The First Genetic Characterization of the SPRN Gene in Pekin Ducks (Anas platyrhynchos domesticus)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Genetic Analysis of Pekin Duck SPRN
2.3. Multiple Sequence Alignment
2.4. 3D Structure Prediction of Duck Sho Protein
2.5. In Silico Evaluation of the Effect of Non-Synonymous SNPs on the Duck Sho Protein
2.6. Statistical Analysis
3. Results
3.1. Identification of the SPRN Gene Sequence of Pekin Ducks
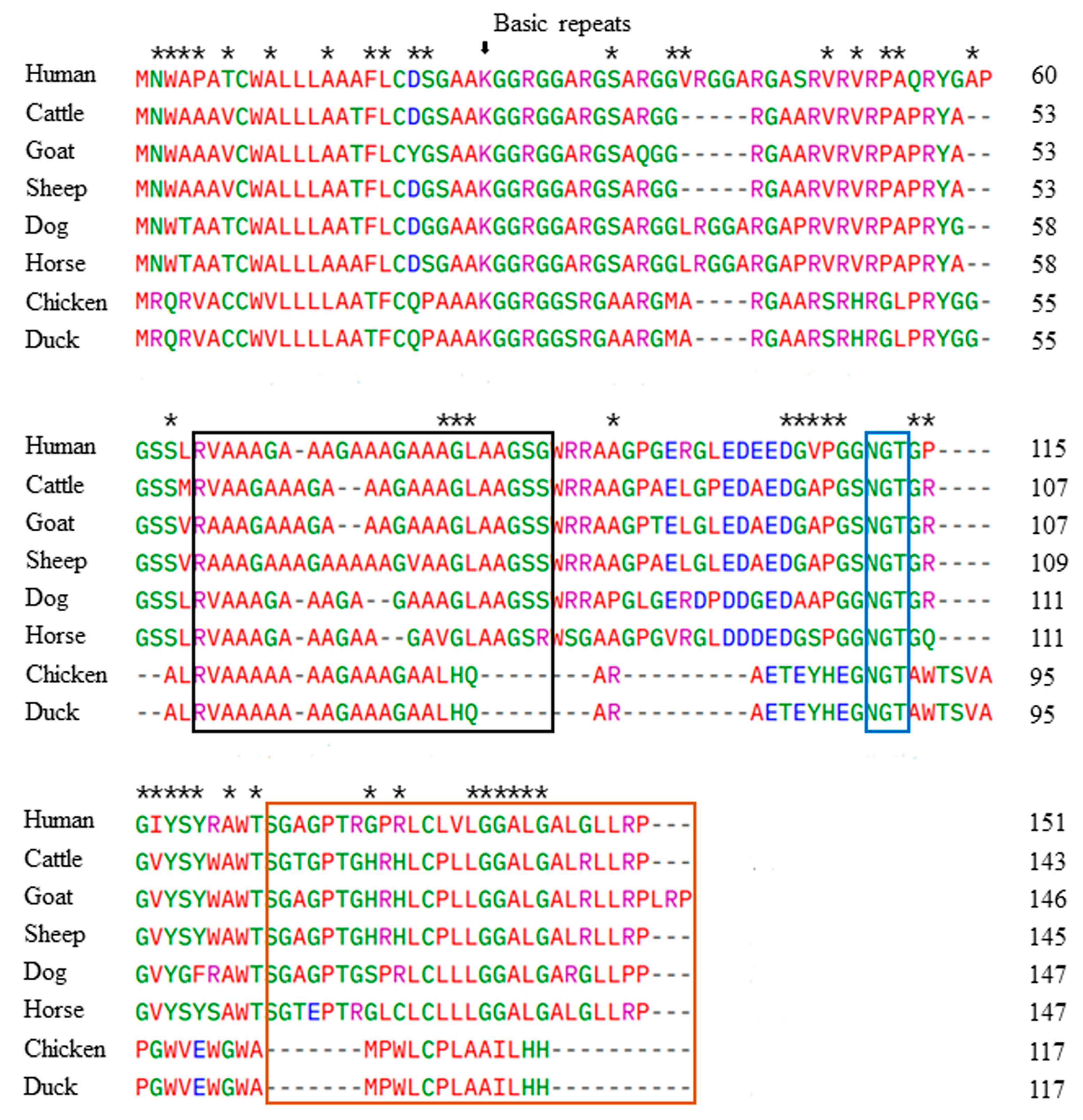
3.2. Comparison of the Duck Sho Protein Sequence with Those of Several Species
3.3. Identification of Novel SNPs for the SPRN Gene in Pekin Ducks
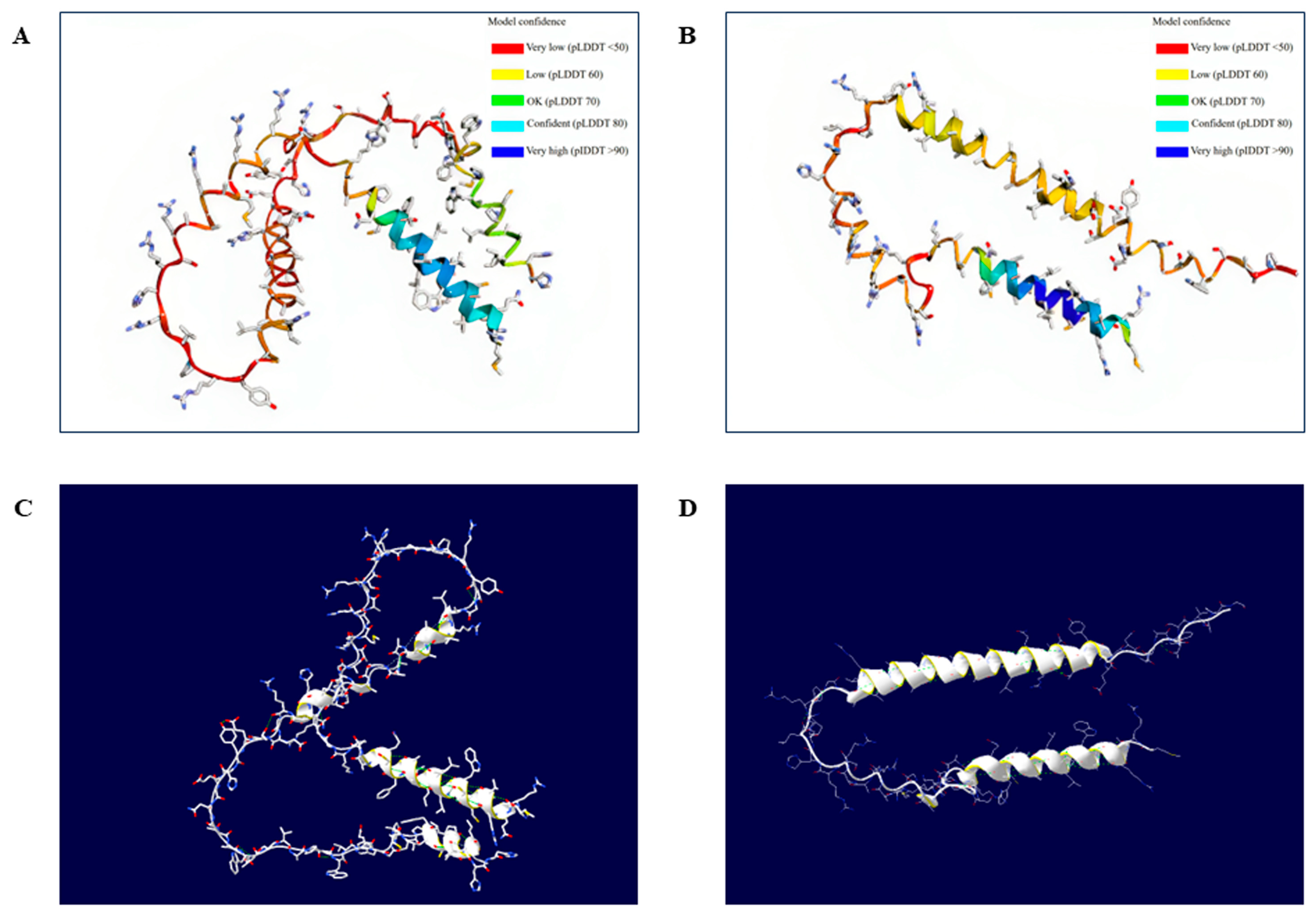
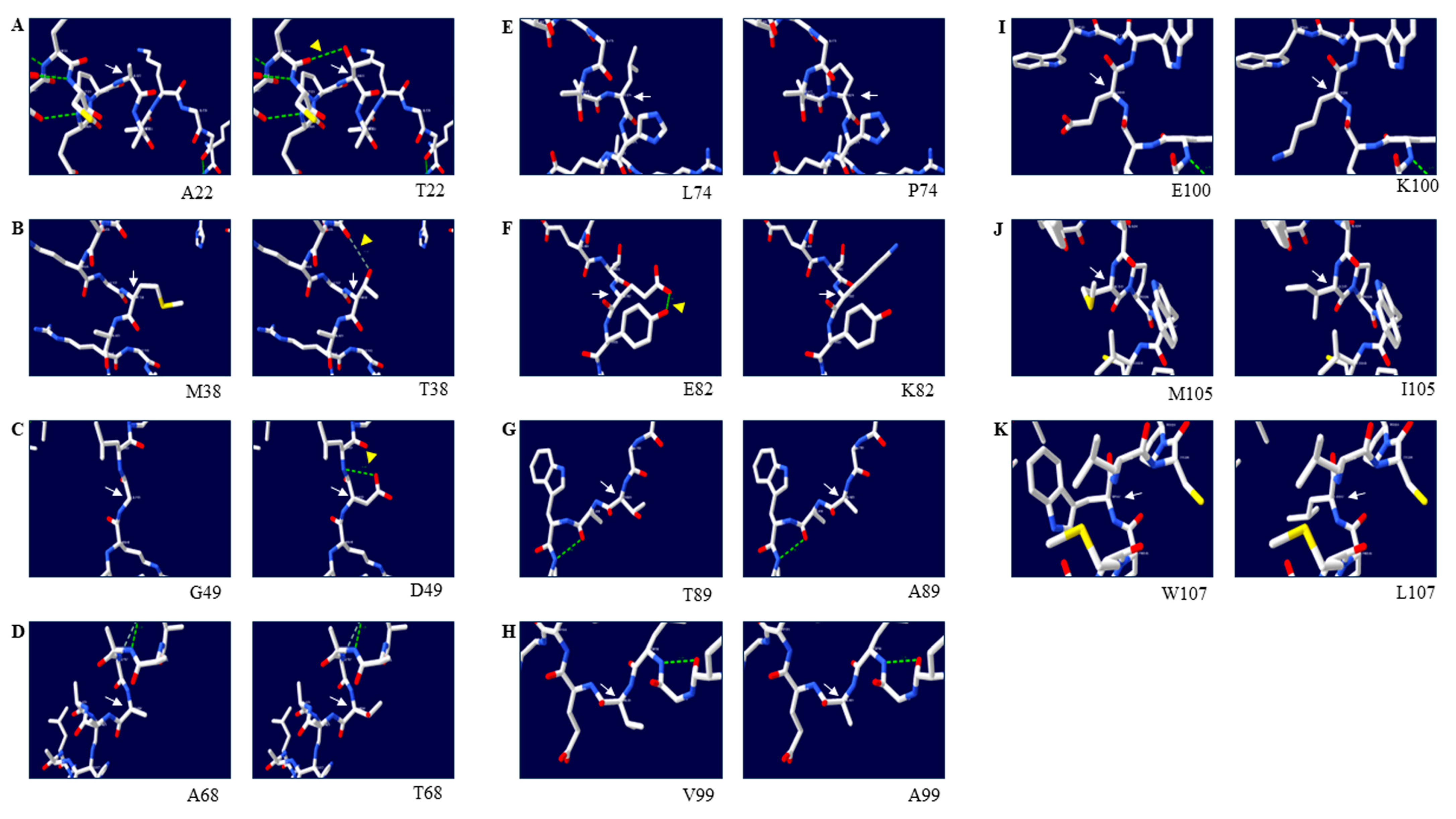
3.4. Effects of Amino Acid Substitutions on the 3D Structure of the Duck Sho Protein
3.5. In Silico Evaluation of the Effects of Non-Synonymous SNPs on the Duck Sho Protein
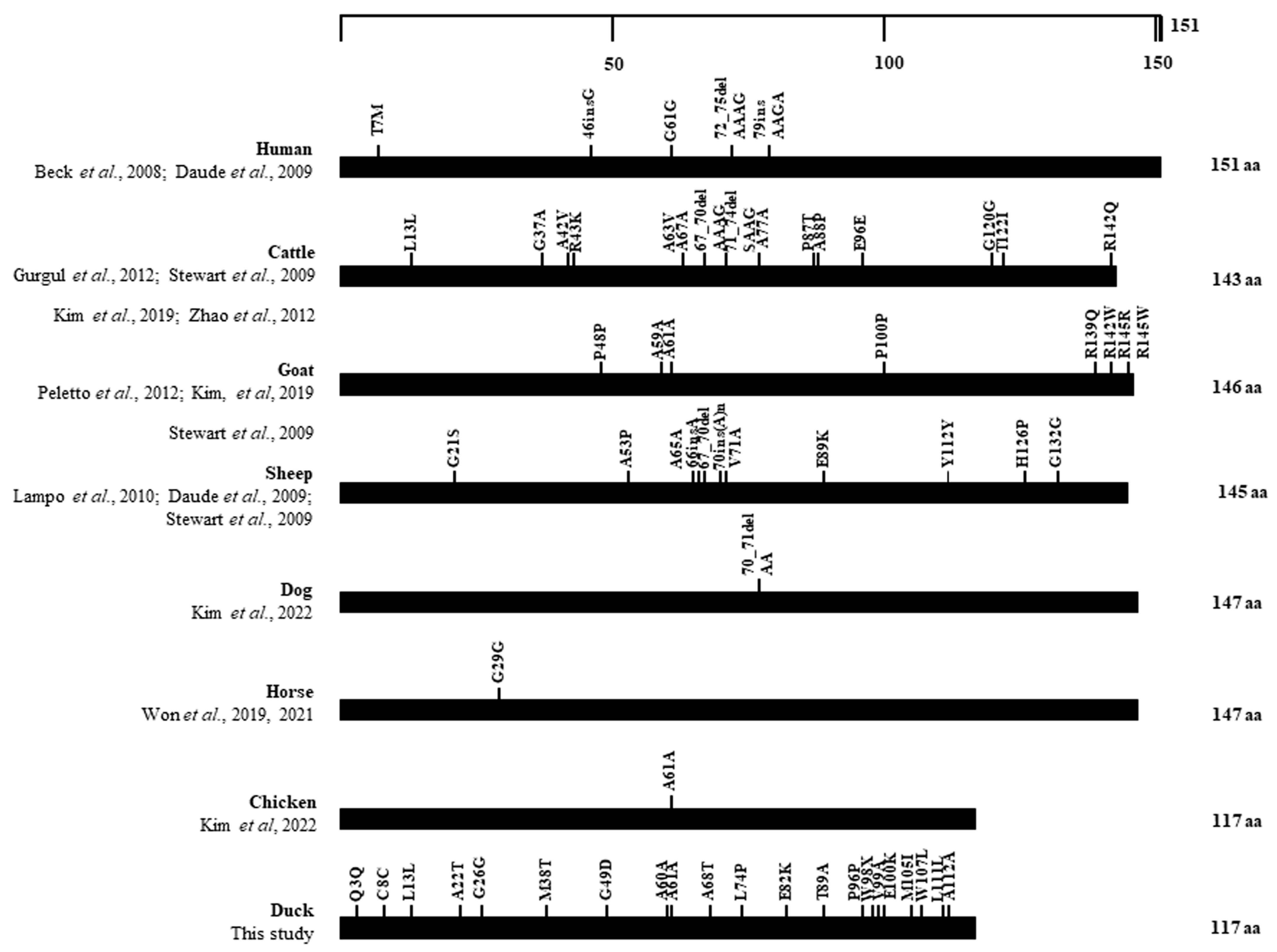
3.6. Distribution of SNPs in the ORF of the SPRN Gene in Several Animals
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prusiner, S.B. Prions. Proc. Natl. Acad. Sci. USA 1998, 95, 13363–13383. [Google Scholar] [CrossRef] [PubMed]
- Brandner, S.; Raeber, A.; Sailer, A.; Blättler, T.; Fischer, M.; Weissmann, C.; Aguzzi, A. Normal host prion protein (PrPC) is required for scrapie spread within the central nervous system. Proc. Natl. Acad. Sci. USA 1996, 93, 13148–13151. [Google Scholar] [CrossRef] [PubMed]
- Prusiner, S.B.; Scott, M.R.; DeArmond, S.J.; Cohen, F.E. Prion protein biology. Cell 1998, 93, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Steele, A.D.; Lindquist, S.; Aguzzi, A. The prion protein knockout mouse: A phenotype under challenge. Prion 2007, 1, 83–93. [Google Scholar] [CrossRef]
- Büeler, H.; Aguzzi, A.; Sailer, A.; Greiner, R.-A.; Autenried, P.; Aguet, M.; Weissmann, C. Mice devoid of PrP are resistant to scrapie. Cell 1993, 73, 1339–1347. [Google Scholar] [CrossRef]
- Prusiner, S.B.; Scott, M.; Foster, D.; Pan, K.-M.; Groth, D.; Mirenda, C.; Torchia, M.; Yang, S.-L.; Serban, D.; Carlson, G.A. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell 1990, 63, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Premzl, M.; Sangiorgio, L.; Strumbo, B.; Marshall Graves, J.A.; Simonic, T.; Gready, J.E. Shadoo, a new protein highly conserved from fish to mammals and with similarity to prion protein. Gene 2003, 314, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Castle, A.R.; Gill, A.C. Physiological functions of the cellular prion protein. Front. Mol. Biosci. 2017, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.C.; Westaway, D. The prion protein family: Diversity, rivalry, and dysfunction. Biochim. Biophys. Acta 2007, 1772, 654–672. [Google Scholar] [CrossRef]
- Watts, J.C.; Drisaldi, B.; Ng, V.; Yang, J.; Strome, B.; Horne, P.; Sy, M.S.; Yoong, L.; Young, R.; Mastrangelo, P. The CNS glycoprotein Shadoo has PrPC-like protective properties and displays reduced levels in prion infections. EMBO J. 2007, 26, 4038–4050. [Google Scholar] [CrossRef]
- Sakthivelu, V.; Seidel, R.P.; Winklhofer, K.F.; Tatzelt, J. Conserved stress-protective activity between prion protein and Shadoo. J. Biol. Chem. 2011, 286, 8901–8908. [Google Scholar] [CrossRef] [PubMed]
- Lampo, E.; Van Poucke, M.; Vandesompele, J.; Erkens, T.; Van Zeveren, A.; Peelman, L.J. Positive correlation between relative mRNA expression of PRNP and SPRN in cerebral and cerebellar cortex of sheep. Mol. Cell. Probes 2009, 23, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Daude, N.; Wohlgemuth, S.; Brown, R.; Pitstick, R.; Gapeshina, H.; Yang, J.; Carlson, G.A.; Westaway, D. Knockout of the prion protein (PrP)-like Sprn gene does not produce embryonic lethality in combination with PrPC-deficiency. Proc. Natl. Acad. Sci. USA 2012, 109, 9035–9040. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, S.E.; Grizenkova, J.; Pota, H.; Collinge, J. Shadoo (Sprn) and prion disease incubation time in mice. Mamm. Genome. 2009, 20, 367–374. [Google Scholar] [CrossRef]
- Zhao, H.; Liu, L.L.; Du, S.H.; Wang, S.Q.; Zhang, Y.P. Comparative analysis of the Shadoo gene between cattle and buffalo reveals significant differences. PLoS ONE 2012, 7, e46601. [Google Scholar] [CrossRef] [PubMed]
- Daude, N.; Westaway, D. Biological properties of the PrP-like Shadoo protein. Front. Biosci. 2011, 16, 1505–1516. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.C.; Stöhr, J.; Bhardwaj, S.; Wille, H.; Oehler, A.; Dearmond, S.J.; Giles, K.; Prusiner, S.B. Protease-resistant prions selectively decrease Shadoo protein. PLoS Pathog. 2011, 7, e1002382. [Google Scholar] [CrossRef] [PubMed]
- Westaway, D.; Genovesi, S.; Daude, N.; Brown, R.; Lau, A.; Lee, I.; Mays, C.E.; Coomaraswamy, J.; Canine, B.; Pitstick, R. Down-regulation of Shadoo in prion infections traces a pre-clinical event inversely related to PrPSc accumulation. PLoS Pathog. 2011, 7, e1002391. [Google Scholar] [CrossRef]
- Ciric, D.; Richard, C.A.; Moudjou, M.; Chapuis, J.; Sibille, P.; Daude, N.; Westaway, D.; Adrover, M.; Béringue, V.; Martin, D.; et al. Interaction between Shadoo and PrP Affects the PrP-Folding Pathway. J. Virol. 2015, 89, 6287–6293. [Google Scholar] [CrossRef]
- Jiayu, W.; Zhu, H.; Ming, X.; Xiong, W.; Songbo, W.; Bocui, S.; Wensen, L.; Jiping, L.; Keying, M.; Zhongyi, L. Mapping the interaction site of prion protein and Sho. Mol. Biol. Rep. 2010, 37, 2295–2300. [Google Scholar] [CrossRef]
- Jeong, B.-H.; Lee, K.-H.; Kim, N.-H.; Jin, J.-K.; Kim, J.-I.; Carp, R.I.; Kim, Y.-S. Association of sporadic Creutzfeldt–Jakob disease with homozygous genotypes at PRNP codons 129 and 219 in the Korean population. Neurogenetics 2005, 6, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Palmer, M.S.; Dryden, A.J.; Hughes, J.T.; Collinge, J. Homozygous prion protein genotype predisposes to sporadic Creutzfeldt–Jakob disease. Nature 1991, 352, 340–342. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-C.; Jeong, B.-H. The First Meta-Analysis of the M129V Single-Nucleotide Polymorphism (SNP) of the Prion Protein Gene (PRNP) with Sporadic Creutzfeldt–Jakob Disease. Cells 2021, 10, 3132. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, S.; Higuchi, J.; Shin, R.W.; Tateishi, J.; Kitamoto, T. Codon 219 Lys allele of PRNP is not found in sporadic Creutzfeldt-Jakob disease. Ann. Neurol. 1998, 43, 826–828. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Garcia, J.; Fernandez-Funez, P. D159 and S167 are protective residues in the prion protein from dog and horse, two prion-resistant animals. Neurobiol. Dis. 2018, 119, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.A.; Campbell, T.A.; Adamson, G.; Poulter, M.; Uphill, J.B.; Molou, E.; Collinge, J.; Mead, S. Association of a null allele of SPRN with variant Creutzfeldt-Jakob disease. J. Genet. Med. 2008, 45, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Peletto, S.; Bertolini, S.; Maniaci, M.G.; Colussi, S.; Modesto, P.; Biolatti, C.; Bertuzzi, S.; Caramelli, M.; Maurella, C.; Acutis, P.L. Association of an indel polymorphism in the 3′UTR of the caprine SPRN gene with scrapie positivity in the central nervous system. J. Gen. Virol. 2012, 93, 1620–1623. [Google Scholar] [CrossRef]
- Lampo, E.; Duchateau, L.; Schepens, B.; Van Poucke, M.; Saelens, X.; Erkens, T.; Van Zeveren, A.; Peelman, L.J. Identification of polymorphisms in the ovine Shadow of prion protein (SPRN) gene and assessment of their effect on promoter activity and susceptibility for classical scrapie. Anim. Genet. 2010, 41, 169–178. [Google Scholar] [CrossRef]
- Imran, M.; Mahmood, S. An overview of animal prion diseases. Virol. J. 2011, 8, 493. [Google Scholar] [CrossRef]
- Moore, J.; Hawkins, S.A.; Austin, A.R.; Konold, T.; Green, R.B.; Blamire, I.W.; Dexter, I.; Stack, M.J.; Chaplin, M.J.; Langeveld, J.P. Studies of the transmissibility of the agent of bovine spongiform encephalopathy to the domestic chicken. BMC Res. Notes 2011, 4, 501. [Google Scholar] [CrossRef]
- Wopfner, F.; Weidenhöfer, G.; Schneider, R.; von Brunn, A.; Gilch, S.; Schwarz, T.F.; Werner, T.; Schätzl, H.M. Analysis of 27 mammalian and 9 avian PrPs reveals high conservation of flexible regions of the prion protein. J. Mol. Biol. 1999, 289, 1163–1178. [Google Scholar] [CrossRef] [PubMed]
- Duck Meat Market by Product and Geography—Forecast and Analysis 2022–2026. Available online: https://www.technavio.com/report/duck-meat-market-industry-analysis (accessed on 21 July 2023).
- Jeong, M.-J.; Kim, Y.-C.; Jeong, B.-H. The first report of the Prion Protein Gene (PRNP) sequence in pekin ducks (Anas platyrhynchos domestica): The potential Prion disease susceptibility in ducks. Genes 2021, 12, 193. [Google Scholar] [CrossRef] [PubMed]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Gao, M.; Skolnick, J. ENTPRISE: An Algorithm for Predicting Human Disease-Associated Amino Acid Substitutions from Sequence Entropy and Predicted Protein Structures. PLoS ONE 2016, 11, e0150965. [Google Scholar] [CrossRef] [PubMed]
- Pejaver, V.; Urresti, J.; Lugo-Martinez, J.; Pagel, K.A.; Lin, G.N.; Nam, H.-J.; Mort, M.; Cooper, D.N.; Sebat, J.; Iakoucheva, L.M. Inferring the molecular and phenotypic impact of amino acid variants with MutPred2. Nat. Commun. 2020, 11, 5918. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, E.; Calabrese, R.; Fariselli, P.; Martelli, P.L.; Altman, R.B.; Casadio, R. WS-SNPs&GO: A web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genom. 2013, 14, S6. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Ardlie, K.G.; Kruglyak, L.; Seielstad, M. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 2002, 3, 299–309. [Google Scholar] [CrossRef]
- Bertoline, L.M.; Lima, A.N.; Krieger, J.E.; Teixeira, S.K. Before and after AlphaFold2: An overview of protein structure prediction. Front. Bioinform. 2023, 3, 1120370. [Google Scholar] [CrossRef]
- Daude, N.; Wohlgemuth, S.; Rogaeva, E.; Farid, A.H.; Heaton, M.; Westaway, D. Frequent missense and insertion/deletion polymorphisms in the ovine Shadoo gene parallel species-specific variation in PrP. PLoS ONE 2009, 4, e6538. [Google Scholar] [CrossRef] [PubMed]
- Gurgul, A.; Polak, M.P.; Larska, M.; Słota, E. PRNP and SPRN genes polymorphism in atypical bovine spongiform encephalopathy cases diagnosed in Polish cattle. J. Appl. Genet. 2012, 53, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.; Shen, C.; Zhao, D.; Goldmann, W. Genetic analysis of the SPRN gene in ruminants reveals polymorphisms in the alanine-rich segment of shadoo protein. J. Gen. Virol. 2009, 90, 2575–2580. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-C.; Kim, S.-K.; Won, S.-Y.; Jeong, B.-H. Polymorphisms of shadow of prion protein gene (SPRN) in Korean native cattle (Hanwoo) and Holstein cattle. Sci. Rep. 2020, 10, 15272. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-C.; Kim, S.-K.; Jeong, B.-H. Scrapie susceptibility-associated indel polymorphism of shadow of prion protein gene (SPRN) in Korean native black goats. Sci. Rep. 2019, 9, 15261. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-C.; Kim, H.-H.; Kim, A.-D.; Jeong, B.-H. Novel insertion/deletion polymorphisms and genetic features of the shadow of prion protein gene (SPRN) in dogs, a prion-resistant animal. Front. Vet. Sci. 2022, 9, 942289. [Google Scholar] [CrossRef] [PubMed]
- Won, S.-Y.; Kim, Y.-C.; Kim, S.-K.; Jeong, B.-H. The first report of genetic and structural diversities in the SPRN gene in the horse, an animal resistant to prion disease. Genes 2019, 11, 39. [Google Scholar] [CrossRef] [PubMed]
- Won, S.-Y.; Kim, Y.-C.; Do, K.; Jeong, B.-H. The first report of genetic polymorphisms of the equine SPRN gene in outbred horses, jeju and halla horses. Animals 2021, 11, 2574. [Google Scholar] [CrossRef]
- Kim, Y.C.; Kim, H.H.; Jeong, B.H. The First Report of Polymorphisms and Genetic Characteristics of the Shadow of Prion Protein (SPRN) in Prion Disease-Resistant Animal, Chickens. Front. Vet. Sci. 2022, 9, 904305. [Google Scholar] [CrossRef]
- Erana, H.; Fernandez-Borges, N.; Elezgarai, S.R.; Harrathi, C.; Charco, J.M.; Chianini, F.; Dagleish, M.P.; Ortega, G.; Millet, O.; Castilla, J. In vitro approach to identify key amino acids in low susceptibility of rabbit prion protein to misfolding. J. Virol. 2017, 91, e01543-17. [Google Scholar] [CrossRef]
- Mo, J.; Youk, S.; Pantin-Jackwood, M.J.; Suarez, D.L.; Lee, D.H.; Killian, M.L.; Bergeson, N.H.; Spackman, E. The pathogenicity and transmission of live bird market H2N2 avian influenza viruses in chickens, Pekin ducks, and guinea fowl. Vet. Microbiol. 2021, 260, 109180. [Google Scholar] [CrossRef] [PubMed]
- Ampuero, F.; Leacy, A.; Pham, P.H.; Che, S.; Jardine, C.; Nagy, E.; Delnatte, P.; Lillie, B.N.; Susta, L. Experimental pathogenesis of aquatic bird bornavirus 1 in Pekin ducks. Sci. Rep. 2023, 13, 18094. [Google Scholar] [CrossRef]
- Goldman, J.S.; Vallabh, S.M. Genetic counseling for prion disease: Updates and best practices. Genet. Med. 2022, 24, 1993–2003. [Google Scholar] [CrossRef] [PubMed]
- Mead, S.; Gandhi, S.; Beck, J.; Caine, D.; Gallujipali, D.; Carswell, C.; Hyare, H.; Joiner, S.; Ayling, H.; Lashley, T.; et al. A novel prion disease associated with diarrhea and autonomic neuropathy. N. Engl. J. Med. 2013, 369, 1904–1914. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, G.J.; Newberry, R.W.; VanVeller, B.; Raines, R.T.; Woolfson, D.N. Interplay of hydrogen bonds and n→ π* interactions in proteins. J. Am. Chem. Soc. 2013, 135, 18682–18688. [Google Scholar] [CrossRef]
- Pace, C.N.; Fu, H.; Lee Fryar, K.; Landua, J.; Trevino, S.R.; Schell, D.; Thurlkill, R.L.; Imura, S.; Scholtz, J.M.; Gajiwala, K. Contribution of hydrogen bonds to protein stability. Protein Sci. 2014, 23, 652–661. [Google Scholar] [CrossRef]

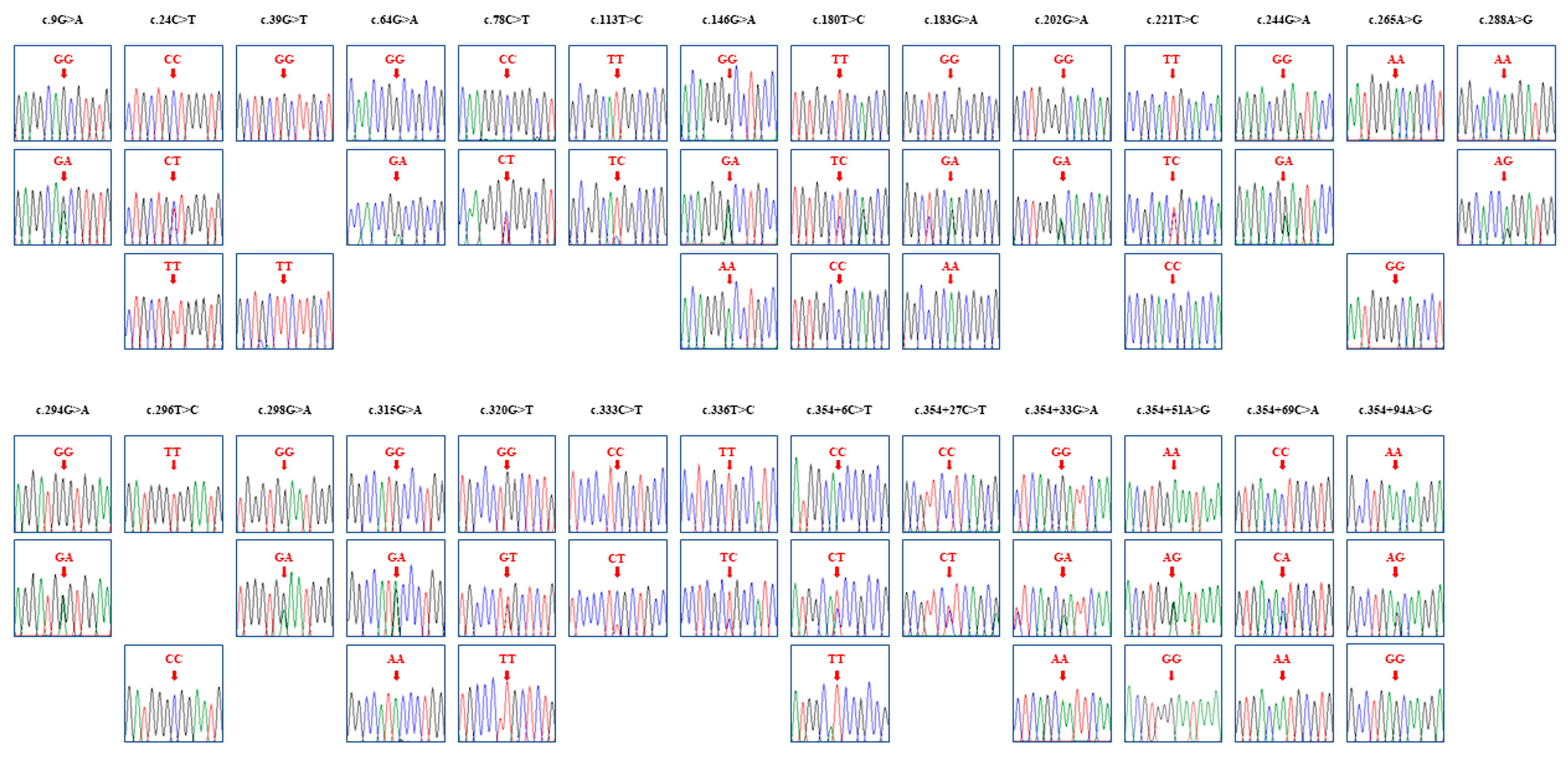
| Polymorphisms | Genotype Frequencies; n (%) | Allele Frequencies; n (%) | HWE p-Value | |||
|---|---|---|---|---|---|---|
| M/M | M/m | m/m | M | m | ||
| c.9G>A (Q3Q) | 84 (45.65) | 100 (54.35) | 0 (0.0) | 268 (72.8) | 100 (27.2) | <0.001 |
| c.24C>T (C8C) | 166 (90.22) | 17 (9.24) | 1 (0.54) | 349 (94.8) | 19 (5.2) | 0.7741 |
| c.39G>T (L13L) | 183 (99.46) | 0 (0.0) | 1 (0.54) | 366 (99.5) | 2 (0.5) | 0.0054 |
| c.64G>A (A22T) | 162 (88.04) | 22 (11.96) | 0 (0.0) | 346 (94.0) | 22 (6.0) | 1.0 |
| c.78C>T (G26G) | 181 (98.37) | 3 (1.63) | 0 (0.0) | 365 (99.2) | 3 (0.8) | 1.0 |
| c.113T>C (M38T) | 181 (98.37) | 3 (1.63) | 0 (0.0) | 365 (99.2) | 3 (0.8) | 1.0 |
| c.146G>A (G49D) | 99 (53.8) | 82 (44.57) | 3 (1.63) | 280 (76.1) | 88 (23.9) | 0.0024 |
| c.180T>C (A60A) | 161 (87.5) | 22 (11.96) | 1 (0.54) | 344 (93.5) | 24 (6.5) | 1 |
| c.183G>A (A61A) | 54 (29.35) | 117 (64.13) | 12 (6.52) | 226 (61.4) | 142 (38.6) | <0.001 |
| c.202G>A (A68T) | 171 (92.93) | 13 (7.07) | 0 (0.0) | 355 (96.5) | 13 (3.5) | 1.0 |
| c.221T>C (L74P) | 170 (92.39) | 13 (7.07) | 1 (0.54) | 353 (95.9) | 15 (4.1) | 0.5141 |
| c.244G>A (E82E) | 179 (97.28) | 5 (2.72) | 0 (0.0) | 363 (98.6) | 5 (1.4) | 1.0 |
| c.265A>G (T89A) | 183 (99.46) | 0 (0.0) | 1 (0.54) | 366 (99.5) | 2 (0.5) | 0.054 |
| c.288A>G(P96P) | 113 (61.41) | 71 (38.59) | 0 (0.0) | 297 (80.7) | 71 (19.3) | <0.001 |
| c.294G>A (W98X) | 171 (92.93) | 13 (7.07) | 0 (0.0) | 355 (96.5) | 13 (3.5) | 1.0 |
| c.296T>C (V99A) | 183 (99.46) | 0 (0.0) | 1 (0.54) | 366 (99.5) | 2 (0.5) | 0.0054 |
| c.298G>A(E100K) | 178 (96.74) | 6 (3.26) | 0 (0.0) | 362 (98.4) | 6 (1.6) | 1.0 |
| c.315G>A (M105I) | 176 (95.65) | 7 (3.8) | 1 (0.54) | 359 (97.6) | 9 (2.4) | 0.1906 |
| c.320G>T (W107L) | 156 (84.78) | 26 (14.13) | 2 (1.09) | 338 (91.8) | 30 (8.2) | 0.6726 |
| c.333C>T (L111L) | 131 (71.2) | 53 (28.8) | 0 (0.0) | 315 (85.6) | 53 (14.4) | 0.0249 |
| c.336T>C (Al12A) | 168 (91.3) | 16 (8.7) | 0 (0.0) | 352 (95.7) | 16 (4.3) | 1.0 |
| c.354+6C>T | 137 (74.46) | 46 (25.0) | 1 (0.54) | 320 (87.0) | 48 (13.0) | 0.2991 |
| c.354+27C>T | 117 (63.59) | 67 (36.41) | 0 (0.0) | 301 (81.8) | 67 (18.2) | 0.0012 |
| c.354+33G>A | 35 (19.02) | 138 (75.0) | 11 (5.98) | 208 (56.5) | 160 (43.5) | <0.001 |
| c.354+51A>G | 72 (39.13) | 111 (60.33) | 1 (0.54) | 255 (69.3) | 113 (30.7) | <0.001 |
| c.354+69C>A | 116 (63.04) | 51 (27.72) | 17 (9.24) | 283 (76.9) | 85 (23.1) | 0.0064 |
| c.354+94A>G | 41 (22.28) | 123 (66.85) | 20 (10.87) | 205 (55.7) | 163 (44.3) | <0.001 |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | - | ||||||||||||||||||||||||||
| 2 | 0.021 | - | |||||||||||||||||||||||||
| 3 | 0.002 | 0.005 | - | ||||||||||||||||||||||||
| 4 | 0.17 | 0.003 | 0 | - | |||||||||||||||||||||||
| 5 | 0.003 | 0.06 | 0 | 0.001 | - | ||||||||||||||||||||||
| 6 | 0.002 | 0 | 0 | 0.05 | 0 | - | |||||||||||||||||||||
| 7 | 0.014 | 0.017 | 0.017 | 0.004 | 0.003 | 0.003 | - | ||||||||||||||||||||
| 8 | 0.026 | 0.004 | 0.017 | 0.005 | 0.001 | 0.001 | 0.14 | - | |||||||||||||||||||
| 9 | 0.234 | 0.061 | 0.009 | 0.04 | 0.013 | 0.001 | 0.006 | 0.111 | - | ||||||||||||||||||
| 10 | 0.014 | 0.001 | 0 | 0.002 | 0 | 0 | 0.002 | 0.008 | 0.009 | - | |||||||||||||||||
| 11 | 0.016 | 0.001 | 0.129 | 0.003 | 0.015 | 0 | 0.007 | 0.001 | 0.023 | 0.391 | - | ||||||||||||||||
| 12 | 0.005 | 0.001 | 0 | 0.001 | 0 | 0 | 0.044 | 0.119 | 0.022 | 0.048 | 0.04 | - | |||||||||||||||
| 13 | 0.002 | 0 | 0 | 0 | 0 | 0 | 0.002 | 0 | 0.003 | 0 | 0 | 0 | - | ||||||||||||||
| 14 | 0.48 | 0.013 | 0.001 | 0.068 | 0.002 | 0.007 | 0.045 | 0.017 | 0.15 | 0.009 | 0.01 | 0.003 | 0.001 | - | |||||||||||||
| 15 | 0.014 | 0.001 | 0 | 0.002 | 0 | 0 | 0.002 | 0 | 0.008 | 0.846 | 0.492 | 0.048 | 0 | 0.009 | - | ||||||||||||
| 16 | 0.002 | 0 | 0 | 0 | 0 | 0 | 0.002 | 0 | 0.003 | 0 | 0 | 0 | 1 | 0.001 | 0 | - | |||||||||||
| 17 | 0.006 | 0.001 | 0 | 0.001 | 0 | 0 | 0.027 | 0.093 | 0.026 | 0.001 | 0.001 | 0.122 | 0 | 0.004 | 0.001 | 0 | - | ||||||||||
| 18 | 0.009 | 0.266 | 0 | 0.002 | 0 | 0 | 0.008 | 0.002 | 0.018 | 0.001 | 0.001 | 0 | 0 | 0.006 | 0.001 | 0 | 0 | - | |||||||||
| 19 | 0.033 | 0.005 | 0.062 | 0.006 | 0.001 | 0.001 | 0.005 | 0.025 | 0.055 | 0.003 | 0.014 | 0 | 0 | 0.021 | 0.003 | 0 | 0 | 0.001 | - | ||||||||
| 20 | 0.423 | 0.001 | 0.001 | 0.153 | 0.001 | 0.014 | 0.01 | 0.012 | 0.106 | 0.006 | 0.007 | 0.002 | 0.001 | 0.303 | 0.006 | 0.001 | 0.003 | 0.004 | 0.015 | - | |||||||
| 21 | 0.122 | 0.001 | 0 | 0.003 | 0 | 0 | 0.014 | 0.003 | 0.029 | 0.002 | 0.002 | 0.001 | 0 | 0.096 | 0.002 | 0 | 0.001 | 0.001 | 0 | 0.09 | - | ||||||
| 22 | 0.094 | 0.008 | 0.001 | 0.05 | 0.001 | 0.001 | 0.145 | 0.001 | 0.094 | 0 | 0.002 | 0.001 | 0.001 | 0.127 | 0 | 0.001 | 0.002 | 0.004 | 0.013 | 0.006 | 0 | - | |||||
| 23 | 0.569 | 0.012 | 0.001 | 0.153 | 0.002 | 0.008 | 0.006 | 0.016 | 0.113 | 0.008 | 0.009 | 0.003 | 0.001 | 0.403 | 0.008 | 0.001 | 0.004 | 0.006 | 0.02 | 0.309 | 0.056 | 0.033 | - | ||||
| 24 | 0.25 | 0.032 | 0.004 | 0.045 | 0.006 | 0.011 | 0.215 | 0.037 | 0.08 | 0.028 | 0.033 | 0.011 | 0.007 | 0.184 | 0.028 | 0.007 | 0.013 | 0.033 | 0.068 | 0.189 | 0 | 0.115 | 0.171 | - | |||
| 25 | 0.642 | 0.024 | 0.002 | 0.143 | 0.004 | 0 | 0.139 | 0.031 | 0.207 | 0.016 | 0.019 | 0.006 | 0.002 | 0.462 | 0.016 | 0.002 | 0.007 | 0.011 | 0.022 | 0.353 | 0.103 | 0.188 | 0.45 | 0.341 | - | ||
| 26 | 0.112 | 0.106 | 0.002 | 0.019 | 0.027 | 0.002 | 0.064 | 0.005 | 0.124 | 0.011 | 0.006 | 0.001 | 0.002 | 0.072 | 0.003 | 0.002 | 0.008 | 0.059 | 0 | 0.051 | 0.014 | 0.045 | 0.067 | 0.135 | 0.133 | - | |
| 27 | 0.225 | 0.046 | 0.004 | 0.08 | 0.01 | 0.007 | 0.085 | 0.041 | 0.066 | 0.029 | 0.007 | 0.011 | 0.004 | 0.221 | 0.029 | 0.004 | 0.013 | 0.015 | 0.029 | 0.134 | 0.057 | 0.106 | 0.169 | 0.456 | 0.242 | 0.125 | - |
| Haplotypes | Frequency, % |
|---|---|
| GCGGCTGTGGTGAAGTGGGCTCCGACA | 24.3 |
| GCGGCTGTAGTGAAGTGGGCTCCGACA | 13.6 |
| GCGGCTGTAGTGAAGTGGGCTCCAAAG | 5.3 |
| GCGGCTGTGGTGAAGTGGGCTCCGAAG | 2.4 |
| GCGGCTACAGTGAAGTGGGCTCCGACA | 2.0 |
| GTGGCTGTAGTGAAGTGAGCTCCAAAG | 1.6 |
| ACGGCTGTGGTGAGGTGGGCTCTAGCG | 1.5 |
| ACGGCTATGGTGAGGTGGGTTCTAGCG | 1.3 |
| GCGGCTGTAGTGAAGTGGTCTCCGGCA | 1.3 |
| ACGGCTGTGGTGAGGTGGGTTCTAGCG | 1.2 |
| ACGGCTATGGTGAGGTGGGCTTCAGCG | 1.1 |
| ACGACTATGGTGAGGTGGGTTTTAGCG | 1.1 |
| Others * | 43.3 |
| Amino Acid Substitutions | MutPred2 | SIFT | SNPs&GO |
|---|---|---|---|
| A22T | Benign (0.045) | Deleterious (0.020) | Neutral (0.019) |
| M38T | Benign (0.041) | Tolerated (0.330) | Neutral (0.031) |
| G49D | Benign (0.043) | Deleterious (0.010) | Neutral (0.076) |
| A68T | Benign (0.049) | Deleterious (0.010) | Neutral (0.055) |
| L74P | Benign (0.112) | Tolerated (0.190) | Neutral (0.110) |
| E82K | Benign (0.045) | Tolerated (0.430) | Neutral (0.067) |
| T89A | Benign (0.029) | Tolerated (0.230) | Neutral (0.051) |
| W98X | Benign (0.276) | NA | NA |
| V99A | Benign (0.032) | Tolerated (0.510) | Neutral (0.033) |
| E100K | Benign (0.053) | Tolerated (0.470) | Neutral (0.059) |
| M105I | Benign (0.030) | Deleterious (0.030) | Neutral (0.077) |
| W107L | Benign (0.087) | Tolerated (0.060) | Neutral (0.126) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, T.-T.-D.; Zayed, M.; Kim, Y.-C.; Jeong, B.-H. The First Genetic Characterization of the SPRN Gene in Pekin Ducks (Anas platyrhynchos domesticus). Animals 2024, 14, 1588. https://doi.org/10.3390/ani14111588
Nguyen T-T-D, Zayed M, Kim Y-C, Jeong B-H. The First Genetic Characterization of the SPRN Gene in Pekin Ducks (Anas platyrhynchos domesticus). Animals. 2024; 14(11):1588. https://doi.org/10.3390/ani14111588
Chicago/Turabian StyleNguyen, Thi-Thuy-Duong, Mohammed Zayed, Yong-Chan Kim, and Byung-Hoon Jeong. 2024. "The First Genetic Characterization of the SPRN Gene in Pekin Ducks (Anas platyrhynchos domesticus)" Animals 14, no. 11: 1588. https://doi.org/10.3390/ani14111588
APA StyleNguyen, T.-T.-D., Zayed, M., Kim, Y.-C., & Jeong, B.-H. (2024). The First Genetic Characterization of the SPRN Gene in Pekin Ducks (Anas platyrhynchos domesticus). Animals, 14(11), 1588. https://doi.org/10.3390/ani14111588







