Comparative Cytogenetics of the Malagasy Ground Geckos of the Paroedura bastardi and Paroedura picta Species Groups
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Molecular Analysis
2.3. Cytogenetic Analysis
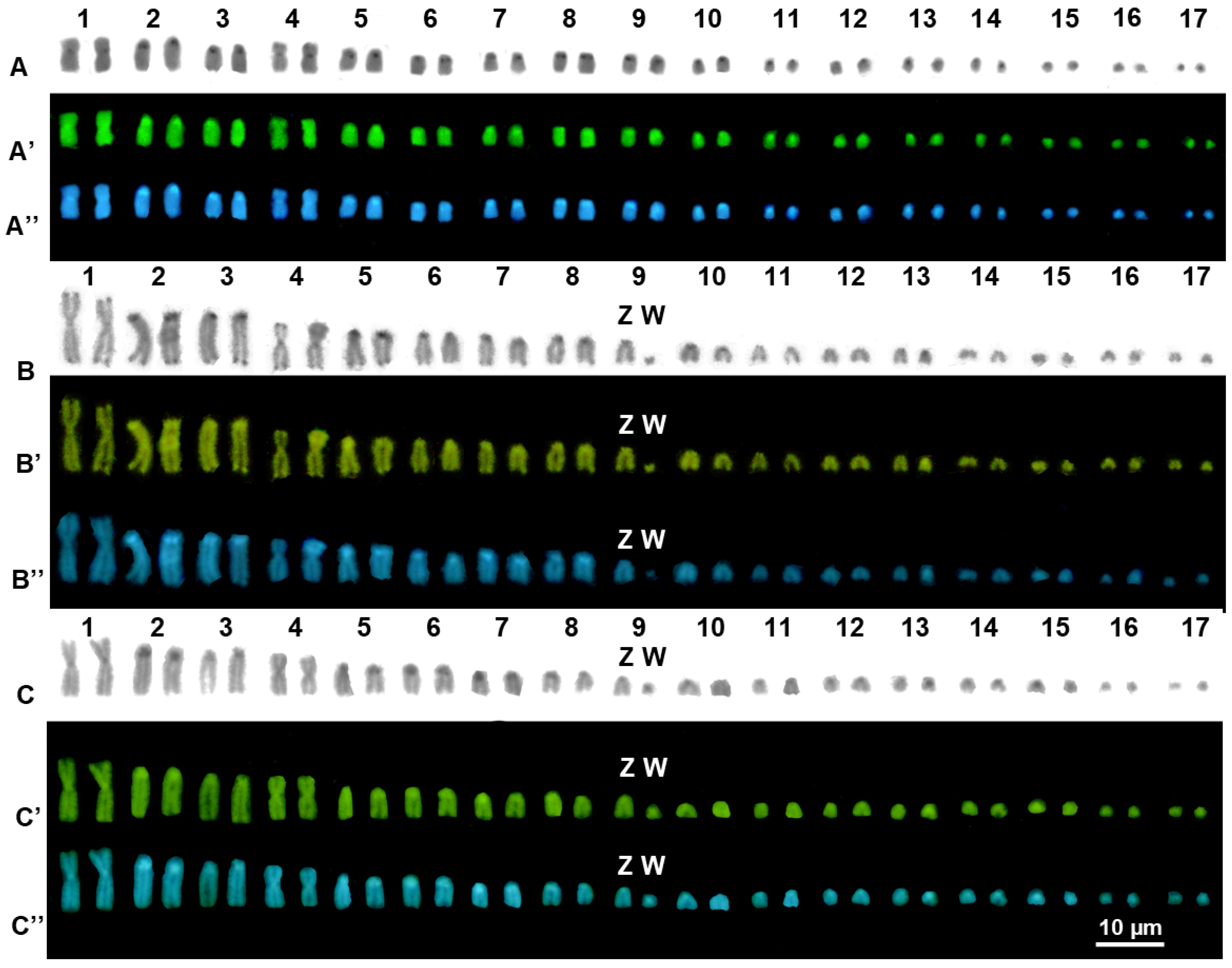
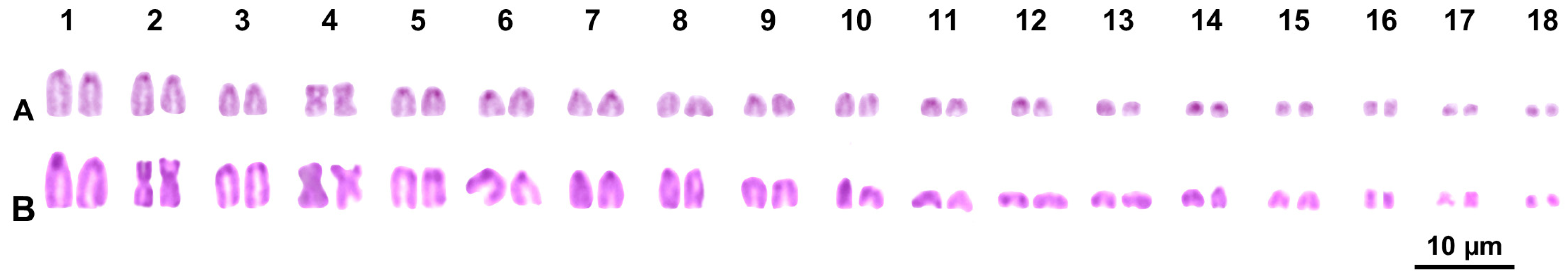
3. Results
3.1. Molecular Analysis
3.2. Cytogenetic Analysis
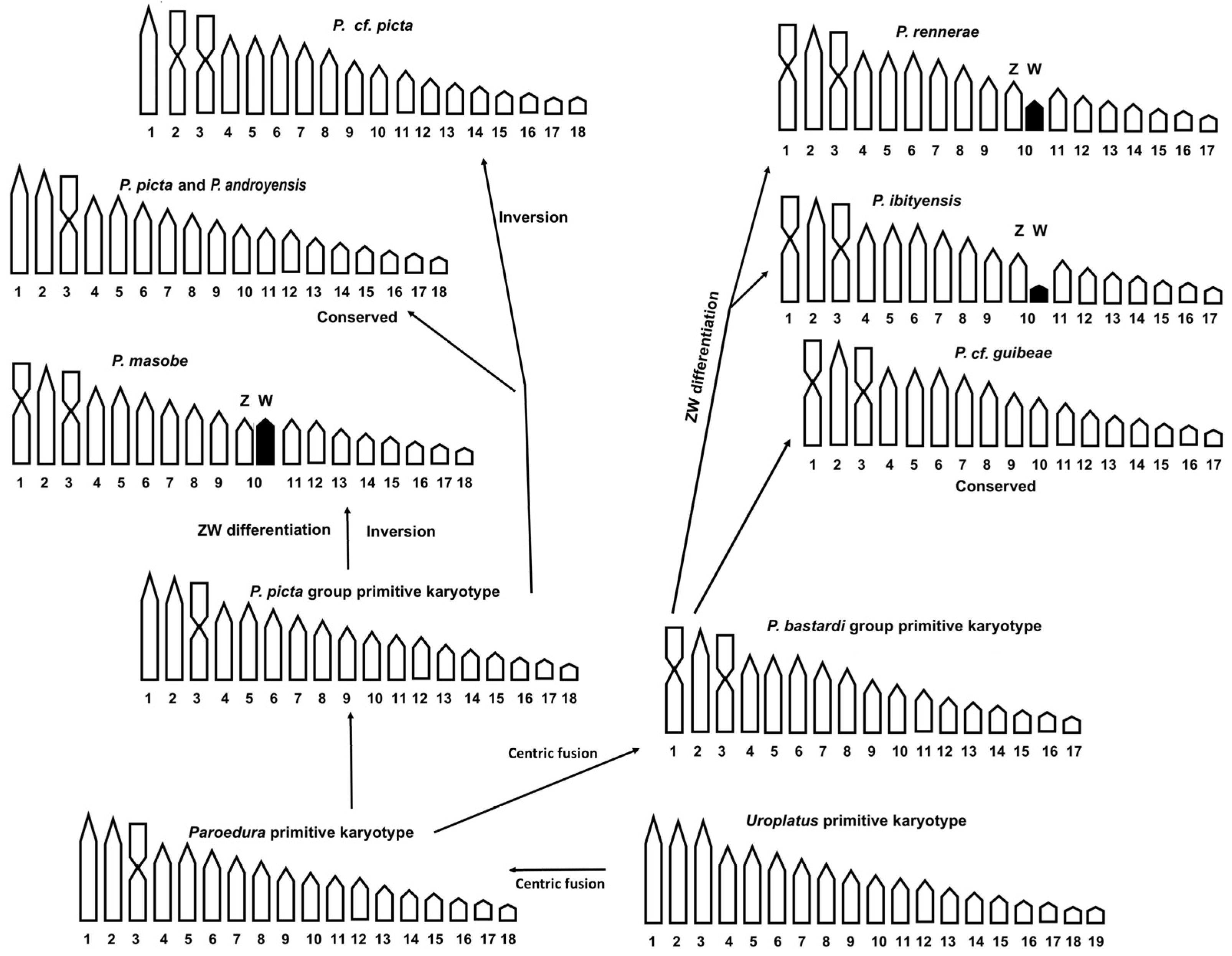
4. Discussion
4.1. Molecular Analysis
4.2. Cytogenetic Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vences, M.; Wollenberg, K.C.; Vieites, D.R.; Lees, D.C. Madagascar as a model region of species diversification. Trends. Ecol. Evol. 2009, 24, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.L.; Cameron, A.; Yoder, A.D.; Vences, M. A necessarily complex model to explain the biogeography of the amphibians and reptiles of Madagascar. Nat. Commun. 2014, 5, 5046. [Google Scholar] [CrossRef] [PubMed]
- Uetz, P.; Freed, P.; Aguilar, R.; Reyes, F.; Hošek, J. (Eds.) The Reptile Database. 2023. Available online: http://reptile-database.org/ (accessed on 10 April 2024).
- IUCN. The IUCN Red List of Threatened Species. Version 2023–1. 2023. Available online: https://www.iucnredlist.org (accessed on 10 April 2024).
- Glaw, F.; Köhler, J.; Vences, M. Three new species of nocturnal geckos of the Paroedura oviceps clade from xeric environments of Madagascar (Squamata: Gekkonidae). Zootaxa 2018, 4433, 305–324. [Google Scholar] [CrossRef]
- Köhler, J.; Vences, M.; Scherz, M.D.; Glaw, F. A new species of nocturnal Gecko, genus Paroedura, from the karstic tsingy de Bemaraha formation in western Madagascar. Salamandra 2019, 55, 73–81. [Google Scholar]
- Miralles, A.; Bruy, T.; Crottini, A.; Rakotoarison, A.; Ratsoavina, F.M.; Scherz, M.D.; Schmidt, R.; Köhler, J.; Glaw, F.; Vences, M. Completing a taxonomic puzzle: Integrative review of geckos of the Paroedura bastari species complex (Squamata, Gekkonidae). Vertebr. Zool. 2021, 71, 27–48. [Google Scholar] [CrossRef]
- Piccoli, C.; Belluardo, F.; Lobón-Rovira, J.; Alves, I.O.; Rasoazanany, M.; Andreone, F.; Rosa, G.M.; Crottini, A. Another step through the crux: A new microendemic rock–dwelling Paroedura (Squamata, Gekkonidae) from south–central Madagascar. ZooKeys 2023, 1181, 125–154. [Google Scholar] [CrossRef] [PubMed]
- Jackman, T.R.; Bauer, A.M.; Greenbaum, E.; Glaw, F.; Vences, M. Molecular phylogenetic relationships among species of the Malagasy-Comoran gecko genus Paroedura (Squamata: Gekkonidae). Mol. Phylogenet. Evol. 2008, 46, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Pyron, R.A.; Burbrink, F.T.; Wiens, J.J. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 2013, 13, 93. [Google Scholar] [CrossRef]
- Glaw, F.; Rösler, H.; Ineich, I.; Gehring, P.-S.; Köhler, J.; Vences, M. A new species of nocturnal gecko (Paroedura) from karstic limestone in northern Madagascar. Zoosyst. Evol. 2014, 90, 249–259. [Google Scholar] [CrossRef]
- Main, H.; Scantlebury, D.P.; Zarkower, D.; Gamble, T. Karyotypes of two species of Malagasy ground gecko (Paroedura: Gekkonidae). Afr. J. Herpetol. 2012, 61, 81–90. [Google Scholar] [CrossRef]
- Aprea, G.; Andreone, F.; Fulgione, D.; Petraccioli, A.; Odierna, G. Chromosomal rearrangements occurred repeatedly and independently during species diversification in Malagasy geckos, genus Paroedura. Afr. Zool. 2013, 48, 96–108. [Google Scholar] [CrossRef]
- Koubová, M.; Johnson Pokorná, M.; Rovatsos, M.; Farkačová, K.; Altmanová, M.; Kratochvíl, L. Sex determination in Madagascar geckos of the genus Paroedura (Squamata: Gekkonidae): Are differentiated sex chromosomes indeed so evolutionary stable? Chromosome Res. 2014, 22, 441–452. [Google Scholar] [CrossRef]
- Rovatsos, M.; Farkačová, K.; Altmanová, M.; Johnson Pokorná, M.; Kratochvíl, L. The rise and fall of differentiated sex chromosomes in geckos. Mol. Ecol. 2019, 28, 3042–3052. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Lab Press: New York, NY, USA, 1989. [Google Scholar]
- Palumbi, S.; Martin, A.; Romano, S.; McMillan, W.O.; Stice, L.; Grabowski, G. The Simple Fool’s Guide to PCR, Version 2.0; Department of Zoology and Kewalo Marine Biology, University of Hawaii: Honolulu, HI, USA, 1991. [Google Scholar]
- Cocca, W.; Rosa, G.M.; Andreone, F.; Aprea, G.; Bergò, P.E.; Mattioli, F.; Mercurio, V.; Randrianirina, J.E.; Rosado, D.; Vences, M.; et al. The herpetofauna (Amphibia, Crocodylia, Squamata, Testudines) of the Isalo massif, southwest Madagascar: Combining morphological, molecular and museum data. Salamandra 2018, 54, 178–200. [Google Scholar]
- Hall, T.A. BioEdit: A user–friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Mezzasalma, M.; Andreone, F.; Aprea, G.; Glaw, F.; Odierna, G.; Guarino, F.M. When can chromosomes drive speciation? The peculiar case of the Malagasy tomato frogs (genus Dyscophus). Zool. Anz. 2017, 268, 41–46. [Google Scholar] [CrossRef]
- Howell, W.M.; Black, D.A. Controlled silver staining of nucleolus organizer regions with a protective colloidal developer: A1–step method. Experientia 1980, 36, 1014–1015. [Google Scholar] [CrossRef]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Andreone, F.; Glaw, F.; Guarino, F.M.; Odierna, G.; Petraccioli, A.; Picariello, O. Changes in heterochromatin content and ancient chromosome fusion in the endemic Malagasy boid snakes Sanzinia and Acrantophis (Squamata: Serpentes). Salamandra 2019, 55, 140–144. [Google Scholar]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Hara, Y.; Takeuchi, M.; Kageyama, Y.; Tatsumi, K.; Hibi, M.; Kiyonari, H.; Kuraku, S. Madagascar ground gecko genome analysis characterizes asymmetric fates of duplicated genes. BMC Biol. 2018, 16, 40. [Google Scholar] [CrossRef] [PubMed]
- Starostová, Z.; Musilová, Z. The complete mitochondrial genome of the Madagascar ground gecko Paroedura picta (Squamata: Gekkonidae). Mitochondrial DNA Part A 2016, 27, 4397–4398. [Google Scholar] [CrossRef] [PubMed]
- Peters, W.C.H. Diagnosen neuer Batrachier, welche zusammen mit der früher (24. Juli und 17. August) gegebenen Übersicht der Schlangen und Eidechsen mitgetheilt warden; Bericht über die zur Bekanntmachung geeigneten Verhandlungen der Königlichen Preußischen Akademie der Wissenschaften zu Berlin; Königl.–Preuss: Berlin, Germany, 1854; pp. 614–628. [Google Scholar]
- Fan, Y.; Linardopoulou, E.; Friedman, C.; Williams, E.; Trask, B.J. Genomic structure and evolution of the ancestral chromosome fusion site in 2q13–2q14.1 and paralogous regions on other human chromosomes. Genome Res. 2002, 12, 1651–1662. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, M.; Barton, N. Chromosome inversions, local adaptation and speciation. Genetics 2006, 173, 419–434. [Google Scholar] [CrossRef] [PubMed]
- Sacerdot, C.; Louis, A.; Bon, C.; Berthelot, C.; Roest Crollius, H. Chromosome evolution at the origin of the ancestral vertebrate genome. Genome Biol. 2018, 19, 166. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.M.; Sarre, S.D.; Gleeson, D.; Georges, A.; Ezaz, T. Did lizards follow unique pathways in sex chromosome evolution? Genes 2018, 9, 239. [Google Scholar] [CrossRef] [PubMed]
- Petraccioli, A.; Guarino, F.M.; Kupriyanova, L.; Mezzasalma, M.; Odierna, G.; Picariello, O.; Capriglione, T. Isolation and Characterization of Interspersed Repeated Sequences in the Common Lizard, Zootoca vivipara, and Their Conservation in Squamata. Cytogenet. Genome Res. 2019, 157, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Schubert, I. Macromutations Yielding Karyotype Alterations (and the Process(es) behind Them) Are the Favored Route of Carcinogenesis and Speciation. Cancers 2024, 16, 554. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Sarre, S.D.; Georges, A.; Matsuda, Y.; Marshall Graves, J.A.; Ezaz, T. Highly Differentiated ZW Sex Microchromosomes in the Australian Varanus Species Evolved through Rapid Amplification of Repetitive Sequences. PLoS ONE 2014, 9, e95226. [Google Scholar] [CrossRef]
- Bachtrog, D.; Mahajan, S.; Bracewell, R. Massive gene amplification on a recently formed Drosophila Y chromosome. Nat. Ecol. Evol. 2019, 3, 1587–1597. [Google Scholar] [CrossRef]
- Rovatsos, M.; Marchal, J.A.; Giagia–Athanasopoulou, E.; Sánchez, A. Molecular Composition of Heterochromatin and Its Contribution to Chromosome Variation in the Microtus thomasi/Microtus atticus Species Complex. Genes 2021, 12, 807. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Brunelli, E.; Odierna, G.; Guarino, F.M. First Insights on the Karyotype Diversification of the Endemic Malagasy Leaf–Toed Geckos (Squamata: Gekkonidae: Uroplatus). Animals 2022, 12, 2054. [Google Scholar] [CrossRef] [PubMed]
- King, M.; Rofe, R. Karyotypic variation in the Australian Gekko Phyllodactylus marmoratus (Gray) (Gekkonidae: Reptilia). Chromosoma 1976, 54, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Mezzasalma, M.; Guarino, F.; Loader, S.; Odierna, G.; Streicher, J.; Cooper, N. First karyological analysis of the endemic Malagasy phantom gecko Matoatoa brevipes (Squamata: Gekkonidae). Acta Herpetol. 2020, 15, 137–141. [Google Scholar]
- Pensabene, E.; Yurchenko, A.; Kratochvíl, L.; Rovatsos, M. Madagascar Leaf–Tail Geckos (Uroplatus spp.) Share Independently Evolved Differentiated ZZ/ZW Sex Chromosomes. Cells 2023, 12, 260. [Google Scholar] [CrossRef] [PubMed]
- Porter, C.A.; Hamilton, M.J.; Sites, J.W., Jr.; Baker, R.J. Location of ribosomal DNA in chromosomes of squamate reptiles: Systematic and evolutionary implications. Herpetologica 1991, 47, 271–280. [Google Scholar]
- Deakin, J.E.; Edwards, M.J.; Patel, H.; O’Meally, D.; Lian, J.; Stenhouse, R.; Ryan, S.; Livernois, A.M.; Azad, B.; Holleley, C.E.; et al. Anchoring genome sequence to chromosomes of the central bearded dragon (Pogona vitticeps) enables reconstruction of ancestral squamate macrochromosomes and identifies sequence content of the Z chromosome. BMC Genom. 2016, 17, 447. [Google Scholar] [CrossRef]
- Olmo, E. Trends in the evolution of reptilian chromosomes. Integr. Comp. Biol. 2008, 48, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Deakin, J.E.; Ezaz, T. Understanding the evolution of reptile chromosomes through applications of combined cytogenetics and genomics approaches. Cytogenet. Genome Res. 2019, 157, 7–20. [Google Scholar] [CrossRef]
- Srikulnath, K.; Uno, Y.; Nishida, C.; Ota, H.; Matsuda, Y. Karyotype Reorganization in the Hokou Gecko (Gekko hokouensis, Gekkonidae): The Process of Microchromosome Disappearance in Gekkota. PLoS ONE 2015, 10, e0134829. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Macirella, R.; Odierna, G.; Brunelli, E. Karyotype Diversification and Chromosome Rearrangements in Squamate Reptiles. Genes 2024, 15, 371. [Google Scholar] [CrossRef] [PubMed]
- Chrostek, G.; Domaradzka, A.; Yurchenko, A.; Kratochvíl, L.; Mazzoleni, S.; Rovatsos, M. Cytogenetic Analysis of Seven Species of Gekkonid and Phyllodactylid Geckos. Genes 2023, 14, 178. [Google Scholar] [CrossRef] [PubMed]

| Species | Specimen | Locality | Sex |
|---|---|---|---|
| P. ibytiensis | GA 388 | Mount Ibity | female |
| P. ibytiensis | GA 389 | Mount Ibity | male |
| P. rennerae | GA 505 | Mandrivazo | female |
| P. rennerae | GA 506 | Mandrivazo | male |
| P. cf. guibeae | FGMV1523 | Toliara | male |
| P. cf. guibeae | FGMV1524 | Toliara | female |
| P. cf. picta | GA 324 | Marofandilia | female |
| P. cf. picta | GA 325 | Marofandilia | female |
| P. picta | FGMV1522 | Toliara region | female |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mezzasalma, M.; Odierna, G.; Macirella, R.; Brunelli, E. Comparative Cytogenetics of the Malagasy Ground Geckos of the Paroedura bastardi and Paroedura picta Species Groups. Animals 2024, 14, 1708. https://doi.org/10.3390/ani14111708
Mezzasalma M, Odierna G, Macirella R, Brunelli E. Comparative Cytogenetics of the Malagasy Ground Geckos of the Paroedura bastardi and Paroedura picta Species Groups. Animals. 2024; 14(11):1708. https://doi.org/10.3390/ani14111708
Chicago/Turabian StyleMezzasalma, Marcello, Gaetano Odierna, Rachele Macirella, and Elvira Brunelli. 2024. "Comparative Cytogenetics of the Malagasy Ground Geckos of the Paroedura bastardi and Paroedura picta Species Groups" Animals 14, no. 11: 1708. https://doi.org/10.3390/ani14111708








