Different Founding Effects Underlie Dominant Blue Eyes (DBE) in the Domestic Cat
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Ethics Statement
2.3. SNP Genotyping, Genome-Wide Association Study, and Genome Sequencing
2.4. PAX3 Sequencing and Variants Genotyping
2.5. Accession Numbers
3. Results
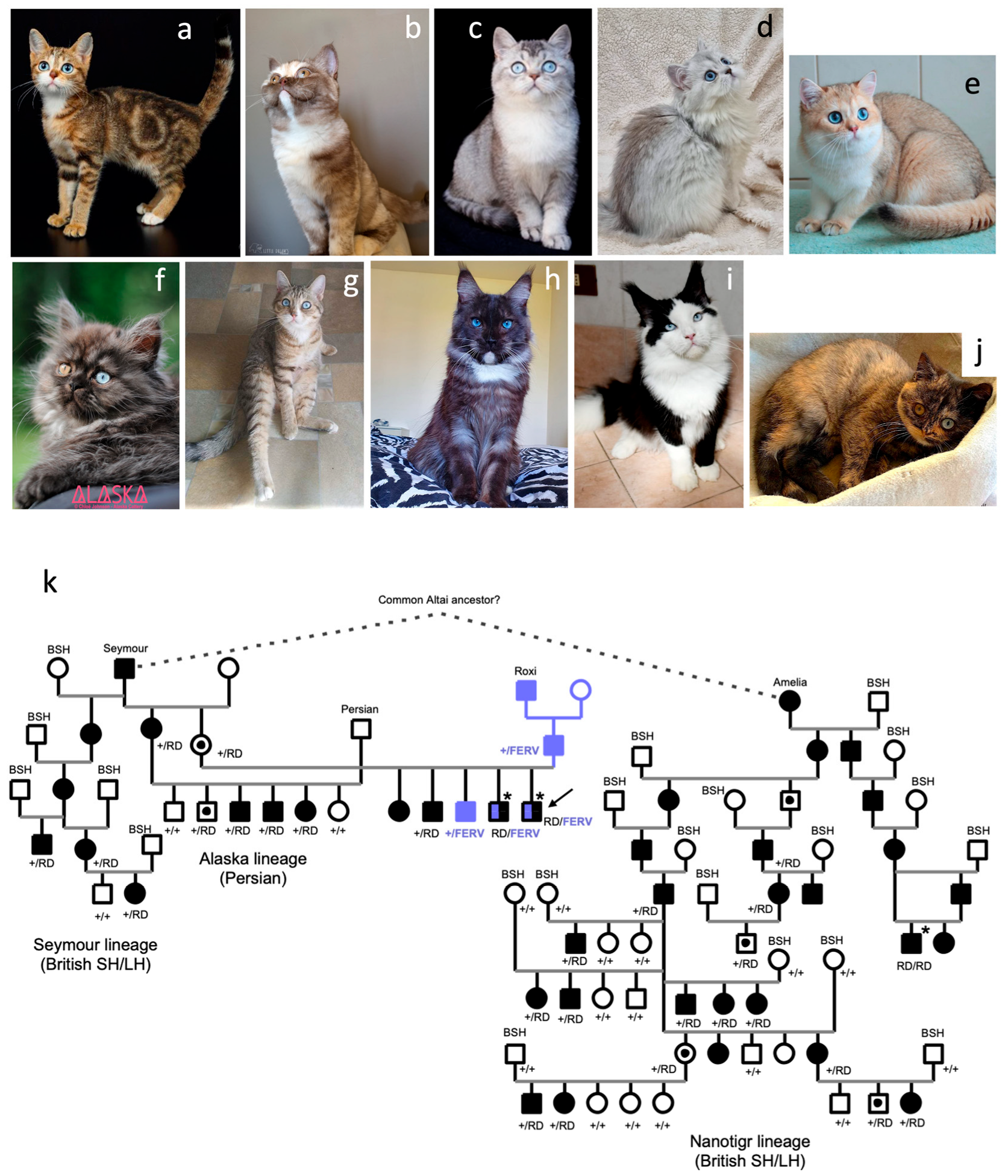
3.1. Several Breeding Lines Segregate DBE
3.2. A Second PAX3 Insertion Is Associated with DBE and Spread in Feline Lines
4. Discussion
4.1. Genetic Heterogeneity of Feline DBE
4.2. Three Spontaneous Variants in the PAX3 Gene
4.3. PAX3-Related DBE in Cats Share Some Common Features but Not All with PAX3-Related Waardenburg Syndrome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gandolfi, B.; Alhaddad, H. Investigation of inherited diseases in cats: Genetic and genomic strategies over three decades. J. Feline Med. Surg. 2015, 17, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Lyons, L.A. DNA mutations of the cat: The good, the bad and the ugly. J. Feline Med. Surg. 2015, 17, 203–219. [Google Scholar] [CrossRef] [PubMed]
- David, V.A.; Menotti-Raymond, M.; Wallace, A.C.; Roelke, M.; Kehler, J.; Leighty, R.; Eizirik, E.; Hannah, S.S.; Nelson, G.; Schäffer, A.A.; et al. Endogenous retrovirus insertion in the KIT oncogene determines white and white spotting in domestic cats. G3 2014, 4, 1881–1891. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Pavan, W.J. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: Do all roads lead to Mitf? Cell Res. 2008, 18, 1163–1176. [Google Scholar] [CrossRef] [PubMed]
- Baxter, L.L.; Watkins-Chow, D.E.; Pavan, W.J.; Loftus, S.K. A curated gene list for expanding the horizons of pigmentation biology. Pigment. Cell Melanoma Res. 2019, 32, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, E.K.; Baranowska, I.; Wade, C.M.; Salmon Hillbertz, N.H.; Zody, M.C.; Anderson, N.; Biagi, T.M.; Patterson, N.; Pielberg, G.R.; Kulbokas, E.J., 3rd; et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat. Genet. 2007, 39, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Yusnizar, Y.; Wilbe, M.; Herlino, A.O.; Sumantri, C.; Noor, R.R.; Boediono, A.; Andersson, L.; Andersson, G. Microphthalmia-associated transcription factor mutations are associated with white-spotted coat color in swamp buffalo. Anim. Genet. 2015, 46, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Hofstetter, S.; Seefried, F.; Häfliger, I.M.; Jagannathan, V.; Leeb, T.; Drögemüller, C. A non-coding regulatory variant in the 5′-region of the MITF gene is associated with white-spotted coat in Brown Swiss cattle. Anim. Genet. 2019, 50, 27–32. [Google Scholar] [CrossRef] [PubMed]
- McFadden, A.; Vierra, M.; Martin, K.; Brooks, S.A.; Everts, R.E.; Lafayette, C. Spotting the Pattern: A Review on White Coat Color in the Domestic Horse. Animals 2024, 14, 451. [Google Scholar] [CrossRef]
- Huang, S.; Song, J.; He, C.; Cai, X.; Yuan, K.; Mei, L.; Feng, Y. Genetic insights, disease mechanisms, and biological therapeutics for Waardenburg syndrome. Gene Ther. 2022, 29, 479–497. [Google Scholar] [CrossRef]
- Rudd Garces, G.; Farke, D.; Schmidt, M.J.; Letko, A.; Schirl, K.; Abitbol, M.; Leeb, T.; Lyons, L.A.; Lühken, G. PAX3 haploinsufficiency in Maine Coon cats with dominant blue eyes and hearing loss resembling the human Waardenburg syndrome. G3, 2024; accepted. [Google Scholar] [CrossRef] [PubMed]
- Abitbol, M.; Couronne, A.; Dufaure de Citres, C.; Gache, V. A PAX3 insertion in the Celestial breed and certain feline breeding lines with dominant blue eyes. Animal Genet. 2024; Online ahead of print. [Google Scholar] [CrossRef]
- Gandolfi, B.; Alhaddad, H.; Abdi, M.; Bach, L.H.; Creighton, E.K.; Davis, B.W.; Decker, J.E.; Dodman, N.H.; Ginns, E.I.; Grahn, J.C.; et al. Applications and efficiencies of the first cat 63K DNA array. Sci. Rep. 2018, 8, 7024. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Källberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C.T. Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 2016, 32, 1220–1222. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef]
- Moore, S.; Ribes, V.; Terriente, J.; Wilkinson, D.; Relaix, F.; Briscoe, J. Distinct regulatory mechanisms act to establish and maintain PAX3 expression in the developing neural tube. PLoS Genet. 2013, 9, e1003811. [Google Scholar] [CrossRef]
- Jern, P.; Coffin, J.M. Effects of retroviruses on host genome function. Annu. Rev. Genet. 2008, 42, 709–732. [Google Scholar] [CrossRef]
- Degenhardt, K.R.; Milewski, R.C.; Padmanabhan, A.; Miller, M.; Singh, M.K.; Lang, D.; Engleka, K.A.; Wu, M.; Li, J.; Zhou, D.; et al. Distinct enhancers at the Pax3 locus can function redundantly to regulate neural tube and neural crest expressions. Dev. Biol. 2010, 339, 519–527. [Google Scholar] [CrossRef]
- Pontius, J.U.; Mullikin, J.C.; Smith, D.R.; Agencourt Sequencing Team; Lindblad-Toh, K.; Gnerre, S.; Clamp, M.; Chang, J.; Stephens, R.; Neelam, B.; et al. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007, 17, 1675–1689. [Google Scholar] [CrossRef]
- Medstrand, P.; van de Lagemaat, L.N.; Mager, D. Retroelement distributions in the human genome: Variations associated with age and proximity to genes. Genome Res. 2002, 12, 1483–1495. [Google Scholar] [CrossRef]
- Cui, Y.Z.; Man, X.Y. Biology of melanocytes in mammals. Front. Cell Dev. Biol. 2023, 11, 1309557. [Google Scholar] [CrossRef]
- Hauswirth, R.; Haase, B.; Blatter, M.; Brooks, S.A.; Burger, D.; Drögemüller, C.; Gerber, V.; Henke, D.; Janda, J.; Jude, R.; et al. Mutations in MITF and PAX3 cause “splashed white” and other white spotting phenotypes in horses. PLoS Genet. 2012, 8, e1002653. [Google Scholar] [CrossRef]
- Hauswirth, R.; Jude, R.; Haase, B.; Bellone, R.R.; Archer, S.; Holl, H.; Brooks, S.A.; Tozaki, T.; Penedo, M.C.; Rieder, S.; et al. Novel variants in the KIT and PAX3 genes in horses with white-spotted coat colour phenotypes. Anim. Genet. 2013, 44, 763–765. [Google Scholar] [CrossRef]
- Epstein, D.J.; Vogan, K.J.; Trasler, D.G.; Gros, P. A mutation within intron 3 of the Pax-3 gene produces aberrantly spliced mRNA transcripts in the splotch (Sp) mouse mutant. Proc. Natl. Acad. Sci. USA 1993, 90, 532–536. [Google Scholar] [CrossRef]
- Zlotogora, J.; Lerer, I.; Bar-David, S.; Ergaz, Z.; Abeliovich, D. Homozygosity for Waardenburg syndrome. Am. J. Hum. Genet. 1995, 56, 1173–1178. [Google Scholar]
- Wollnik, B.; Tukel, T.; Uyguner, O.; Ghanbari, A.; Kayserili, H.; Emiroglu, M.; Yuksel-Apak, M. Homozygous and heterozygous inheritance of PAX3 mutations causes different types of Waardenburg syndrome. Am. J. Med. Genet. A 2003, 122A, 42–45. [Google Scholar] [CrossRef] [PubMed]
- Salah, S.; Meiner, V.; Abumayaleh, A.; Asafra, A.; Al-Sharif, T.; Al-Fallah, O.; Hasasneh, B.; Zlotogora, J. Biallelic variants in PAX3 cause Klein syndrome. Clin. Genet. 2022, 102, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, R.; Sato, E.; Igarashi, T.; Miyazawa, T. Characterization of RD-114 virus isolated from a commercial canine vaccine manufactured using CRFK cells. J. Clin. Microbiol. 2010, 48, 3366–3369. [Google Scholar] [CrossRef]

| Lineage or Breed | Founder Cats | Genetic Background | White Spotting | Eye Phenotype | Hearing Phenotype in Heterozygous DBE Cats | Inheritance Characteristics | Homozygous DBE Cats | Sources and References |
|---|---|---|---|---|---|---|---|---|
| Altai | Fyodor (DSH) | Mixed | Minimal | Heterochromia or two blue eyes | Assumed to be normal (no BAER test available) | Autosomal dominant, incomplete penetrance, variable expressivity | Van-like * or white | Studied in this paper |
| Topaz | Roxi and Seymour (DSH) | Mixed | Minimal | Heterochromia or two blue eyes | Assumed to be normal (no BAER test available) | Autosomal dominant, incomplete penetrance, variable expressivity and pleiotropy | Bicolour **, van-like, or white; some of them are deaf; this group included homozygous and compound heterozygous cats | The line is probably lost (http://messybeast.com/DBE-topaz.htm, accessed on 11 April 2024) Not studied |
| Celestial | Roxi (DSH) | British shorthair/longhair, domestic shorthair/longhair | Minimal | Heterochromia or two blue eyes | Normal (BAER tested) | Autosomal dominant, incomplete penetrance, variable expressivity and pleiotropy | White; a single kitten was produced and died at birth | [12] Studied in this paper |
| British DBE (Seymour line) | Seymour (DSH) | British shorthair/longhair | Minimal | Heterochromia or two blue eyes | Assumed to be normal (no BAER test available) | Autosomal dominant, incomplete penetrance, variable expressivity and pleiotropy | Unknown, no homozygous cat was produced | Studied in this paper |
| British DBE (Nanotigr line) | Oliver (DSH) and his daughter named Amelia | British shorthair/longhair | Minimal | Heterochromia or two blue eyes | Assumed to be normal (no BAER test available) | Autosomal dominant, incomplete penetrance, variable expressivity | Van-like or white, some of them are deaf | Studied in this paper |
| British DBE (Igor line) | Igor Azur Dream (DSH) | British shorthair/longhair | Minimal | Heterochromia or two blue eyes | Deaf cats were reported (no BAER test available) | Autosomal dominant, incomplete penetrance, variable expressivity and pleiotropy | Unknown | Studied in this paper |
| British DBE (spontaneous variant, Nadeya line) | Nadeya Ermine Trace (BSH) | British shorthair/longhair | Minimal | Heterochromia or two blue eyes | Deaf cats were reported (no BAER test available) | Autosomal dominant, incomplete penetrance, variable expressivity and pleiotropy | Unknown | Studied in this paper |
| Persian DBE (Alaska line) | Seymour (DSH) | Persian | Minimal | Heterochromia or two blue eyes | Assumed to be normal (no BAER test available) | Autosomal dominant, incomplete penetrance, variable expressivity | Unknown, no homozygous cat was produced | Studied in this paper |
| Persian DBE (Cyrridwen line) | Marusya (DSH) | Persian and Exotic | Minimal | Heterochromia or two blue eyes | Assumed to be normal (no BAER test available) | Autosomal dominant, incomplete penetrance, variable expressivity | Unknown | Studied in this paper |
| Ragdoll DBE | Seymour (DSH) | Ragdoll | Minimal to bicolour $ | Heterochromia or two blue eyes | Assumed to be normal (no BAER test available) | Autosomal dominant, incomplete penetrance, variable expressivity | Unknown | Studied in this paper |
| Sphynx DBE | Unknown | Sphynx | Minimal | Heterochromia or two blue eyes | Assumed to be normal (no BAER test available) | Autosomal dominant, incomplete penetrance, variable expressivity | Unknown | Studied in this paper |
| Siberian DBE | Unknown | Siberian | Minimal to bicolour $ | Heterochromia or two blue eyes | Assumed to be normal (no BAER test available) | Autosomal dominant, incomplete penetrance, variable expressivity | Unknown | [12] Studied in this paper |
| Maine Coon DBE (Topaz line) | Roxi and Seymour (DSH) | Maine Coon | Minimal to bicolour $ | Heterochromia or two blue eyes | Assumed to be normal (no BAER test available) | Autosomal dominant, incomplete penetrance, variable expressivity | Unknown, no homozygous cat was reported # | Studied in this paper |
| Maine Coon DBE (spontaneous variant, Dutch line) | Rociri Elvis (MCO) | Maine Coon | Minimal to bicolour $ | Heterochromia or two blue eyes | Deaf cats were reported (BAER tested) | Autosomal dominant, incomplete penetrance, variable expressivity and pleiotropy | Unknown, no homozygous cat was reported # | [11] Studied in this paper |
| Maine Coon (Pillowtalk line) | Unknown | Maine Coon | Minimal to bicolour $ | Heterochromia or two blue eyes | Unknown | Autosomal dominant | Unknown | Studied in this paper |
| Maine Coon (Nahal line) | Nahal (domestic cat from Russia) | Maine Coon | No data available | No data available | No data available | No data available | No data available | Not studied |
| Phenotype | Lineage * | Breed | Founder Cats | Number | Genotype |
|---|---|---|---|---|---|
| DBE | Altai | Altai | Fyodor (outbred cat from Kazakhstan) | 15 | Heterozygous |
| White $ | Altai | Altai | Fyodor | 2 | Homozygous |
| DBE | Seymour | British (=6), Persian (=3, Alaska line), Ragdoll (=4) | Seymour (outbred cat from Russia) | 13 | Heterozygous |
| Latent | Seymour | Mixed breed (=1) and Persian (=1, Alaska line) | Seymour | 2 | Heterozygous |
| DBE | Nanotigr | British SH/LH | Unknown (Russia) | 13 | Heterozygous |
| White $ | Nanotigr | British SH/LH | Unknown (Russia) | 1 | Homozygous |
| Latent | Nanotigr | British SH/LH | Unknown (Russia) | 3 | Heterozygous |
| DBE | Unknown | Sphynx | Unknown | 3 | Heterozygous |
| DBE | Mixed (Seymour and Roxi) | Chinese Tank | Roxi and Seymour (outbred cats from Kazakhstan and Russia) | 1 | Heterozygous |
| White $ | Mixed (Seymour and Roxi) | Mixed breed | Roxi and Seymour | 2 | Heterozygous |
| DBE | Mixed (Seymour and Roxi) | Mixed breed | Roxi and Seymour | 1 | Heterozygous |
| DBE | Mixed (Seymour and Roxi) | Mixed breed | Roxi and Seymour | 1 | WT |
| DBE | Topaz (Seymour and Roxi) | Maine Coon | Roxi and Seymour | 10 | WT |
| DBE | Roxi | Celestial | Roxi (outbred cat from Kazakhstan) | 20 | WT |
| White $ | Roxi | Celestial | Roxi | 1 | WT |
| Latent | Roxi | Celestial | Roxi | 1 | WT |
| DBE | Dutch Maine Coon | Maine Coon | Purebred Maine Coon cat from the Netherlands | 13 | WT |
| DBE | Pillowtalk | Maine Coon | Unknown | 2 | WT |
| DBE | Unknown | Siberian | Unknown | 4 | WT |
| DBE | Igor | British SH/LH | Unknown (Russia) | 6 | WT |
| DBE | Nadeya | British SH/LH | Purebred British cat from Russia | 2 | WT |
| DBE | Cyrridwen | Exotic/Persian | Marusya (outbred cat from Russia) | 1 | WT |
| Total | 117 | ||||
| Control | British SH/LH | 22 | WT | ||
| Control | Celestial | 2 | WT | ||
| Control | Maine Coon | 3 | WT | ||
| Control | Siberian | 2 | WT | ||
| Control | Turkish Angora | 1 | WT | ||
| Control | Birman | 1 | WT | ||
| Control | Chartreux | 2 | WT | ||
| Control | Bengal | 1 | WT | ||
| Control | Devon Rex | 1 | WT | ||
| Control | Donskoy | 1 | WT | ||
| Control | Persian | 2 | WT | ||
| Control | Siamese | 2 | WT | ||
| Control | Sphynx | 9 | WT | ||
| Control | Domestic shorthair | 2 | WT | ||
| Control | Mixed breed | 9 | WT | ||
| Total | 60 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abitbol, M.; Dufaure de Citres, C.; Rudd Garces, G.; Lühken, G.; Lyons, L.A.; Gache, V. Different Founding Effects Underlie Dominant Blue Eyes (DBE) in the Domestic Cat. Animals 2024, 14, 1845. https://doi.org/10.3390/ani14131845
Abitbol M, Dufaure de Citres C, Rudd Garces G, Lühken G, Lyons LA, Gache V. Different Founding Effects Underlie Dominant Blue Eyes (DBE) in the Domestic Cat. Animals. 2024; 14(13):1845. https://doi.org/10.3390/ani14131845
Chicago/Turabian StyleAbitbol, Marie, Caroline Dufaure de Citres, Gabriela Rudd Garces, Gesine Lühken, Leslie A. Lyons, and Vincent Gache. 2024. "Different Founding Effects Underlie Dominant Blue Eyes (DBE) in the Domestic Cat" Animals 14, no. 13: 1845. https://doi.org/10.3390/ani14131845





