Comprehensive Metagenomic Analysis of Veterinary Probiotics in Broiler Chickens
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Conditions of the Animal Experiment
2.2. The Probiotic Products Tested and Their Dosage
2.3. Measurement of Animal Body Weight and Sampling
2.4. Analysis of Phenotypic Expression
2.5. Next Generation Sequencing (NGS)
2.6. Bioinformatics Data Analysis
3. Results
3.1. Animal Body Weight Gain
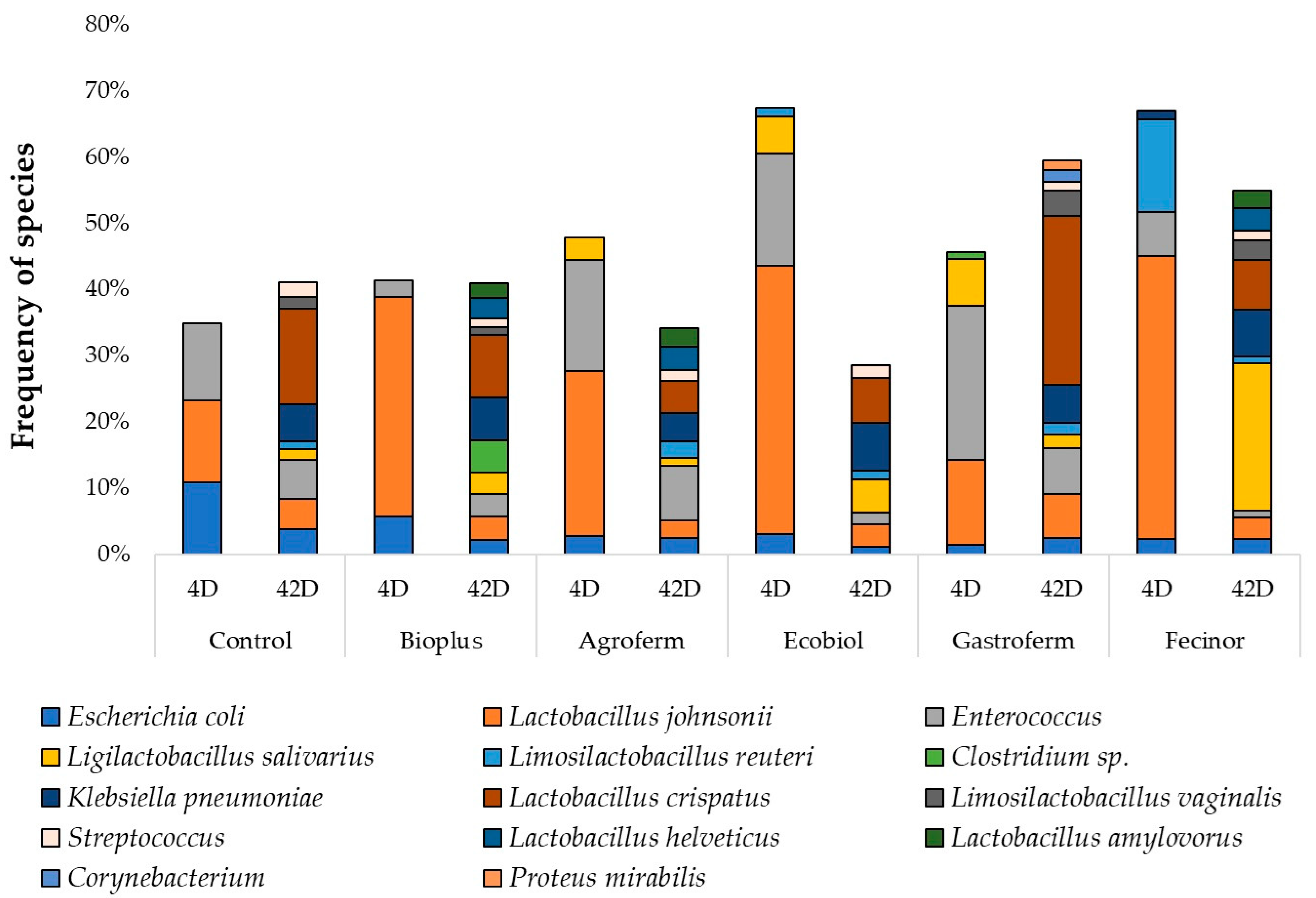
3.2. Results of NGS Regarding the Probiotic Products
3.3. Phenotypic Analysis of Probiotic Strains
3.4. Gut Microbiome Composition and Antimicrobial Resistance Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Macpherson, A.J.; Uhr, T. Induction of Protective IgA by Intestinal Dendritic Cells Carrying Commensal Bacteria. Science 2004, 303, 1662–1665. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, K.; Ota, H.; Akamatsu, T.; Sugiyama, A.; Katsuyama, T. Histochemistry of the Surface Mucous Gel Layer of the Human Colon. Gut 1997, 40, 782–789. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Meriluoto, J.; Salminen, S. Role of Commercial Probiotic Strains against Human Pathogen Adhesion to Intestinal Mucus. Lett. Appl. Microbiol. 2007, 45, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Mack, D.R. Extracellular MUC3 Mucin Secretion Follows Adherence of Lactobacillus Strains to Intestinal Epithelial Cells in Vitro. Gut 2003, 52, 827–833. [Google Scholar] [CrossRef] [PubMed]
- Asahara, T.; Shimizu, K.; Nomoto, K.; Hamabata, T.; Ozawa, A.; Takeda, Y. Probiotic Bifidobacteria Protect Mice from Lethal Infection with Shiga Toxin-Producing Escherichia Coli O157:H7. Infect. Immun. 2004, 72, 2240–2247. [Google Scholar] [CrossRef] [PubMed]
- Maqueda, M.; Sánchez-Hidalgo, M.; Fernández, M.; Montalbán-López, M.; Valdivia, E.; Martínez-Bueno, M. Genetic Features of Circular Bacteriocins Produced by Gram-Positive Bacteria. FEMS Microbiol. Rev. 2008, 32, 2–22. [Google Scholar] [CrossRef] [PubMed]
- Kovács, L.; Nagy, D.; Könyves, L.; Jerzsele, Á.; Kerek, Á. Antimicrobial Properties of Essential Oils—Animal Health Aspects. Hung. Vet. J. 2023, 145, 497–510. [Google Scholar]
- Sebők, C.; Márton, R.A.; Meckei, M.; Neogrády, Z.; Mátis, G. Antimicrobial Peptides as New Tools to Combat Infectious Diseases. Hung. Vet. J. 2024, 146, 181–191. [Google Scholar]
- Olasz, Á.; Jerzsele, Á.; Balta, L.; Dobra, P.F.; Kerek, Á. In Vivo Efficacy of Different Extracts of Propolis in Broiler Salmonellosis. Hung. Vet. J. 2023, 145, 461–475. [Google Scholar]
- Kerek, Á.; Csanády, P.; Jerzsele, Á. Antibacterial Efficiency of Propolis—Part 1. Hung. Vet. J. 2022, 144, 285–298. [Google Scholar]
- Kerek, Á.; Csanády, P.; Jerzsele, Á. Antiprotozoal and Antifungal Efficiency of Propolis—Part 2. Hung. Vet. J. 2022, 144, 691–704. [Google Scholar]
- Kerek, Á.; Csanády, P.; Tuska-Szalay, B.; Kovács, L.; Jerzsele, Á. In Vitro Efficacy of Hungarian Propolis against Bacteria, Yeast, and Trichomonas Gallinae Isolated from Pigeons—A Possible Antibiotic Alternative? Resources 2023, 12, 101. [Google Scholar] [CrossRef]
- Fredenucci, I.; Chomarat, M.; Boucaud, C.; Flandrois, J.P. Saccharomyces Boulardii Fungemia in a Patient Receiving Ultra-Levure Therapy. Clin. Infect. Dis. 1998, 27, 222–223. [Google Scholar] [CrossRef]
- Henry, S.; D’Hondt, L.; André, M.; Holemans, X.; Canon, J.L. Saccharomyces Cerevisiae Fungemia in a Head and Neck Cancer Patient: A Case Report and Review of the Literature. Acta Clin. Belg. 2004, 59, 220–222. [Google Scholar] [CrossRef] [PubMed]
- Lolis, N.; Veldekis, D.; Moraitou, H.; Kanavaki, S.; Velegraki, A.; Triandafyllidis, C.; Tasioudis, C.; Pefanis, A.; Pneumatikos, I. Saccharomyces Boulardii Fungaemia in an Intensive Care Unit Patient Treated with Caspofungin. Crit. Care 2008, 12, 414. [Google Scholar] [CrossRef]
- Muñoz, P.; Bouza, E.; Cuenca-Estrella, M.; Eiros, J.M.; Pérez, M.J.; Sánchez-Somolinos, M.; Rincón, C.; Hortal, J.; Peláez, T. Saccharomyces Cerevisiae Fungemia: An Emerging Infectious Disease. Clin. Infect. Dis. 2005, 40, 1625–1634. [Google Scholar] [CrossRef] [PubMed]
- Niault, M.; Thomas, F.; Prost, J.; Ansari, F.H.; Kalfon, P. Fungemia Due to Saccharomyces Species in a Patient Treated with Enteral Saccharomyces Boulardii. Clin. Infect. Dis. 1999, 28, 930. [Google Scholar] [CrossRef]
- Land, M.H.; Rouster-Stevens, K.; Woods, C.R.; Cannon, M.L.; Cnota, J.; Shetty, A.K. Lactobacillus Sepsis Associated with Probiotic Therapy. Pediatrics 2005, 115, 178–181. [Google Scholar] [CrossRef]
- Oggioni, M.R.; Pozzi, G.; Valensin, P.E.; Galieni, P.; Bigazzi, C. Recurrent Septicemia in an Immunocompromised Patient Due to Probiotic Strains of Bacillus Subtilis. J. Clin. Microbiol. 1998, 36, 325–326. [Google Scholar] [CrossRef]
- Ohishi, A.; Takahashi, S.; Ito, Y.; Ohishi, Y.; Tsukamoto, K.; Nanba, Y.; Ito, N.; Kakiuchi, S.; Saitoh, A.; Morotomi, M.; et al. Bifidobacterium Septicemia Associated with Postoperative Probiotic Therapy in a Neonate with Omphalocele. J. Pediatr. 2010, 156, 679–681. [Google Scholar] [CrossRef] [PubMed]
- Zein, E.F.; Karaa, S.; Chemaly, A.; Saidi, I.; Daou-Chahine, W.; Rohban, R. Lactobacillus rhamnosus septicemia in a diabetic patient associated with probiotic use: A case report. Ann. Biol. Clin. 2008, 66, 195–198. [Google Scholar] [CrossRef]
- Vaarala, O. Immunological Effects of Probiotics with Special Reference to Lactobacilli. Clin. Exp. Allergy 2003, 33, 1634–1640. [Google Scholar] [CrossRef] [PubMed]
- Veckman, V.; Miettinen, M.; Pirhonen, J.; Sirén, J.; Matikainen, S.; Julkunen, I. Streptococcus Pyogenes and Lactobacillus Rhamnosus Differentially Induce Maturation and Production of Th1-Type Cytokines and Chemokines in Human Monocyte-Derived Dendritic Cells. J. Leukoc. Biol. 2004, 75, 764–771. [Google Scholar] [CrossRef]
- Rijkers, G.T.; de Vos, W.M.; Brummer, R.-J.; Morelli, L.; Corthier, G.; Marteau, P. Health Benefits and Health Claims of Probiotics: Bridging Science and Marketing. Br. J. Nutr. 2011, 106, 1291–1296. [Google Scholar] [CrossRef] [PubMed]
- Sniffen, J.C.; McFarland, L.V.; Evans, C.T.; Goldstein, E.J.C. Choosing an Appropriate Probiotic Product for Your Patient: An Evidence-Based Practical Guide. PLoS ONE 2018, 13, e0209205. [Google Scholar] [CrossRef] [PubMed]
- Hempel, S.; Newberry, S.; Ruelaz, A.; Wang, Z.; Miles, J.N.V.; Suttorp, M.J.; Johnsen, B.; Shanman, R.; Slusser, W.; Fu, N.; et al. Safety of Probiotics Used to Reduce Risk and Prevent or Treat Disease. Evid. Rep. Technol. Assess. (Full Rep.) 2011, 200, 1–645. [Google Scholar]
- Harbarth, S.; Balkhy, H.H.; Goossens, H.; Jarlier, V.; Kluytmans, J.; Laxminarayan, R.; Saam, M.; Van Belkum, A.; Pittet, D.; for the World Healthcare-Associated Infections Resistance Forum participants. Antimicrobial Resistance: One World, One Fight! Antimicrob. Resist. Infect. Control 2015, 4, 49. [Google Scholar] [CrossRef]
- Baquero, F. Threats of Antibiotic Resistance: An Obliged Reappraisal. Int. Microbiol. 2021, 24, 499–506. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Gandra, S.; Ashok, A.; Caudron, Q.; Grenfell, B.T.; Levin, S.A.; Laxminarayan, R. Global Antibiotic Consumption 2000 to 2010: An Analysis of National Pharmaceutical Sales Data. Lancet Infect Dis 2014, 14, 742–750. [Google Scholar] [CrossRef]
- Zheng, M.; Zhang, R.; Tian, X.; Zhou, X.; Pan, X.; Wong, A. Assessing the Risk of Probiotic Dietary Supplements in the Context of Antibiotic Resistance. Front. Microbiol. 2017, 8, 908. [Google Scholar] [CrossRef]
- Sanders, M.E.; Akkermans, L.M.A.; Haller, D.; Hammerman, C.; Heimbach, J.; Hörmannsperger, G.; Huys, G.; Levy, D.D.; Lutgendorff, F.; Mack, D.; et al. Safety Assessment of Probiotics for Human Use. Gut Microbes 2010, 1, 164–185. [Google Scholar] [CrossRef] [PubMed]
- Vankerckhoven, V.; Huys, G.; Vancanneyt, M.; Vael, C.; Klare, I.; Romond, M.-B.; Entenza, J.M.; Moreillon, P.; Wind, R.D.; Knol, J.; et al. Biosafety Assessment of Probiotics Used for Human Consumption: Recommendations from the EU-PROSAFE Project. Trends Food Sci. Technol. 2008, 19, 102–114. [Google Scholar] [CrossRef]
- Kastner, S.; Perreten, V.; Bleuler, H.; Hugenschmidt, G.; Lacroix, C.; Meile, L. Antibiotic Susceptibility Patterns and Resistance Genes of Starter Cultures and Probiotic Bacteria Used in Food. Syst. Appl. Microbiol. 2006, 29, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Huys, G.; D’Haene, K.; Swings, J. Genetic Basis of Tetracycline and Minocycline Resistance in Potentially Probiotic Lactobacillus Plantarum Strain CCUG 43738. Antimicrob. Agents Chemother. 2006, 50, 1550–1551. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.-C.; Chen, J.-Y.; Cai, Y.-F.; Huang, C.-B.; Liao, X.-D.; Mi, J.-D. Bacillus Coagulans R11 Consumption Influenced the Abundances of Cecum Antibiotic Resistance Genes in Lead-Exposed Laying Hens. Environ. Pollut. 2021, 274, 116562. [Google Scholar] [CrossRef] [PubMed]
- Zavišić, G.; Popović, M.; Stojkov, S.; Medić, D.; Gusman, V.; Jovanović Lješković, N.; Jovanović Galović, A. Antibiotic Resistance and Probiotics: Knowledge Gaps, Market Overview and Preliminary Screening. Antibiotics 2023, 12, 1281. [Google Scholar] [CrossRef] [PubMed]
- Merenstein, D.; Pot, B.; Leyer, G.; Ouwehand, A.C.; Preidis, G.A.; Elkins, C.A.; Hill, C.; Lewis, Z.T.; Shane, A.L.; Zmora, N.; et al. Emerging Issues in Probiotic Safety: 2023 Perspectives. Gut Microbes 2023, 15, 2185034. [Google Scholar] [CrossRef]
- Gareau, M.G.; Sherman, P.M.; Walker, W.A. Probiotics and the Gut Microbiota in Intestinal Health and Disease. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 503–514. [Google Scholar] [CrossRef]
- Simon, O. Micro-Organisms as Feed Additives—Probiotics. Adv. Pork Prod. 2005, 16, 161–167. [Google Scholar]
- Arias, C.A.; Contreras, G.A.; Murray, B.E. Management of Multidrug-Resistant Enterococcal Infections. Clin. Microbiol. Infect. 2010, 16, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Murray, B.E. The Rise of the Enterococcus: Beyond Vancomycin Resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Howden, B.P.; Stinear, T.P. Evolution of Virulence in Enterococcus Faecium, a Hospital-Adapted Opportunistic Pathogen. Curr. Opin. Microbiol. 2018, 41, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Doron, S.; Snydman, D.R. Risk and Safety of Probiotics. Clin. Infect. Dis. 2015, 60, S129–S134. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-F.; Liu, P.-Y.; Chen, Y.-Y.; Nong, B.-R.; Huang, I.-F.; Hsieh, K.-S.; Chen, K.-T. Three-Combination Probiotics Therapy in Children With Salmonella and Rotavirus Gastroenteritis. J. Clin. Gastroenterol. 2014, 48, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Poloni, V.; Salvato, L.; Pereyra, C.; Oliveira, A.; Rosa, C.; Cavaglieri, L.; Keller, K.M. Bakery By-Products Based Feeds Borne-Saccharomyces Cerevisiae Strains with Probiotic and Antimycotoxin Effects plus Antibiotic Resistance Properties for Use in Animal Production. Food Chem. Toxicol. 2017, 107, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Charteris, W.P.; Kelly, P.M.; Morelli, L.; Collins, J.K. Development and Application of an in Vitro Methodology to Determine the Transit Tolerance of Potentially Probiotic Lactobacillus and Bifidobacterium Species in the Upper Human Gastrointestinal Tract. J. Appl. Microbiol. 1998, 84, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Charteris, W.P.; Kelly, P.M.; Morelli, L.; Collins, J.K. Gradient Diffusion Antibiotic Susceptibility Testing of Potentially Probiotic Lactobacilli. J. Food Prot. 2001, 64, 2007–2014. [Google Scholar] [CrossRef]
- Nawaz, M.; Wang, J.; Zhou, A.; Ma, C.; Wu, X.; Moore, J.E.; Cherie Millar, B.; Xu, J. Characterization and Transfer of Antibiotic Resistance in Lactic Acid Bacteria from Fermented Food Products. Curr. Microbiol. 2011, 62, 1081–1089. [Google Scholar] [CrossRef]
- Belletti, N.; Gatti, M.; Bottari, B.; Neviani, E.; Tabanelli, G.; Gardini, F. Antibiotic Resistance of Lactobacilli Isolated from Two Italian Hard Cheeses. J. Food Prot. 2009, 72, 2162–2169. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Yu, Y.-H. Bacillus Licheniformis-Fermented Products and Enramycin Differentially Modulate Microbiota and Antibiotic Resistome in the Cecal Digesta of Broilers. Poult. Sci. 2022, 101, 102010. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, Y.; Liu, F.; Cao, J.; Lv, N.; Zhu, B.; Zhang, G.; Gao, G.F. Integrated Metagenomic and Metatranscriptomic Profiling Reveals Differentially Expressed Resistomes in Human, Chicken, and Pig Gut Microbiomes. Environ. Int. 2020, 138, 105649. [Google Scholar] [CrossRef]
- Reiczigel, J.; Harnos, A.; Solymosi, N. Biostatisztika Nem Statisztikusoknak, Javított Utánnyomás; Pars Kft.: Budapest, Hungary, 2014. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Volume CLSI Standards M07. In Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018.
- Sahin-Tóth, J.; Kovács, E.; Tóthpál, A.; Juhász, J.; Forró, B.; Bányai, K.; Havril, K.; Horváth, A.; Ghidán, Á.; Dobay, O. Whole Genome Sequencing of Coagulase Positive Staphylococci from a Dog-and-Owner Screening Survey. PLoS ONE 2021, 16, e0245351. [Google Scholar] [CrossRef]
- Modi, A.; Vai, S.; Caramelli, D.; Lari, M. The Illumina Sequencing Protocol and the NovaSeq 6000 System. Methods Mol. Biol. 2021, 2242, 15–42. [Google Scholar] [CrossRef] [PubMed]
- Muzzey, D.; Evans, E.A.; Lieber, C. Understanding the Basics of NGS: From Mechanism to Variant Calling. Curr. Genet. Med. Rep. 2015, 3, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Mihály, Z.; Győrffy, B. Következő generációs szekvenálási technológiák kifejlődése és alkalmazásai = Next generation sequencing technologies (NGST) development and applications. Orvosi Hetil. 2011, 152, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 25 April 2022).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Benoit, G.; Lavenier, D.; Lemaitre, C.; Rizk, G. Bloocoo, a Memory Efficient Read Corrector. In Proceedings of the European Conference on Computational Biology (ECCB), Strasbourg, France, 7 September 2014. [Google Scholar]
- Krueger, F. Trim Galore 2022. Available online: https://github.com/FelixKrueger/TrimGalore (accessed on 25 April 2022).
- Dinghua MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics. Oxford Academic. Available online: https://academic.oup.com/bioinformatics/article/31/10/1674/177884 (accessed on 25 April 2022).
- Vicedomini, R.; Vezzi, F.; Scalabrin, S.; Arvestad, L.; Policriti, A. GAM-NGS: Genomic Assemblies Merger for next Generation Sequencing. BMC Bioinform. 2013, 14 (Suppl. S7), S6. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI Reference Sequences (RefSeq): A Curated Non-Redundant Sequence Database of Genomes, Transcripts and Proteins. Nucleic Acids Res. 2007, 35, D61–D65. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Lahti, L.; Shetty, S. Microbiome R Package. 2012. Available online: https://www.bioconductor.org/packages/release/bioc/html/microbiome.html (accessed on 25 April 2022).
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. MetaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of Mobile Genetic Elements Associated with Antibiotic Resistance in Salmonella Enterica Using a Newly Developed Web Tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, P.S.; Lipinski, L.; Dziembowski, A. PlasFlow: Predicting Plasmid Sequences in Metagenomic Data Using Genome Signatures. Nucleic Acids Res. 2018, 46, e35. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: Mining Viral Signal from Microbial Genomic Data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef] [PubMed]
- FEEDAP. Guidance on the Assessment of Bacterial Susceptibility to Antimicrobials of Human and Veterinary Importance. EFSA J. 2012, 10. [Google Scholar] [CrossRef]
- Xu, Y.; Yu, Y.; Shen, Y.; Li, Q.; Lan, J.; Wu, Y.; Zhang, R.; Cao, G.; Yang, C. Effects of Bacillus Subtilis and Bacillus Licheniformis on Growth Performance, Immunity, Short Chain Fatty Acid Production, Antioxidant Capacity, and Cecal Microflora in Broilers. Poult. Sci. 2021, 100, 101358. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, W.; Zhang, H.; Wang, J.; Zhang, W.; Gao, J.; Wu, S.; Qi, G. Supplemental Bacillus Subtilis DSM 32315 Manipulates Intestinal Structure and Microbial Composition in Broiler Chickens. Sci. Rep. 2018, 8, 15358. [Google Scholar] [CrossRef] [PubMed]
- Trela, J.; Kierończyk, B.; Hautekiet, V.; Józefiak, D. Combination of Bacillus Licheniformis and Salinomycin: Effect on the Growth Performance and GIT Microbial Populations of Broiler Chickens. Animals 2020, 10, 889. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Gong, L.; Zhou, Y.; Tang, L.; Zeng, Z.; Wang, Q.; Zou, P.; Yu, D.; Li, W. Probiotic Paenibacillus Polymyxa 10 and Lactobacillus Plantarum 16 Enhance Growth Performance of Broilers by Improving the Intestinal Health. Anim. Nutr. 2021, 7, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Reuben, R.C.; Sarkar, S.L.; Ibnat, H.; Roy, P.C.; Jahid, I.K. Novel Mono- and Multi-Strain Probiotics Supplementation Modulates Growth, Intestinal Microflora Composition and Haemato-Biochemical Parameters in Broiler Chickens. Vet. Med. Sci. 2022, 8, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhen, W.; Geng, Y.; Wang, Z.; Guo, Y. Effects of Dietary Enterococcus Faecium NCIMB 11181 Supplementation on Growth Performance and Cellular and Humoral Immune Responses in Broiler Chickens. Poult. Sci 2019, 98, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Zhang, X.L.; Wang, D.W.; Li, L.Y.; Liu, G.L.; Li, A.K.; Zhao, Y.X. Effects of Microencapsulated Enterococcus Fecalis CG1.0007 on Growth Performance, Antioxidation Activity, and Intestinal Microbiota in Broiler Chickens. J. Anim. Sci. 2013, 91, 4374–4382. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Hou, Q.; Kwok, L.-Y.; Huo, D.; Feng, S.; Zhang, H. Effect of Feeding Lactobacillus Plantarum P-8 on the Faecal Microbiota of Broiler Chickens Exposed to Lincomycin. Sci. Bull. 2017, 62, 105–113. [Google Scholar] [CrossRef]
- Sureshkumar, S.; Jung, S.K.; Kim, D.; Oh, K.B.; Yang, H.; Lee, H.C.; Jo, Y.J.; Lee, H.S.; Lee, S.; Byun, S.J. Administration of L. Salivarius Expressing 3D8 ScFv as a Feed Additive Improved the Growth Performance, Immune Homeostasis, and Gut Microbiota of Chickens. Anim. Sci. J. 2020, 91, e13399. [Google Scholar] [CrossRef]
- Khan, S.; Chousalkar, K.K. Short-Term Feeding of Probiotics and Synbiotics Modulates Caecal Microbiota during Salmonella Typhimurium Infection but Does Not Reduce Shedding and Invasion in Chickens. Appl. Microbiol. Biotechnol. 2020, 104, 319–334. [Google Scholar] [CrossRef]
- Ahmed, M.; Lyass, L.; Markham, P.N.; Taylor, S.S.; Vázquez-Laslop, N.; Neyfakh, A.A. Two Highly Similar Multidrug Transporters of Bacillus Subtilis Whose Expression Is Differentially Regulated. J. Bacteriol. 1995, 177, 3904–3910. [Google Scholar] [CrossRef]
- Neyfakh, A.A.; Bidnenko, V.E.; Chen, L.B. Efflux-Mediated Multidrug Resistance in Bacillus Subtilis: Similarities and Dissimilarities with the Mammalian System. Proc. Natl. Acad. Sci. USA 1991, 88, 4781–4785. [Google Scholar] [CrossRef] [PubMed]
- Klyachko, K.A.; Schuldiner, S.; Neyfakh, A.A. Mutations Affecting Substrate Specificity of the Bacillus Subtilis Multidrug Transporter Bmr. J. Bacteriol. 1997, 179, 2189–2193. [Google Scholar] [CrossRef] [PubMed]
- Podlesek, Z.; Comino, A.; Herzog-Velikonja, B.; Zgur-Bertok, D.; Komel, R.; Grabnar, M. Bacillus Licheniformis Bacitracin-Resistance ABC Transporter: Relationship to Mammalian Multidrug Resistance. Mol. Microbiol. 1995, 16, 969–976. [Google Scholar] [CrossRef]
- Jack, D.L.; Storms, M.L.; Tchieu, J.H.; Paulsen, I.T.; Saier, M.H. A Broad-Specificity Multidrug Efflux Pump Requiring a Pair of Homologous SMR-Type Proteins. J. Bacteriol. 2000, 182, 2311–2313. [Google Scholar] [CrossRef] [PubMed]
- Agersø, Y.; Bjerre, K.; Brockmann, E.; Johansen, E.; Nielsen, B.; Siezen, R.; Stuer-Lauridsen, B.; Wels, M.; Zeidan, A.A. Putative Antibiotic Resistance Genes Present in Extant Bacillus Licheniformis and Bacillus Paralicheniformis Strains Are Probably Intrinsic and Part of the Ancient Resistome. PLoS ONE 2019, 14, e0210363. [Google Scholar] [CrossRef]
- Pawlowski, A.C.; Stogios, P.J.; Koteva, K.; Skarina, T.; Evdokimova, E.; Savchenko, A.; Wright, G.D. The Evolution of Substrate Discrimination in Macrolide Antibiotic Resistance Enzymes. Nat. Commun. 2018, 9, 112. [Google Scholar] [CrossRef]
- Hachmann, A.-B.; Sevim, E.; Gaballa, A.; Popham, D.L.; Antelmann, H.; Helmann, J.D. Reduction in Membrane Phosphatidylglycerol Content Leads to Daptomycin Resistance in Bacillus Subtilis. Antimicrob. Agents Chemother. 2011, 55, 4326–4337. [Google Scholar] [CrossRef]
- Hachmann, A.-B.; Angert, E.R.; Helmann, J.D. Genetic Analysis of Factors Affecting Susceptibility of Bacillus Subtilis to Daptomycin. Antimicrob. Agents Chemother. 2009, 53, 1598–1609. [Google Scholar] [CrossRef] [PubMed]
- Ernst, C.M.; Staubitz, P.; Mishra, N.N.; Yang, S.-J.; Hornig, G.; Kalbacher, H.; Bayer, A.S.; Kraus, D.; Peschel, A. The Bacterial Defensin Resistance Protein MprF Consists of Separable Domains for Lipid Lysinylation and Antimicrobial Peptide Repulsion. PLoS Pathog. 2009, 5, e1000660. [Google Scholar] [CrossRef]
- Urshev, Z.; Yungareva, T. Initial Safety Evaluation of Enterococcus Faecium LBB.E81. Biotechnol. Biotechnol. Equip. 2021, 35, 11–17. [Google Scholar] [CrossRef]
- Hassen, B.; Abbassi, M.S.; Ruiz-Ripa, L.; Mama, O.M.; Hassen, A.; Torres, C.; Hammami, S. High Prevalence of Mcr-1 Encoding Colistin Resistance and First Identification of BlaCTX-M-55 in ESBL/CMY-2-Producing Escherichia Coli Isolated from Chicken Faeces and Retail Meat in Tunisia. Int. J. Food Microbiol. 2020, 318, 108478. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Héritier, C.; Podglajen, I.; Sougakoff, W.; Gutmann, L.; Nordmann, P. Emergence in Klebsiella Pneumoniae of a Chromosome-Encoded SHV Beta-Lactamase That Compromises the Efficacy of Imipenem. Antimicrob. Agents Chemother. 2003, 47, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Di Conza, J.A.; Badaracco, A.; Ayala, J.; Rodríguez, C.; Famiglietti, A.; Gutkind, G.O. β-Lactamases Produced by Amoxicillin-Clavulanate-Resistant Enterobacteria Isolated in Buenos Aires, Argentina: A New BlaTEM Gene. Rev. Argent. Microbiol. 2014, 46, 210–217. [Google Scholar] [CrossRef]



| No. | Product | Probiotic Strain | Tribal Number | CFU/g | Target Animal |
|---|---|---|---|---|---|
| 1. | BioPlus YC | Bacillus licheniformis | DSMZ5749 | 1.6 × 109 | poultry, swine, cattle |
| Bacillus subtilis | DSMZ5750 | 1.6 × 109 | |||
| 2. | Agroferm M+C | Enterococcus faecium | DSM7134 | 1 × 109 | broiler, goose, turkey, duck |
| Lactobacillus plantarum | DSM12837 | ||||
| Pediococcus acidilactici | DSM16243 | ||||
| 3. | Ecobiol WX | Bacillus amyloliquefacieus | CECT5940 | 1 × 1010 | poultry |
| 4. | Gastroferm M+C | Lactobacillus plantarum | DSM12837 | 1 × 109 | poultry |
| Pediococcus acidilactici | DSM16243 | ||||
| Enterococcus faecium | DSM7134 | ||||
| 5. | Fecinor Soluble Plus | Enterococcus faecium | CECT4515 | 1 × 1010 | poultry, swine |
| Product | Sequence % | ARG | Mechanism | Resistance | |
|---|---|---|---|---|---|
| Coverage | Identity | ||||
| BioPlus YC | 100 | 99.25 | blt | efflux pump | fluoroquinolones |
| 100 | 98.91 | vmlR | target mutation | macrolides, lincosamides, pleuromutilinek, phenicols, tetracyclines, streptogramin, oxazolidone | |
| 100 | 98.59 | aadK | enzymatic inactivation | aminoglycosides | |
| 100 | 99.49 | tmrB | permeability reduction | nucleosides | |
| 100 | 99.79 | lmrB | efflux pump | lincosamides | |
| 100 | 98.68 | mphK | enzymatic inactivation | macrolides | |
| 100 | 100 | mprF | target mutation | peptide antibiotics | |
| 100 | 99.74 | bmr | efflux pump | fluoroquinolones, phenicols, nucleosides | |
| 86.15 | 35.52 | dfrA43 | target mutation | diaminopyrimidines | |
| 100 | 99.65 | ermD | target mutation | macrolides, lincosamides | |
| 100 | 98.03 | bcrC | target mutation | peptide antibiotics | |
| 100 | 98.56 | bcrB | efflux pump | peptide antibiotics | |
| 96.08 | 99.32 | bcrA | efflux pump | peptide antibiotics | |
| 100 | 100 | ykkC | efflux pump | aminoglycosides, tetracyclines, phenicols | |
| 100 | 99.05 | ykkD | efflux pump | aminoglycosides, tetracyclines, phenicols | |
| Agroferm M+C | 100 | 100 | efmA | efflux pump | macrolides, fluoroquinolones |
| 100 | 98.9 | AAC(6’)-Ii | enzymatic inactivation | aminoglycosides | |
| 100 | 97.15 | msrC | target mutation | macrolides, lincosamides, pleuromutilinek, phenicols, tetracyclines, streptogramin, oxazolidone | |
| 100 | 99 | eatAv | target mutation | ||
| Ecobiol WX | 100 | 99.43 | clbA | target mutation | phenicols, lincosamides, pleuromutilinek, streptogramin, oxazolidone |
| 82.56 | 35.75 | dfrA43 | target mutation | diaminopyrimidines | |
| Gastroferm M+C | 100 | 100 | efmA | efflux pump | macrolides, fluoroquinolones |
| 100 | 97.15 | msrC | target mutation | macrolides, lincosamides, pleuromutilines, phenicols, tetracyclines, streptogramin, oxazolidone | |
| 100 | 98.9 | AAC(6’)-Ii | enzymatic inactivation | aminoglycosides | |
| 100 | 99 | eatAv | target mutation | macrolides, lincosamides, pleuromutilines, phenicols, tetracyclines, streptogramin, oxazolidone | |
| Fecinor Soluble Plus | 100 | 98.9 | AAC(6’)-Ii | enzymatic inactivation | aminoglycosides |
| 100 | 97.15 | msrC | target mutation | macrolides, lincosamides, pleuromutilines, phenicols, tetracyclines, streptogramin, oxazolidone | |
| 100 | 99 | eatAv | target mutation | ||
| No. | Probiotic Strain | PEN | AM | AMC | CTR | GEN | OTC | DOX | TIL |
|---|---|---|---|---|---|---|---|---|---|
| µg/mL | |||||||||
| 1. | Bacillus licheniformis | 0.12 | 1 | 2 | 16 | 8 | 8 | 0.06 | 0.5 |
| Bacillus subtilis | 1 | 0.5 | 0.5 | 0.25 | 16 | 0.125 | 32 | 0.5 | |
| 2. | Enterococcus faecium | 8 | 1 | 1 | 16 | 32 | 0.25 | 0.125 | 1 |
| Lactobacillus plantarum | 1 | 16 | 4 | 16 | 8 | 8 | 4 | 2 | |
| Pediococcus acidilactici | 1 | 16 | 8 | 16 | 4 | 4 | 0.5 | 0.125 | |
| 3. | Bacillus amyloliquefacieus | 0.06 | 1 | 2 | 4 | 32 | 0.25 | 0.25 | 0.5 |
| 4. | Lactobacillus plantarum | 4 | 2 | 0.5 | 16 | 32 | 0.5 | 0.125 | 4 |
| Pediococcus acidilactici | 1 | 16 | 4 | 16 | 32 | 32 | 16 | 4 | |
| Enterococcus faecium | 8 | 1 | 1 | 16 | 32 | 0.25 | 0.125 | 1 | |
| 5. | Enterococcus faecium | 4 | 1 | 1 | 16 | 32 | 0.25 | 0.125 | 4 |
| No. | Probiotic Strain | FLO | CLI | TIA | VAN | GAT | PSA | SUL | TRI |
|---|---|---|---|---|---|---|---|---|---|
| µg/mL | |||||||||
| 1. | Bacillus licheniformis | 2 | 32 | 64 | 0.25 | 0.03 | 4 | 64 | 0.25 |
| Bacillus subtilis | 2 | 2 | 64 | 0.25 | 0.03 | 2 | 64 | 0.5 | |
| 2. | Enterococcus faecium | 8 | 4 | 64 | 2 | 0.5 | 128 | 128 | 128 |
| Lactobacillus plantarum | 4 | 0.06 | 64 | 32 | 2 | 128 | 128 | 64 | |
| Pediococcus acidilactici | 4 | 0.06 | 1 | 32 | 0.5 | 128 | 64 | 128 | |
| 3. | Bacillus amyloliquefacieus | 2 | 0.5 | 64 | 1 | 0.03 | 4 | 128 | 0.5 |
| 4. | Lactobacillus plantarum | 8 | 4 | 64 | 32 | 1 | 128 | 128 | 64 |
| Pediococcus acidilactici | 8 | 0.06 | 64 | 32 | 8 | 128 | 128 | 128 | |
| Enterococcus faecium | 8 | 4 | 64 | 1 | 2 | 128 | 128 | 128 | |
| 5. | Enterococcus faecium | 8 | 4 | 64 | 2 | 1 | 128 | 128 | 128 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kerek, Á.; Román, I.L.; Szabó, Á.; Papp, M.; Bányai, K.; Kardos, G.; Kaszab, E.; Bali, K.; Makrai, L.; Jerzsele, Á. Comprehensive Metagenomic Analysis of Veterinary Probiotics in Broiler Chickens. Animals 2024, 14, 1927. https://doi.org/10.3390/ani14131927
Kerek Á, Román IL, Szabó Á, Papp M, Bányai K, Kardos G, Kaszab E, Bali K, Makrai L, Jerzsele Á. Comprehensive Metagenomic Analysis of Veterinary Probiotics in Broiler Chickens. Animals. 2024; 14(13):1927. https://doi.org/10.3390/ani14131927
Chicago/Turabian StyleKerek, Ádám, István László Román, Ábel Szabó, Márton Papp, Krisztián Bányai, Gábor Kardos, Eszter Kaszab, Krisztina Bali, László Makrai, and Ákos Jerzsele. 2024. "Comprehensive Metagenomic Analysis of Veterinary Probiotics in Broiler Chickens" Animals 14, no. 13: 1927. https://doi.org/10.3390/ani14131927






