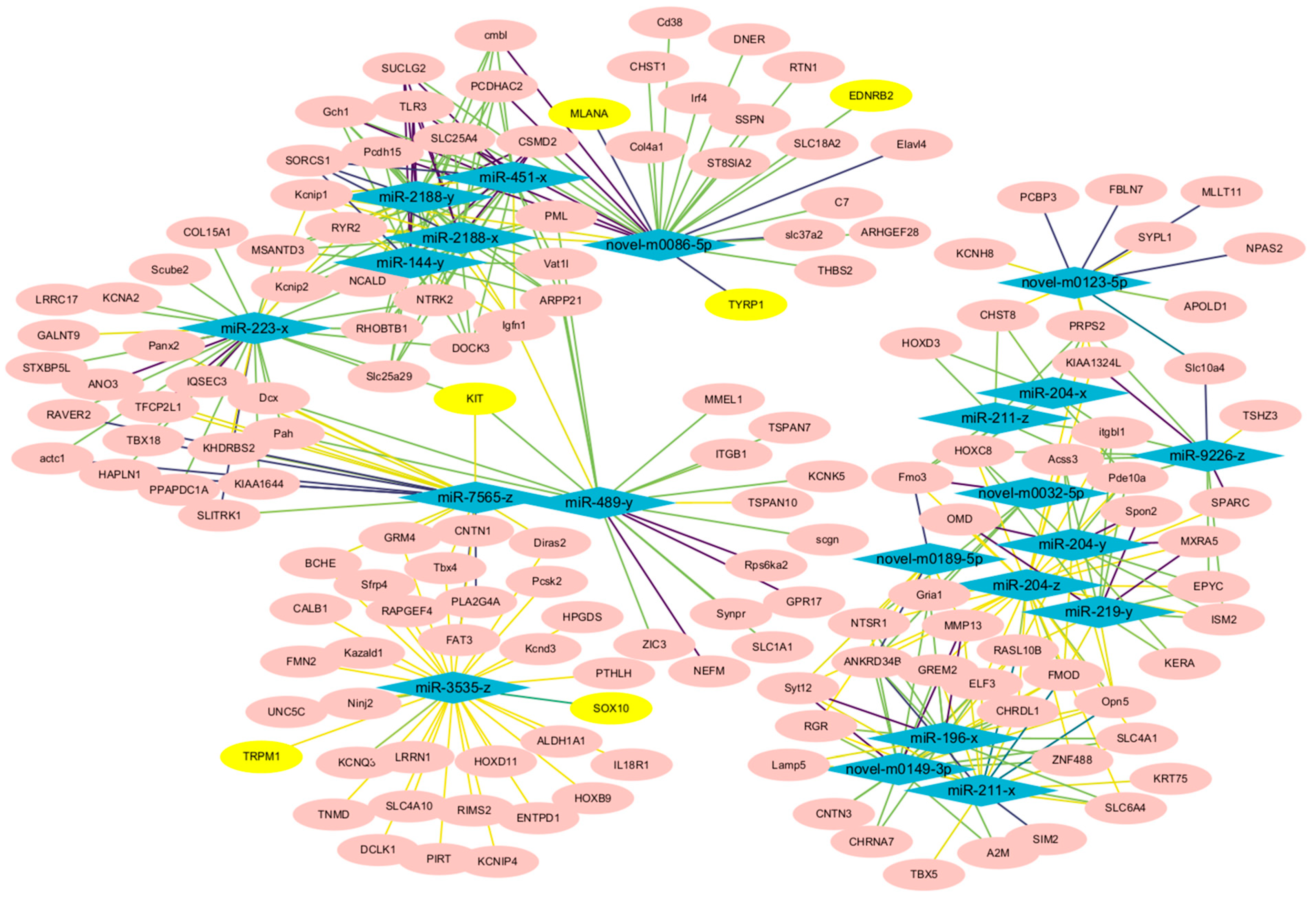
3.4. Analysis of the Constructed miRNA and mRNA Network
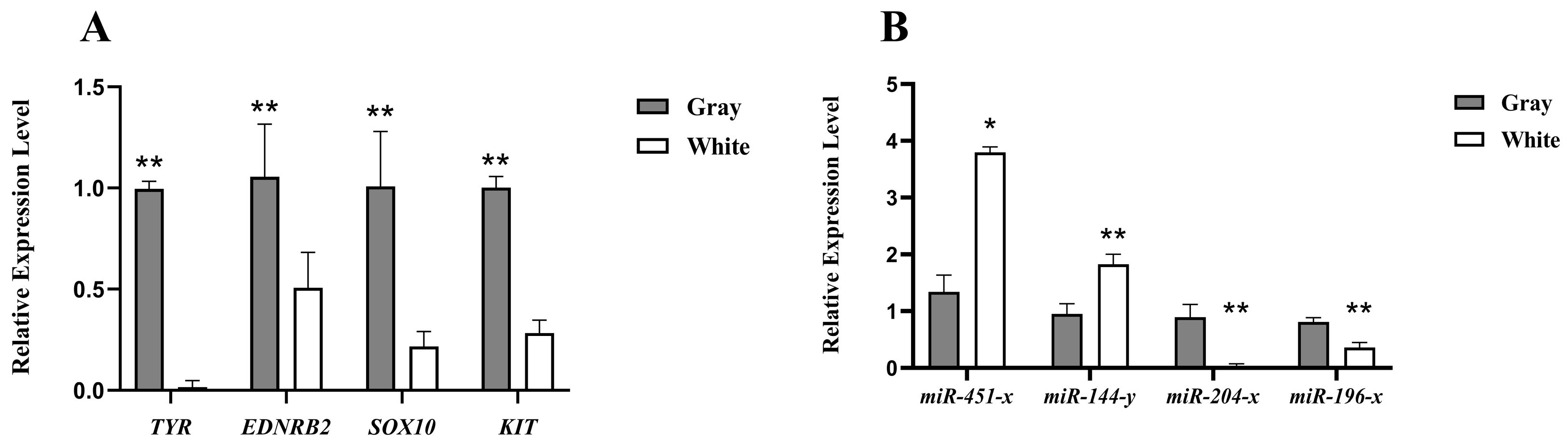
3.5. Validation of RNA and miRNA Expression Results by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paxton, E.H. The Utility of Plumage Coloration for Taxonomic and Ecological Studies. Open Ornithol. J. 2009, 2, 17–23. [Google Scholar] [CrossRef]
- Roulin, A.; Ducrest, A.L. Genetics of colouration in birds. Semin. Cell Dev. Biol. 2013, 24, 594–608. [Google Scholar] [CrossRef]
- Gong, Y.; Yang, Q.; Li, S.; Feng, Y.; Gao, C.; Tu, G.; Peng, X. Grey plumage colouration in the duck is genetically determined by the alleles on two different, interacting loci. Anim. Genet. 2010, 41, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, C.; Yu, W.; Zhao, S.; Gong, Y. Identification of Genes Related to White and Black Plumage Formation by RNA-Seq from White and Black Feather Bulbs in Ducks. PLoS ONE 2012, 7, e36592. [Google Scholar] [CrossRef]
- Xi, Y.; Wang, L.; Liu, H.; Ma, S.; He, H. A 14-bp insertion in endothelin receptor B-like (EDNRB2) is associated with white plumage in Chinese geese. BMC Genom. 2020, 21, 162. [Google Scholar] [CrossRef]
- Zhao, Q.; Chen, J.; Zhang, X.; Xu, Z.; Lin, Z.; Li, H.; Lin, W.; Xie, Q. Genome-Wide Association Analysis Reveals Key Genes Responsible for Egg Production of Lion Head Goose. Front. Genet. 2019, 10, 1391. [Google Scholar] [CrossRef]
- Cieslak, M.; Reissmann, M.; Hofreiter, M.; Hofreiter, M.; Ludwig, A.; Ludwig, A. Colours of domestication. Biol. Rev. Camb. Philos. Soc. 2011, 86, 885–899. [Google Scholar] [CrossRef]
- Ye, W.; Siming, L.; Jing, H.; Shiyi, C.; Yiping, L. Mutations of TYR and MITF genes are associated with plumage colour phenotypes in geese. Asian-Australas. J. Anim. Sci. 2014, 27, 778–783. [Google Scholar]
- Yang, Y.; Wang, H.; Li, G.; Liu, Y.; Wang, C.; Qiu, S.; Wang, X.; Yao, J.; Zhu, L.; He, D. Using comparative genomics to detect mutations regulating plumage variations in graylag (A. anser) and swan geese (A. cygnoides). Gene 2022, 834, 146612. [Google Scholar] [CrossRef]
- Ouyang, J.; Zheng, S.; Huang, M.; Tang, H.; Qiu, X.; Chen, S.; Wang, Z.; Zhou, Z.; Gao, Y.; Xiong, Y. Chromosome-level genome and population genomics reveal evolutionary characteristics and conservation status of Chinese indigenous geese. Commun. Biol. 2022, 5, 1191. [Google Scholar] [CrossRef]
- Ren, S.; Lyu, G.; Irwin, D.M.; Liu, X.; Feng, C.; Luo, R.; Zhang, J.; Sun, Y.; Shang, S.; Zhang, S. Pooled Sequencing Analysis of Geese (Anser cygnoides) Reveals Genomic Variations Associated with Feather Color. Front. Genet. 2021, 12, 650013. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Shao, P.; Chen, Y.; Wang, L.; Lv, X.; Yang, W.; Jia, Y.; Jiang, Z.; Zhu, B.; Qu, L. Genomic scan revealed KIT gene underlying white/gray plumage color in Chinese domestic geese. Anim. Genet. 2021, 52, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Hushcha, Y.; Blo, I.; Oton-Gonzalez, L.A.-O.; Mauro, G.A.-O.; Martini, F.; Tognon, M.; Mattei, M.A.-O. microRNAs in the Regulation of Melanogenesis. Int. J. Mol. Sci. 2021, 22, 6104. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Du, B.; Zhang, P.; Zhang, J.; Zhu, Z.; Liu, B.; Fan, R.A.-O. miR-380-3p regulates melanogenesis by targeting SOX6 in melanocytes from alpacas (Vicugna pacos). BMC Genom. 2019, 20, 962. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Li, J.; Ma, T.; Li, J.; Zhang, Q. MiR-27a regulates WNT3A and KITLG expression in Cashmere goats with different coat colors. Anim. Biotechnol. 2019, 32, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Luo, M.; Wang, L.; Yin, H.; Fu, J. MicroRNA-206 Regulation of Skin Pigmentation in Koi Carp (Cyprinus carpio L.). Front. Genet. 2020, 11, 47. [Google Scholar] [CrossRef]
- Li, J.; Ba, X.; Li, J.; Li, Y.; Wu, S.; Jiang, H.; Zhang, Q. MicroRNA-200a regulates skin pigmentation by targeting WNT5A and FZD4 in Cashmere goats. Res. Vet. Sci. 2022, 147, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.A.-O. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.A.-O. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; McCue, K.; Schaeffer, L.; Schaeffer, L.; Wold, B.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Ostell, J.; Pruitt, K.D.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2020, 48, D84–D86. [Google Scholar] [PubMed]
- Kalvari, I.; Nawrocki, E.P.; Ontiveros-Palacios, N.; Argasinska, J.; Lamkiewicz, K.; Marz, M.; Griffiths-Jones, S.; Toffano-Nioche, C.; Gautheret, D.; Weinberg, Z.; et al. Rfam 14: Expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021, 49, D192–D200. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, M.R.; Chen, W.; Adamidi, C.; Adamidi, C.; Maaskola, J.; Maaskola, J.; Einspanier, R.; Einspanier, R.; Knespel, S.; Knespel, S.; et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008, 26, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human MicroRNA targets. PLoS Biol. 2005, 2, e363. [Google Scholar] [CrossRef]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Ozier, O.; Baliga, N.S.; Baliga, N.S.; Wang, J.T.; Wang, J.T.; Ramage, D.; Ramage, D.; et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Raposo, G.; Marks, M.S. Melanosomes–dark organelles enlighten endosomal membrane transport. Nat. Rev. Mol. Cell Biol. 2007, 8, 786–797. [Google Scholar] [CrossRef]
- Baxter, L.L.; Hou, L.; Loftus, S.K.; Pavan, W.J.J.P.C.R. Spotlight on Spotted Mice: A Review of White Spotting Mouse Mutants and Associated Human Pigmentation Disorders. Pigment. Cell Res. 2010, 17, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zhang, B.; Zhang, Y.; Zhong, H.; Nie, R.; Li, J.; Zhang, H.A.-O.; Wu, C. Transcriptome analysis of feather follicles reveals candidate genes and pathways associated with pheomelanin pigmentation in chickens. Sci. Rep. 2020, 10, 12088. [Google Scholar] [CrossRef]
- Pla, P.; Alberti, C.; Solov’eva, O.; Solov’eva, O.; Pasdar, M.; Pasdar, M.; Kunisada, T.; Kunisada, T.; Larue, L.; Larue, L. Ednrb2 orients cell migration towards the dorsolateral neural crest pathway and promotes melanocyte differentiation. Pigment. Cell Res. 2005, 18, 181–187. [Google Scholar] [CrossRef]
- Harris, M.L.; Baxter, L.L.; Loftus, S.K.; Loftus, S.K.; Pavan, W.J.; Pavan, W.J. Sox proteins in melanocyte development and melanoma. Pigment. Cell Melanoma Res. 2010, 23, 496–513. [Google Scholar] [CrossRef] [PubMed]
- Wehrle-Haller, B. The role of Kit-ligand in melanocyte development and epidermal homeostasis. Pigment. Cell Res. 2003, 16, 287–296. [Google Scholar] [CrossRef]
- Miwa, M.; Inoue-Murayama, M.; Kobayashi, N.; Kayang, B.B.; Mizutani, M.; Takahashi, H.; Ito, S. Mapping of panda plumage color locus on the microsatellite linkage map of the Japanese quail. BMC Genet. 2006, 7, 2. [Google Scholar] [CrossRef]
- Miwa, M.; Inoue-Murayama, M.; Aoki, H.; Kunisada, T.; Hiragaki, T.; Mizutani, M.; Ito, S. Endothelin receptor B2 (EDNRB2) is associated with the panda plumage colour mutation in Japanese quail. Anim. Genet. 2007, 38, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, K.; Akiyama, T.; Mizutani, M.; Shinomiya, A.; Ishikawa, A.; Younis, H.H.; Tsudzuki, M.; Namikawa, T.; Matsuda, Y. Endothelin receptor B2 (EDNRB2) is responsible for the tyrosinase-independent recessive white (mo(w)) and mottled (mo) plumage phenotypes in the chicken. PLoS ONE 2014, 9, e86361. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, D.; Liu, L.; Li, S.; Feng, Y.; Peng, X.; Gong, Y. Endothelin Receptor B2 (EDNRB2) Gene Is Associated with Spot Plumage Pattern in Domestic Ducks (Anas platyrhynchos). PLoS ONE 2015, 10, e0125883. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Jiang, Y.; Wang, Z.; Bi, Y.; Chen, G.; Bai, H.A.-O.; Chang, G.A.-O.X. Genome-Wide Analysis Identifies Candidate Genes Encoding Feather Color in Ducks. Genes 2022, 13, 1249. [Google Scholar] [CrossRef]
- Gunnarsson, U.; Kerje, S.; Bed’Hom, B.; Sahlqvist, A.S.; Ekwall, O.; Tixier-Boichard, M.; Kmpe, O.; Andersson, L. The Dark brown plumage color in chickens is caused by an 8.3-kb deletion upstream of SOX10. Pigment. Cell Melanoma Res. 2011, 24, 268–274. [Google Scholar] [CrossRef]
- Zhu, T.; Liu, M.; Peng, S.; Zhang, X.; Chen, Y.; Lv, X.; Yang, W.; Li, K.; Zhang, J.; Wang, H. A Deletion Upstream of SOX10 Causes Light Yellow Plumage Colour in Chicken. Genes 2022, 13, 327. [Google Scholar] [CrossRef]
- Zhang, L.; Tang, J.; Haines, C.J.; Feng, H.; Lai, L.; Teng, X.; Han, Y. c-kit expression profile and regulatory factors during spermatogonial stem cell differentiation. BMC Dev. Biol. 2013, 13, 38. [Google Scholar] [CrossRef]
- Johansson Moller, M.; Chaudhary, R.; Hellmén, E.; Höyheim, B.; Chowdhary, B.; Andersson, L. Pigs with the dominant white coat color phenotype carry a duplication of the KIT gene encoding the mast/stem cell growth factor receptor. Mamm. Genome 1996, 7, 822–830. [Google Scholar] [CrossRef]
- Anello, M.; Daverio, M.S.; Silbestro, M.B.; Vidal-Rioja, L.; Di Rocco, F. Characterization and expression analysis of KIT and MITF-M genes in llamas and their relation to white coat color. Anim. Genet. 2019, 50, 143–149. [Google Scholar] [CrossRef]
- Wu, Z.; Deng, Z.; Huang, M.; Hou, Y.; Zhang, H.; Chen, H.; Ren, J. Whole-Genome Resequencing Identifies KIT New Alleles That Affect Coat Color Phenotypes in Pigs. Front. Genet. 2019, 10, 218. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, C.; Xu, H.; Xia, X.; Luo, X.; Li, K.; Han, J.; Lei, C.A.-O.; Chen, N.A.-O.; Yue, X.A.-O.X. Genomic analysis reveals a KIT-related chromosomal translocation associated with the white coat phenotype in yak. J. Anim. Breed. Genet. 2023, 140, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Pavan, W.J.; Sturm, R.A. The Genetics of Human Skin and Hair Pigmentation. Annu. Rev. Genom. Hum. Genet. 2019, 20, 41–72. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Urabe, K.; Winder, A.; Jimenezcervantes, C.; Hearing, V.J.E.J. Tyrosinase related protein 1 (TRP1) functions as a DHICA oxidase in melanin biosynthesis. EMBO J. 1994, 13, 5818–5825. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Imokawa, G.; Bennett, D.C.; Hearing, V.J. Tyrosinase Stabilization by Tyrp1 (the brown Locus Protein). J. Biol. Chem. 1998, 273, 31801–31805. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.M.; Coville, J.L.; Coquerelle, G.; Gourichon, D.; Oulmouden, A.; Tixier-Boichard, M. Complete association between a retroviral insertion in the tyrosinase gene and the recessive white mutation in chickens. BMC Genom. 2006, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, S.M.; Berryere, T.G.; Goldfinch, A.D. TYRP1 and MC1R genotypes and their effects on coat color in dogs. Mamm. Genome 2002, 13, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Mao, H.; Zhang, Z.; Xiao, S.; Ding, N.; Huang, L. A 6-bp deletion in the TYRP1 gene causes the brown colouration phenotype in Chinese indigenous pigs. Heredity 2011, 106, 862–868. [Google Scholar] [CrossRef] [PubMed]
- Becker, D.; Otto, M.; Ammann, P.; Keller, I.; Leeb, T. The brown coat colour of Coppernecked goats is associated with a non-synonymous variant at the TYRP1 locus on chromosome 8. Anim. Genet. 2015, 46, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Bedhom, B.; Marthey, S.; Boichard, M.T. A missense mutation in TYRP1 causes the chocolate plumage color in chicken and alters melanosome structure. Pigment. Cell Melanoma Res. 2018, 32, 381–390. [Google Scholar] [CrossRef]
- Corbin, L.A.-O.; Pope, J.; Sanson, J.; Antczak, D.F.; Miller, D.; Sadeghi, R.; Brooks, S.A.-O. An Independent Locus Upstream of ASIP Controls Variation in the Shade of the Bay Coat Colour in Horses. Genes 2020, 11, 606. [Google Scholar] [CrossRef]
- Trigo, B.B.; Utsunomiya, A.T.H.; Fortunato, A.; Milanesi, M.; Torrecilha, R.B.P.; Lamb, H.; Nguyen, L.; Ross, E.M.; Hayes, B.; Padula, R.C.M.; et al. Variants at the ASIP locus contribute to coat color darkening in Nellore cattle. Genet. Sel. Evol. 2021, 53, 40. [Google Scholar] [CrossRef]
- Nadeau, N.J.; Minvielle, F.; Ito, S.; Inoue-Murayama, M.; Gourichon, D.; Follett, S.A.; Burke, T.; Mundy, N.I. Characterization of Japanese quail yellow as a genomic deletion upstream of the avian homolog of the mammalian ASIP (agouti) gene. Genetics 2008, 178, 777–786. [Google Scholar] [CrossRef]
- Sturm, R.A. Molecular genetics of human pigmentation diversity. Hum. Mol. Genet. 2009, 18, R9–R17. [Google Scholar] [CrossRef] [PubMed]
- Bruders, R.A.-O.; Van Hollebeke, H.; Osborne, E.A.-O.; Kronenberg, Z.A.-O.; Maclary, E.; Yandell, M.; Shapiro, M.A.-O. A copy number variant is associated with a spectrum of pigmentation patterns in the rock pigeon (Columba livia). PLoS Genet. 2020, 16, e1008274. [Google Scholar] [CrossRef] [PubMed]
- Oancea, E.; Vriens, J.; Brauchi, S.; Jun, J.; Splawski, I.; Clapham, D.E. TRPM1 forms ion channels associated with melanin content in melanocytes. Sci. Signal. 2009, 2, ra21. [Google Scholar] [CrossRef] [PubMed]
- Gunnarsson, U.; Hellström, A.R.; Tixier-Boichard, M.; Minvielle, F.; Bed’hom, B.; Ito, S.; Jensen, P.; Rattink, A.; Vereijken, A.; Andersson, L. Mutations in SLC45A2 cause plumage color variation in chicken and Japanese quail. Genetics 2007, 175, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Mack, M.; Kowalski, E.; Grahn, R.; Bras, D.; Penedo, M.C.T.; Bellone, R.A.-O. Two Variants in SLC24A5 Are Associated with “Tiger-Eye” Iris Pigmentation in Puerto Rican Paso Fino Horses. G3 (Bethesda) 2017, 7, 2799–2806. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; You, B.; Xu, K.; Zhang, X.; Xie, Y.; Li, Y. GPR143 genotypic and ocular phenotypic characterisation in a Chinese cohort with ocular albinism. Ophthalmic Genet. 2021, 42, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Caduff, M.; Bauer, A.; Jagannathan, V.; Leeb, T.A.-O. OCA2 splice site variant in German Spitz dogs with oculocutaneous albinism. PLoS ONE 2017, 12, e0185944. [Google Scholar] [CrossRef]
- Dong, C.; Wang, H.; Xue, L.; Dong, Y.; Yang, L.; Fan, R.; Yu, X.; Tian, X.; Ma, S.; Smith, G.W. Coat color determination by miR-137 mediated down-regulation of microphthalmia-associated transcription factor in a mouse model. RNA 2012, 18, 1679–1686. [Google Scholar] [CrossRef]
- Li, J.; Liu, L.; Zhang, J.; Cheng, L.; Ren, L.; Zhao, Y.A.-O. The expression of miR-129-5p and its target genes in the skin of goats. Anim. Biotechnol. 2021, 32, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xiong, G.; Zeng, D.; Zhang, J.; Ge, L.; Liu, L.; Wang, X.; Hu, Y. Comparative transcriptome and miRNA analysis of skin pigmentation during embryonic development of Chinese soft-shelled turtle (Pelodiscus sinensis). BMC Genom. 2022, 23, 801. [Google Scholar] [CrossRef] [PubMed]
- Xiang, B.; Li, Y.; Li, J.; Li, J.; Jiang, H.; Zhang, Q.A.-O. MiR-19 3b regulated the formation of coat colors by targeting WNT10A and GNAI2 in Cashmere goats. Anim. Biotechnol. 2023, 34, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Wang, R.; Wang, Z.-X.; Wang, Z.X. The potential role of miR-451 in cancer diagnosis, prognosis, and therapy. Mol. Cancer Ther. 2013, 12, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, H.; Li, L.H.; Tang, J.B.; Sheng, Y.L. Mir-451 inhibits proliferation and migration of non-small cell lung cancer cells via targeting LKB1/AMPK. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 274–280. [Google Scholar] [PubMed]
- Wu, C.; Liu, X.; Li, B.; Sun, G.; Peng, C.; Xiang, D. miR-451 suppresses the malignant characteristics of colorectal cancer via targeting SAMD4B. Mol. Med. Rep. 2021, 24, 557. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.R.; Reverendo, M.; Pereira, P.M.; Nivelles, O.; Pendeville, H.; Bezerra, A.R.; Moura, G.R.; Struman, I.; Santos, M.A.S. Dre-miR-2188 targets Nrp2a and mediates proper intersegmental vessel development in zebrafish embryos. PLoS ONE 2012, 7, e39417. [Google Scholar] [CrossRef] [PubMed]
- Mabrouk, I.; Zhou, Y.; Wang, S.; Song, Y.; Fu, X.; Xu, X.; Liu, T.; Wang, Y.; Feng, Z.; Fu, J.; et al. Transcriptional Characteristics Showed That miR-144-y/FOXO3 Participates in Embryonic Skin and Feather Follicle Development in Zhedong White Goose. Animals 2022, 12, 2099. [Google Scholar] [CrossRef] [PubMed]
- Jia, N.; Tong, H.; Zhang, Y.; Katayama, H.; Wang, Y.; Lu, W.; Zhang, S.; Wang, J. CeRNA Expression Profiling Identifies KIT-Related circRNA-miRNA-mRNA Networks in Gastrointestinal Stromal Tumour. Front. Genet. 2019, 10, 825. [Google Scholar] [CrossRef]
- Kok, M.G.M.; de Ronde, M.W.J.; Moerland, P.D.; Ruijter, J.M.; Creemers, E.E.; Pinto-Sietsma, S.J. Small sample sizes in high-throughput miRNA screens: A common pitfall for the identification of miRNA biomarkers. Biomol. Detect. Quantif. 2017, 15, 1–5. [Google Scholar] [CrossRef]
| Sample | Raw Reads | Clean Reads | Clean Bases (bp) | GC_Content (%) | Q30_Value (%) | Total_Mapped (%) | Unique_Mapped (%) |
|---|---|---|---|---|---|---|---|
| G1 | 43,590,714 | 43,270,040 | 6,450,918,824 | 50.18 | 93.70 | 82.18 | 78.74 |
| G2 | 445,30,290 | 44,220,580 | 6,591,098,695 | 50.26 | 94.17 | 83.01 | 79.02 |
| G3 | 43,000,870 | 42,707,118 | 6,369,209,432 | 50.06 | 93.58 | 82.52 | 79.43 |
| W1 | 43,590,238 | 43,295,454 | 6,455,337,938 | 51.32 | 93.87 | 80.89 | 76.74 |
| W2 | 44,149,770 | 43,872,124 | 6,539,210,760 | 51.51 | 94.25 | 80.70 | 75.88 |
| W3 | 43,824,664 | 43,536,374 | 6,501,179,292 | 50.86 | 93.49 | 81.12 | 77.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, P.; Chen, J.; Luo, L.; Zhang, X.; Li, X.; Huang, Y.; Wu, Z.; Tian, Y. Identification of Differentially Expressed Genes and microRNAs in the Gray and White Feather Follicles of Shitou Geese. Animals 2024, 14, 1508. https://doi.org/10.3390/ani14101508
Guo P, Chen J, Luo L, Zhang X, Li X, Huang Y, Wu Z, Tian Y. Identification of Differentially Expressed Genes and microRNAs in the Gray and White Feather Follicles of Shitou Geese. Animals. 2024; 14(10):1508. https://doi.org/10.3390/ani14101508
Chicago/Turabian StyleGuo, Pengyun, Junpeng Chen, Lei Luo, Xumeng Zhang, Xiujin Li, Yunmao Huang, Zhongping Wu, and Yunbo Tian. 2024. "Identification of Differentially Expressed Genes and microRNAs in the Gray and White Feather Follicles of Shitou Geese" Animals 14, no. 10: 1508. https://doi.org/10.3390/ani14101508
APA StyleGuo, P., Chen, J., Luo, L., Zhang, X., Li, X., Huang, Y., Wu, Z., & Tian, Y. (2024). Identification of Differentially Expressed Genes and microRNAs in the Gray and White Feather Follicles of Shitou Geese. Animals, 14(10), 1508. https://doi.org/10.3390/ani14101508







