Homology in Sex Determination in Two Distant Spiny Frogs, Nanorana quadranus and Quasipaa yei
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Preparation
2.2. Genomic Library Preparation
2.3. Filtering and SNP Calling
2.4. Screening Sex-Linked Markers
2.5. Confirmation of Sex-Specific Markers
2.6. PCR Validation
2.7. Assigning Sex-Linked Markers onto Sex Chromosome
2.8. Predicting the Sex-Linked Markers Involved in Genes for Sex Determination
3. Results
3.1. GBS Data Analyses and SNP Calling
3.2. Sex-Linked Marker Screening
3.3. Validation of the Sex-Linked Markers
3.4. PCR Validation
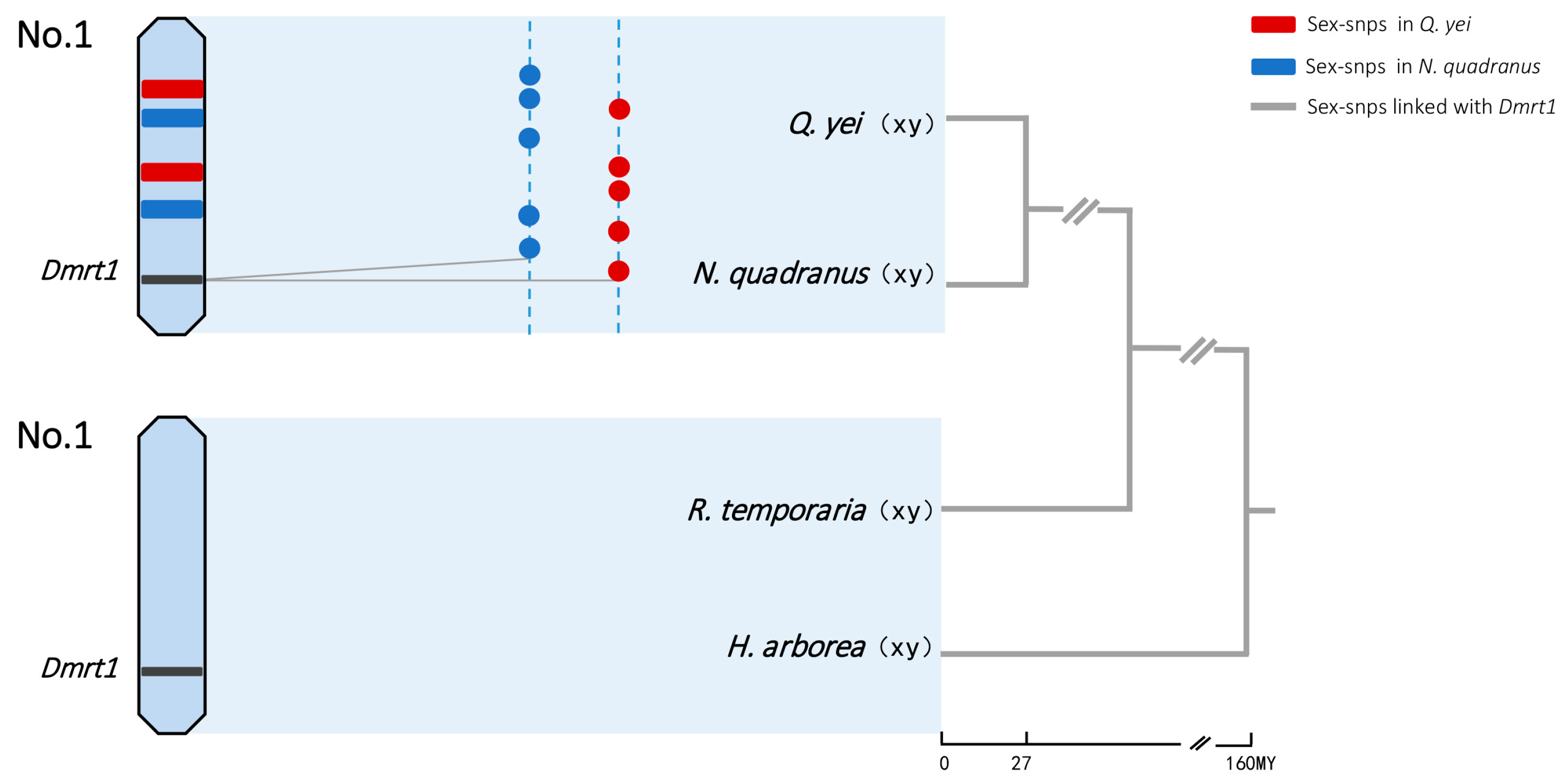
3.5. Identifying the Sex Chromosome
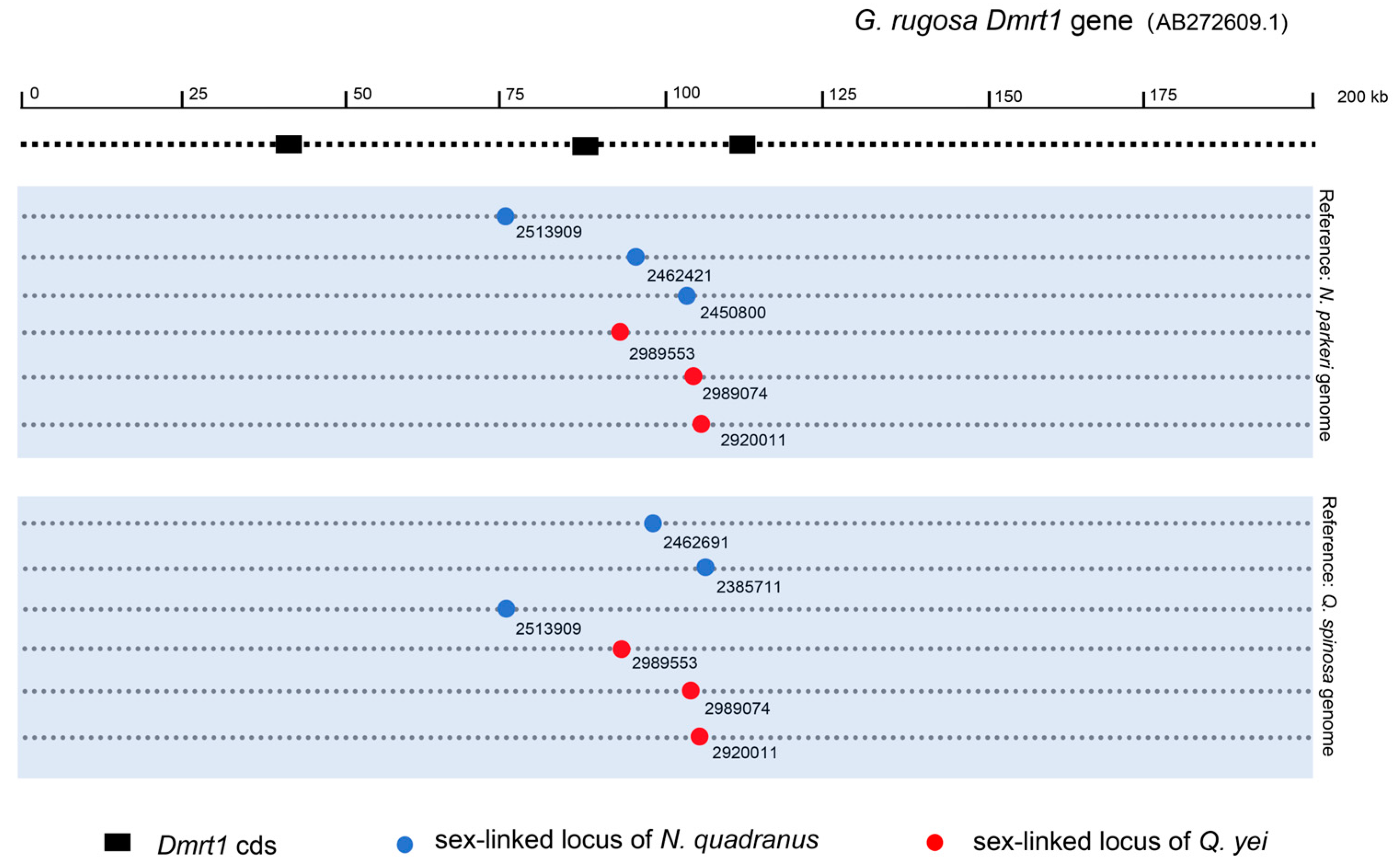
3.6. Identifying Potential Sex-Determining Genes
4. Discussion
4.1. Sex Determination Is Limited to Homologous Chromosomes
4.2. Sex Determination May Pertain to the DMRT1 Gene
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beukeboom, L.W.; Perrin, N. Evolution of Sex Determination; Oxford University Press: Oxford, UK, 2014. [Google Scholar]
- Bachtrog, D.; Mank, J.E.; Peichel, C.L.; Kirkpatrick, M.; Otto, S.P.; Ashman, T.L.; Hahn, M.W.; Kitano, J.; Mayrose, I.; Ming, R.; et al. Tree of Sex Consortium. Sex determination: Why so many ways of doing it? PLoS Biol. 2014, 12, e1001899. [Google Scholar] [CrossRef]
- Nakamura, M. Sex determination in amphibians. Semin. Cell Dev. Biol. 2009, 20, 271–282. [Google Scholar] [CrossRef]
- Schmid, M.; Steinlein, C.; Bogart, J.P.; Feichtinger, W.; León, P.; La Marca, E.; Díaz, L.M.; Sanz, A.; Chen, S.H.; Hedges, S.B. The chromosomes of terraranan frogs. Insights into vertebrate cytogenetics. Cytogenet. Genome Res. 2010, 130–131, 1–14. [Google Scholar] [CrossRef]
- Schmid, M.; Steinlein, C. Sex chromosomes, sex-linked genes, and sex determination in the vertebrate class amphibia. EXS 2001, 91, 143–176. [Google Scholar]
- Malcom, J.W.; Kudra, R.S.; Malone, J.H. The sex chromosomes of frogs: Variability and tolerance offer clues to genome evolution and function. J. Genom. 2014, 20, 68–76. [Google Scholar] [CrossRef]
- Uno, Y.; Nishida, C.; Takagi, C.; Igawa, T.; Ueno, N.; Sumida, M.; Matsuda, Y. Extraordinary diversity in the origins of sex chromosomes in anurans inferred from comparative gene mapping. Cytogenet. Genome Res. 2015, 145, 218–229. [Google Scholar] [CrossRef]
- Miura, I. An evolutionary witness: The frog Rana rugosa underwent change of heterogametic sex from XY male to ZW female. Sex. Dev. 2007, 1, 323–331. [Google Scholar] [CrossRef]
- Rodrigues, N.; Vuille, Y.; Brelsford, A.; Merilä, J.; Perrin, N. The genetic contribution to sex determination and number of sex chromosomes vary among populations of common frogs (Rana temporaria). Heredity 2016, 117, 25–32. [Google Scholar] [CrossRef]
- Furman, B.L.S.; Cauret, C.M.S.; Knytl, M.; Song, X.Y.; Premachandra, T.; Ofori-Boateng, C.; Jordan, D.C.; Horb, M.E.; Evans, B.J. A frog with three sex chromosomes that co-mingle together in nature: Xenopus tropicalis has a degenerate W and a Y that evolved from a Z chromosome. PLoS Genet. 2020, 16, e1009121. [Google Scholar] [CrossRef]
- Song, X.Y.; Furman, B.L.S.; Premachandra, T.; Knytl, M.; Cauret, C.M.S.; Wasonga, D.V.; Measey, J.; Dworkin, I.; Evans, B.J. Sex chromosome degeneration, turnover, and sex-biased expression of sex-linked transcripts in African clawed frogs (Xenopus). Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 20200095. [Google Scholar] [CrossRef]
- Wilkins, A.S. Moving up the hierarchy: A hypothesis on the evolution of a genetic sex determination pathway. Bioessays 1995, 17, 71–77. [Google Scholar] [CrossRef]
- Schartl, M. Sex chromosome evolution in non-mammalian vertebrates. Curr. Opin. Genet. Dev. 2004, 14, 634–641. [Google Scholar] [CrossRef]
- Volff, J.N.; Nanda, I.; Schmid, M.; Schartl, M. Governing sex determination in fish: Regulatory putsches and ephemeral dictators. Sex. Dev. 2007, 1, 85–99. [Google Scholar] [CrossRef]
- Graves, J.A. How to evolve new vertebrate sex determining genes. Dev. Dyn. 2013, 242, 354–359. [Google Scholar] [CrossRef]
- Charlesworth, D.; Mank, J.E. The birds and the bees and the flowers and the trees: Lessons from genetic mapping of sex determination in plants and animals. Genetics 2010, 186, 9–31. [Google Scholar] [CrossRef]
- Hillis, D.M.; Green, D.M. Evolutionary changes of heterogametic sex in the phylogenetic history of amphibians. J. Evol. Biol. 1990, 3, 349–364. [Google Scholar] [CrossRef]
- Jeffries, D.L.; Lavanchy, G.; Sermier, R.; Sredl, M.J.; Miura, I.; Borzée, A.; Barrow, L.N.; Canestrelli, D.; Crochet, P.A.; Dufresnes, C.; et al. A rapid rate of sex-chromosome turnover and non-random transitions in true frogs. Nat. Commun. 2018, 9, 4088. [Google Scholar] [CrossRef]
- Ma, W.J.; Veltsos, P. The diversity and evolution of sex chromosomes in frogs. Genes. 2021, 12, 483. [Google Scholar] [CrossRef]
- Marshall Graves, J.A.; Peichel, C.L. Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol. 2010, 11, 205. [Google Scholar] [CrossRef]
- O’Meally, D.; Ezaz, T.; Georges, A.; Sarre, S.D.; Graves, J.A. Are some chromosomes particularly good at sex? insights from amniotes. Chromosome Res. 2012, 20, 7–19. [Google Scholar] [CrossRef]
- Wright, D.A.; Richards, C.M.; Frost, J.S.; Camozzi, A.M.; Kunz, B.J. Genetic mapping in amphibians. Isozymes Curr. Top. Biol. Med. Res. 1983, 10, 287–311. [Google Scholar]
- Wright, D.A.; Richards, C.M. Two sex-linked loci in the leopard frog, Rana pipiens. Genetics 1983, 103, 249–261. [Google Scholar] [CrossRef]
- Sumida, M.; Nishioka, M. A pronounced sex difference when two linked loci of the Japanese brown frog Rana japonica are recombined. Biochem. Genet. 1994, 32, 361–369. [Google Scholar] [CrossRef]
- Sumida, M.; Nishioka, M. Sex-linked genes and linkage maps in amphibians. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2000, 126, 257–270. [Google Scholar] [CrossRef]
- Luo, W.; Xia, Y.; Yue, B.S.; Zeng, X.M. Assigning the Sex-Specific Markers via Genotyping-by-Sequencing onto the Y Chromosome for a torrent frog Amolops mantzorum. Genes 2020, 11, 727. [Google Scholar] [CrossRef]
- Miura, I. Sex determination and sex chromosomes in amphibia. Sex. Dev. 2017, 11, 298–306. [Google Scholar] [CrossRef]
- Roco, Á.S.; Olmstead, A.W.; Degitz, S.J.; Amano, T.; Zimmerman, L.B.; Bullejos, M. Coexistence of Y, W, and Z sex chromosomes in Xenopus tropicalis. Proc. Natl. Acad. Sci. USA 2015, 112, E4752–E4761. [Google Scholar] [CrossRef]
- Cauret, C.M.S.; Gansauge, M.T.; Tupper, A.S.; Furman, B.L.S.; Knytl, M.; Song, X.Y.; Greenbaum, E.; Meyer, M.; Evans, B.J. Developmental Systems drift and the drivers of sex chromosome evolution. Mol. Biol. Evol. 2020, 37, 799–810. [Google Scholar] [CrossRef]
- Miura, I.; Ohtani, H.; Nakamura, M.; Ichikawa, Y.; Saitoh, K. The origin and differentiation of the heteromorphic sex chromosomes Z, W, X, and Y in the frog Rana rugosa, inferred from the sequences of a sex-linked gene, ADP/ATP translocase. Mol. Biol. Evol. 1998, 15, 1612–1619. [Google Scholar] [CrossRef]
- Uno, Y.; Nishida, C.; Yoshimoto, S.; Ito, M.; Oshima, Y.; Yokoyama, S.; Nakamura, M.; Matsuda, Y. Diversity in the origins of sex chromosomes in anurans inferred from comparative mapping of sexual differentiation genes for three species of the Raninae and Xenopodinae. Chromosome Res. 2008, 16, 999–1011. [Google Scholar] [CrossRef]
- Oshima, Y.; Kato, T.; Wang, D.; Murakami, T.; Matsuda, Y.; Nagahama, Y.; Nakamura, M. Promoter activity and chromosomal location of the Rana rugosa P450 aromatase (CYP19) gene. Zoolog Sci. 2006, 23, 79–85. [Google Scholar] [CrossRef]
- Sakurai, N.; Maruo, K.; Haraguchi, S.; Uno, Y.; Oshima, Y.; Tsutsui, K.; Matsuda, Y.; Do Rego, J.L.; Pelletier, G.; Vaudry, H.; et al. Immunohistochemical detection and biological activities of CYP17 (P450c17) in the indifferent gonad of the frog Rana rugosa. J. Steroid Biochem. Mol. Biol. 2008, 112, 5–12. [Google Scholar] [CrossRef]
- Yoshimoto, S.; Okada, E.; Umemoto, H.; Tamura, K.; Uno, Y.; Nishida-Umehara, C.; Matsuda, Y.; Takamatsu, N.; Shiba, T.; Ito, M. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc. Natl. Acad. Sci. USA 2008, 105, 2469–2474. [Google Scholar] [CrossRef]
- Kuhl, H.; Tan, W.H.; Klopp, C.; Kleiner, W.; Koyun, B.; Ciorpac, M.; Feron, R.; Knytl, M.; Kloas, W.; Schartl, M.; et al. A candidate sex determination locus in amphibians which evolved by structural variation between X- and Y-chromosomes. Nat. Commun. 2024, 15, 4781. [Google Scholar] [CrossRef]
- Brelsford, A.; Stöck, M.; Betto-Colliard, C.; Dubey, S.; Dufresnes, C.; Jourdan-Pineau, H.; Rodrigues, N.; Savary, R.; Sermier, R.; Perrin, N. Homologous sex chromosomes in three deeply divergent anuran species. Evolution. 2013, 67, 2434–2440. [Google Scholar] [CrossRef]
- Che, J.; Zhou, W.W.; Hu, J.S.; Yan, F.; Papenfuss, T.J.; Wake, D.B.; Zhang, Y.P. Spiny frogs (Paini) illuminate the history of the Himalayan region and Southeast Asia. Proc. Natl. Acad. Sci. USA 2010, 107, 13765–13770. [Google Scholar] [CrossRef]
- Fei, L.; Hu, S.Q.; Ye, C.Y.; Huang, Y. Fauna sinica. Amphibia, Anura Ranidae; Science Press: Beijing, China, 2009; Volume 3. (In Chinese) [Google Scholar]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Catchen, J.M.; Amores, A.; Hohenlohe, P.; Cresko, W.; Postlethwait, J.H. Stacks: Building and genotyping Loci de novo from short-read sequences. G3 2011, 1, 171–182. [Google Scholar] [CrossRef]
- Brelsford, A.; Lavanchy, G.; Sermier, R.; Rausch, A.; Perrin, N. Identifying Homomorphic Sex Chromosomes from Wild-Caught adults with limited genomic resources. Mol. Ecol. Resour. 2017, 17, 752–759. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statisticalcomputing. R Foundation for Statistical Computing, Vienna, Austria. 2022. Available online: https://www.R-project.org/ (accessed on 16 June 2021).
- Stovall, W.R.; Taylor, H.R.; Black, M.; Grosser, S.; Rutherford, K.; Gemmell, N.J. Genetic sex assignment in wild populations using genotyping-by-sequencing data: A statistical threshold approach. Mol. Ecol. Resour. 2018, 18, 179–190. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Hu, X.X.; Jiang, Z.Y.; Ming, Y.; Jian, J.B.; Jiang, S.J.; Zhang, D.D.; Zhang, J.Y.; Zheng, S.J.; Fang, X.D.; Yang, Y.L.; et al. A chromosomal level genome sequence for Quasipaa spinosa (Dicroglossidae) reveals chromosomal evolution and population diversity. Mol. Ecol. Resour. 2022, 22, 1545–1558. [Google Scholar] [CrossRef]
- Fu, T.T.; Sun, Y.B.; Gao, W.; Long, C.B.; Yang, C.H.; Yang, X.W.; Zhang, Y.; Lan, X.Q.; Huang, S.; Jin, J.Q.; et al. The highest-elevation frog provides insights into mechanisms and evolution of defenses against high UV radiation. Proc. Natl. Acad. Sci. USA 2022, 119, e2212406119. [Google Scholar] [CrossRef]
- Brelsford, A.; Dufresnes, C.; Perrin, N. Trans-species variation in Dmrt1 is associated with sex determination in four European tree-frog species. Evolution 2016, 70, 840–847. [Google Scholar] [CrossRef]
- Yuan, Z.Y.; Zhang, B.L.; Raxworthy, C.J.; Weisrock, D.W.; Hime, P.M.; Jin, J.Q.; Lemmon, E.M.; Lemmon, A.R.; Holland, S.D.; Kortyna, M.L.; et al. Natatanuran frogs used the Indian Plate to step-stone disperse and radiate across the Indian Ocean. Natl. Sci. Rev. 2019, 6, 10–14. [Google Scholar] [CrossRef]
- Dufresnes, C.; Borzée, A.; Horn, A.; Stöck, M.; Ostini, M.; Sermier, R.; Wassef, J.; Litvinchuck, S.N.; Kosch, T.A.; Waldman, B.; et al. Sex-Chromosome homomorphy in palearctic tree frogs results from both turnovers and X-Y recombination. Mol. Biol. Evol. 2015, 32, 2328–2337. [Google Scholar] [CrossRef]
- Dufresnes, C.; Brelsford, A.; Baier, F.; Perrin, N. When sex chromosomes recombine only in the heterogametic sex: Heterochiasmy and heterogamety in Hyla tree frogs. Mol. Biol. Evol. 2021, 38, 192–200. [Google Scholar] [CrossRef]
- Brelsford, A.; Dufresnes, C.; Perrin, N. High-density sex-specific linkage maps of a European tree frog (Hyla arborea) identify the sex chromosome without information on offspring sex. Heredity 2016, 116, 177–181. [Google Scholar] [CrossRef]
- Tamschick, S.; Rozenblut-Kościsty, B.; Bonato, L.; Dufresnes, C.; Lymberakis, P.; Kloas, W.; Ogielska, M.; Stöck, M. Sex chromosome conservation, DMRT1 phylogeny and gonad morphology in diploid Palearctic green toads (Bufo viridis subgroup). Cytogenet. Genome Res. 2014, 144, 315–324. [Google Scholar] [CrossRef]
- Ping, J.; Xia, Y.; Ran, J.H.; Zeng, X.M. Heterogeneous evolution of sex chromosomes in the torrent frog Genus Amolops. Int. J. Mol. Sci. 2022, 23, 11146. [Google Scholar] [CrossRef]
- Blaser, O.; Grossen, C.; Neuenschwander, S.; Perrin, N. Sex-chromosome turnovers induced by deleterious mutation load. Evolution. 2013, 67, 635–645. [Google Scholar] [CrossRef] [PubMed]
- van Doorn, G.S.; Kirkpatrick, M. Turnover of sex chromosomes induced by sexual conflict. Nature 2007, 449, 909–912. [Google Scholar] [CrossRef] [PubMed]
- Toups, M.A.; Rodrigues, N.; Perrin, N.; Kirkpatrick, M. A reciprocal translocation radically reshapes sex-linked inheritance in the common frog. Mol. Ecol. 2019, 28, 1877–1889. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.B.; Xiong, Z.J.; Xiang, X.Y.; Liu, S.P.; Zhou, W.W.; Tu, X.L.; Zhong, L.; Wang, L.; Wu, D.D.; Zhang, B.L.; et al. Whole-genome sequence of the Tibetan frog Nanorana parkeri and the comparative evolution of tetrapod genomes. Proc. Natl. Acad. Sci. USA 2015, 112, E1257–E1262. [Google Scholar] [CrossRef]
- Palomar, G.; Ahmad, F.; Vasemägi, A.; Matsuba, C.; Nicieza, A.G.; Cano, J.M. Comparative high-density linkage mapping reveals conserved genome structure but variation in levels of heterochiasmy and location of recombination cold spots in the common frog. G3 2017, 7, 637–645. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, Y.; Liao, G.; Luo, W.; Xia, Y.; Zeng, X. Homology in Sex Determination in Two Distant Spiny Frogs, Nanorana quadranus and Quasipaa yei. Animals 2024, 14, 1849. https://doi.org/10.3390/ani14131849
Xiao Y, Liao G, Luo W, Xia Y, Zeng X. Homology in Sex Determination in Two Distant Spiny Frogs, Nanorana quadranus and Quasipaa yei. Animals. 2024; 14(13):1849. https://doi.org/10.3390/ani14131849
Chicago/Turabian StyleXiao, Yu, Guangjiong Liao, Wei Luo, Yun Xia, and Xiaomao Zeng. 2024. "Homology in Sex Determination in Two Distant Spiny Frogs, Nanorana quadranus and Quasipaa yei" Animals 14, no. 13: 1849. https://doi.org/10.3390/ani14131849





