Assessment of Encapsulated Probiotic Lactococcus lactis A12 Viability Using an In Vitro Digestion Model for Tilapia
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Microorganisms
2.3. Preparation of Culture Medium and Fermentation Conditions
2.4. Encapsulation of Probiotic Bacteria
2.4.1. Preparation of Feed Solution
2.4.2. Spray Drying of Probiotic Bacteria
2.4.3. Viability of Probiotic Spray-Dried Powder
2.5. In Vitro Assays
2.5.1. Preparation of Tilapia Enzymatic Extract
2.5.2. First Set of Assays: One Factor at a Time
2.5.3. Second Set of Assays: RSM Design
2.5.4. Model Validation
2.5.5. Antibacterial Activity (AA) of In Vitro Digestion of Probiotic-Supplemented Feed
2.6. Statistical Analysis
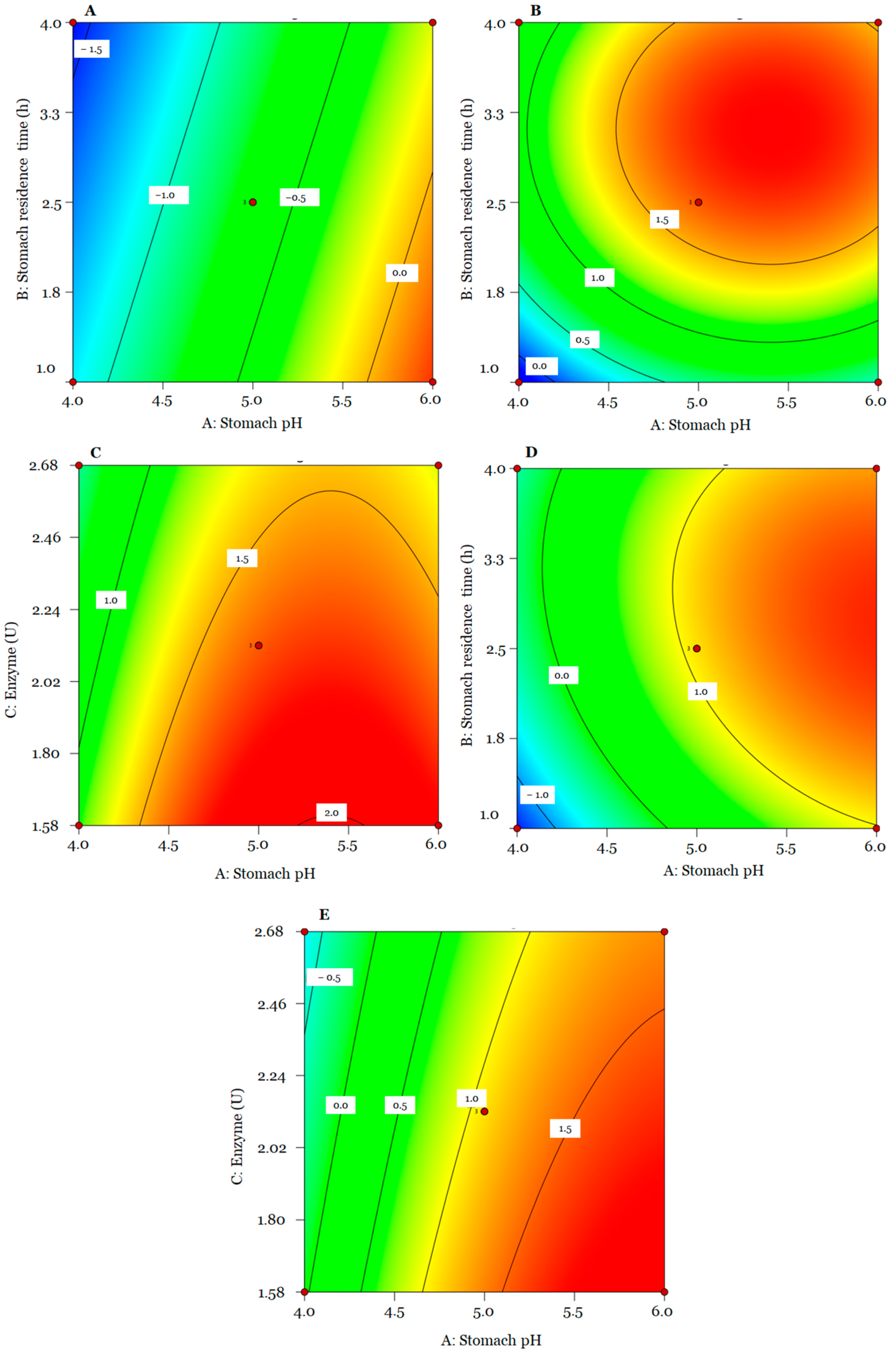
3. Results
3.1. OFAT Experiments
3.2. Box Behnken Design (BBD) of In Vitro Assays
3.3. Validation of the Model
3.4. Antibacterial Activity of In Vitro Digestion
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture 2024; FAO: Rome, Italy, 2024; ISBN 978-92-5-138763-4. [Google Scholar]
- Karvonen, A.; Räihä, V.; Klemme, I.; Ashrafi, R.; Hyvärinen, P.; Sundberg, L.-R. Quantity and Quality of Aquaculture Enrichments Influence Disease Epidemics and Provide Ecological Alternatives to Antibiotics. Antibiotics 2021, 10, 335. [Google Scholar] [CrossRef]
- Zarantoniello, M.; Bortoletti, M.; Olivotto, I.; Ratti, S.; Poltronieri, C.; Negrato, E.; Caberlotto, S.; Radaelli, G.; Bertotto, D. Salinity, Temperature and Ammonia Acute Stress Response in Sea Bream (Sparus aurata) Juveniles: A Multidisciplinary Study. Animals 2021, 11, 97. [Google Scholar] [CrossRef] [PubMed]
- del Valle, J.C.; Bonadero, M.C.; Fernández-Gimenez, A.V. Saccharomyces cerevisiae as Probiotic, Prebiotic, Synbiotic, Postbiotics and Parabiotics in Aquaculture: An Overview. Aquaculture 2023, 569, 739342. [Google Scholar] [CrossRef]
- Ibrahim, M.; Ahmad, F.; Yaqub, B.; Ramzan, A.; Imran, A.; Afzaal, M.; Mirza, S.A.; Mazhar, I.; Younus, M.; Akram, Q.; et al. Current Trends of Antimicrobials Used in Food Animals and Aquaculture. In Antibiotics and Antimicrobial Resistance Genes in the Environment; Elsevier: Amsterdam, The Netherlands, 2020; pp. 39–69. [Google Scholar] [CrossRef]
- World Health Organization. Food and Agriculture Organization of the United Nations Probiotics in Food Health and Nutritional Properties and Guidelines for Evaluation. In FAO Food and Nutrition Paper; World Health Organization and Food and Agriculture Organization of the United Nations: Rome, Italy, 2006; Volume 85, pp. 1–50. ISBN 92-5-105513-0. [Google Scholar]
- Alayande, K.A.; Aiyegoro, O.A.; Ateba, C.N. Probiotics in Animal Husbandry: Applicability and Associated Risk Factors. Sustainability 2020, 12, 1087. [Google Scholar] [CrossRef]
- Kuebutornye, F.K.A.; Abarike, E.D.; Sakyi, M.E.; Lu, Y.; Wang, Z. Modulation of Nutrient Utilization, Growth, and Immunity of Nile Tilapia, Oreochromis niloticus: The Role of Probiotics. Aquac. Int. 2020, 28, 277–291. [Google Scholar] [CrossRef]
- El-Saadony, M.T.; Alagawany, M.; Patra, A.K.; Kar, I.; Tiwari, R.; Dawood, M.A.O.; Dhama, K.; Abdel-Latif, H.M.R. The Functionality of Probiotics in Aquaculture: An Overview. Fish Shellfish Immunol. 2021, 117, 36–52. [Google Scholar] [CrossRef] [PubMed]
- Ringø, E.; Van Doan, H.; Lee, S.H.; Soltani, M.; Hoseinifar, S.H.; Harikrishnan, R.; Song, S.K. Probiotics, Lactic Acid Bacteria and Bacilli: Interesting Supplementation for Aquaculture. J. Appl. Microbiol. 2020, 129, 116–136. [Google Scholar] [CrossRef] [PubMed]
- Merrifield, D.L.; Carnevali, O. Probiotic Modulation of the Gut Microbiota of Fish. In Aquaculture Nutrition; John Wiley & Sons, Ltd.: Chichester, UK, 2014; pp. 185–222. [Google Scholar] [CrossRef]
- Kaktcham, P.M.; Tchamani Piame, L.; Sandjong Sileu, G.M.; Foko Kouam, E.M.; Temgoua, J.B.; Zambou Ngoufack, F.; de Lourdes Pérez-Chabela, M. Bacteriocinogenic Lactococcus lactis subsp. lactis 3MT Isolated from Freshwater Nile Tilapia: Isolation, Safety Traits, Bacteriocin Characterisation, and Application for Biopreservation in Fish Pâté. Arch. Microbiol. 2019, 201, 1249–1258. [Google Scholar] [CrossRef] [PubMed]
- Melo-Bolívar, J.F.; Pardo, R.; Junca, R.Y.; Sidjabat, H.; Cano-Lozano, H.E.; Yolanda, R.; Junca, H.; Sidjabat, H.E.; Cano-Lozano, J.A.; Villamil Díaz, L.M. Competitive Exclusion Bacterial Culture Derived from the Gut Microbiome of Nile Tilapia (Oreochromis niloticus) as a Resource to Efficiently Recover Probiotic Strains: Taxonomic, Genomic, and Functional Proof of Concept. Microorganisms 2022, 10, 1376. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Lu, M.; Chen, G.; Cao, J.; Gao, F.; Wang, M.; Liu, Z.; Zhang, D.; Zhu, H.; Yi, M. Effects of Dietary Lactobacillus rhamnosus JCM1136 and Lactococcus lactis subsp. lactis JCM5805 on the Growth, Intestinal Microbiota, Morphology, Immune Response and Disease Resistance of Juvenile Nile Tilapia, Oreochromis niloticus. Fish Shellfish Immunol. 2018, 76, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Cao, J.; Wang, M.; Lu, M.; Chen, G.; Gao, F.; Liu, Z.; Zhang, D.; Ke, X.; Yi, M. Effects of Lactococcus lactis subsp. lactis JCM5805 on Colonization Dynamics of Gut Microbiota and Regulation of Immunity in Early Ontogenetic Stages of Tilapia. Fish Shellfish Immunol. 2019, 86, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Yu, E.; Lu, M.; Xie, J. Effects of Probiotic Supplementation on Gut Microbiota as Well as Metabolite Profiles within Nile Tilapia, Oreochromis niloticus. Aquaculture 2020, 527, 735428. [Google Scholar] [CrossRef]
- Melo-Bolívar, J.F.; Ruiz Pardo, R.Y.; Quintanilla-Carvajal, M.X.; Díaz, L.E.; Alzate, J.F.; Junca, H.; Rodríguez Orjuela, J.A.; Villamil Diaz, L.M. Evaluation of Dietary Single Probiotic Isolates and Probiotic Multistrain Consortia in Growth Performance, Gut Histology, Gut Microbiota, Immune Regulation, and Infection Resistance of Nile Tilapia, Oreochromis niloticus, Shows Superior Monostrain Performance. Fish Shellfish Immunol. 2023, 140, 108928. [Google Scholar] [CrossRef] [PubMed]
- Jahangiri, L.; Esteban, M. Administration of Probiotics in the Water in Finfish Aquaculture Systems: A Review. Fishes 2018, 3, 33. [Google Scholar] [CrossRef]
- Kumaree, K.K.; Akbar, A.; Anal, A.K. Bioencapsulation and Application of Lactobacillus plantarum Isolated from Catfish Gut as an Antimicrobial Agent and Additive in Fish Feed Pellets. Ann. Microbiol. 2015, 65, 1439–1445. [Google Scholar] [CrossRef]
- Sribounoy, U.; Pirarat, N.; Solval, K.M.; Sathivel, S.; Chotiko, A. Development of Pelleted Feed Containing Probiotic Lactobacillus rhamnosus GG and Jerusalem Artichoke for Nile Tilapia and Its Biocompatibility Studies. 3 Biotech 2021, 11, 279. [Google Scholar] [CrossRef] [PubMed]
- Pinpimai, K.; Rodkhum, C.; Chansue, N.; Katagiri, T.; Maita, M.; Pirarat, N. The Study on the Candidate Probiotic Properties of Encapsulated Yeast, Saccharomyces cerevisiae JCM 7255, in Nile Tilapia (Oreochromis niloticus). Res. Vet. Sci. 2015, 102, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Pirarat, N.; Pinpimai, K.; Rodkhum, C.; Chansue, N.; Ooi, E.L.; Katagiri, T.; Maita, M. Viability and Morphological Evaluation of Alginate-Encapsulated Lactobacillus rhamnosus GG under Simulated Tilapia Gastrointestinal Conditions and Its Effect on Growth Performance, Intestinal Morphology and Protection against Streptococcus agalactiae. Anim. Feed. Sci. Technol. 2015, 207, 93–103. [Google Scholar] [CrossRef]
- Kahieshesfandiari, M.; Nami, Y.; Lornezhad, G.; Kiani, A.; Javanmard, A.; Jaymand, M.; Haghshenas, B. Herbal Hydrogel-Based Encapsulated Enterococcus faecium ABRIINW.N7 Improves the Resistance of Red Hybrid Tilapia against Streptococcus iniae. J. Appl. Microbiol. 2021, 131, 2516–2527. [Google Scholar] [CrossRef] [PubMed]
- Masoomi Dezfooli, S.; Gutierrez-Maddox, N.; Alfaro, A.; Seyfoddin, A. Encapsulation for Delivering Bioactives in Aquaculture. Rev. Aquac. 2019, 11, 631–660. [Google Scholar] [CrossRef]
- Mota de Carvalho, N.; Oliveira, D.L.; Saleh, M.A.D.; Pintado, M.E.; Madureira, A.R. Importance of Gastrointestinal in vitro Models for the Poultry Industry and Feed Formulations. Anim. Feed. Sci. Technol. 2021, 271, 114730. [Google Scholar] [CrossRef]
- Gilannejad, N.; Martínez-Rodríguez, G.; Yúfera, M.; Moyano, F.J. Modelling Digestive Hydrolysis of Nutrients in Fish Using Factorial Designs and Desirability Function. PLoS ONE 2018, 13, e0206556. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Antequera, F.P.; López-Ruiz, R.; Martos-Sitcha, J.A.; Mancera, J.M.; Moyano, F.J. Assessing Differences in the Bioaccessibility of Phenolics Present in Two Wine By-Products Using an In-Vitro Model of Fish Digestion. Front. Vet. Sci. 2023, 10, 1151045. [Google Scholar] [CrossRef]
- Aragón-Rojas, S.; Hernández-Álvarez, A.J.; Mainville, I.; Arcand, Y.; Quintanilla-Carvajal, M.X. Effect of the Carrier Material, Drying Technology and Dissolution Media on the Viability of: Lactobacillus fermentum K73 during Simulated Gastrointestinal Transit. Food Funct. 2020, 11, 2339–2348. [Google Scholar] [CrossRef] [PubMed]
- Naissinger da Silva, M.; Tagliapietra, B.L.; Flores, V.d.A.; Pereira dos Santos Richards, N.S. In Vitro Test to Evaluate Survival in the Gastrointestinal Tract of Commercial Probiotics. Curr. Res. Food Sci. 2021, 4, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Kazlauskaite, R.; Cheaib, B.; Heys, C.; Ijaz, U.Z.; Connelly, S.; Sloan, W.; Russel, J.; Rubio, L.; Sweetman, J.; Kitts, A.; et al. SalmoSim: The Development of a Three-Compartment In Vitro Simulator of the Atlantic Salmon GI Tract and Associated Microbial Communities. Microbiome 2021, 9, 179. [Google Scholar] [CrossRef] [PubMed]
- Rosolen, M.D.; Bordini, F.W.; de Oliveira, P.D.; Conceição, F.R.; Pohndorf, R.S.; Fiorentini, Â.M.; da Silva, W.P.; Pieniz, S. Symbiotic Microencapsulation of Lactococcus lactis subsp. lactis R7 Using Whey and Inulin by Spray Drying. LWT 2019, 115, 108411. [Google Scholar] [CrossRef]
- de Oliveira, C.G.; López-Olmeda, J.F.; Costa, L.S.; do Espirito Santo, A.H.; dos Santos, F.A.C.; Luz, R.K.; Ribeiro, P.A.P. Gastrointestinal Emptying and Daily Patterns of Activity of Proteinolytic Enzymes in Nile Tilapia (Oreochromis niloticus). Aquaculture 2022, 546, 737338. [Google Scholar] [CrossRef]
- Anukoolpra, T.; Srinuansom, K.; Rukdontri, T.; Nonkhukhet, S.; Petkam, R. Postprandial In Vitro Protease-Specific Activity of Nile Tilapia (Oreochromis niloticus L.) Digestive Organs. Pak. J. Nutr. 2019, 18, 125–133. [Google Scholar] [CrossRef]
- Uscanga, A.; Moyano, F.J.; Alvarez, C.A. Assessment of Enzymatic Efficiency on Protein Digestion in the Tilapia Oreochromis niloticus. Fish Physiol. Biochem. 2010, 36, 1079–1085. [Google Scholar] [CrossRef] [PubMed]
- Gilannejad, N.; Martínez-Rodríguez, G.; Yúfera, M.; Moyano, F.J. Estimating the Effect of Different Factors on the Digestive Bioaccessibility of Protein by the Senegalese Sole (Solea senegalensis); Combination of Response Surface Methodology and in vitro Assays. Aquaculture 2017, 477, 28–34. [Google Scholar] [CrossRef]
- Melo-Bolívar, J.F.; Ruiz Pardo, R.Y.; Hume, M.E.; Nisbet, D.J.; Rodríguez-Villamizar, F.; Alzate, J.F.; Junca, H.; Villamil Díaz, L.M. Establishment and Characterization of a Competitive Exclusion Bacterial Culture Derived from Nile tilapia (Oreochromis niloticus) Gut Microbiomes Showing Antibacterial Activity against Pathogenic Streptococcus agalactiae. PLoS ONE 2019, 14, e0215375. [Google Scholar] [CrossRef]
- Valle-Vargas, M.F.; Ruiz-Pardo, R.Y.; Villamil-Díaz, L.; Quintanilla-Carvajal, M.X. Production of a Potential Multistrain Probiotic in Co-Culture Conditions Using Agro-Industrial by-Products-Based Medium for Fish Nutrition. BMC Biotechnol. 2023, 23, 54. [Google Scholar] [CrossRef] [PubMed]
- Norizan, N.A.B.M.; Halim, M.; Tan, J.S.; Abbasiliasi, S.; Mat Sahri, M.; Othman, F.; Ariff, A.B. Enhancement of β-Mannanase Production by Bacillus subtilis ATCC11774 through Optimization of Medium Composition. Molecules 2020, 25, 3516. [Google Scholar] [CrossRef] [PubMed]
- Kunitz, M. Crystalline Soybean Trypsin Inhibitor. J. Gen. Physiol. 1947, 30, 291–310. [Google Scholar] [CrossRef] [PubMed]
- Walter, H.E. Proteinases: Methods with Haemoglobin, Casein and Azocoll as Substrates. In Methods of Enzymatic Analysis; Bergmeyer, H.U., Ed.; Weinheim Verlag Chemie: Deerfield Beach, FL, USA, 1984; Volume 5, pp. 270–277. [Google Scholar] [CrossRef]
- Shinke, R.; Mizuno, S.; Aoki, K.; Nishira, H. Effects of Metal Ions on Determination of β-Amylase Activity with 3,5-Dinitrosalicylic Acid. Agricutural Biol. Chem. 1978, 42, 2393–2394. [Google Scholar] [CrossRef]
- Moyano, F.J.; Saénz de Rodrigáñez, M.A.; Díaz, M.; Tacon, A.G.J. Application of In Vitro Digestibility Methods in Aquaculture: Constraints and Perspectives. Rev. Aquac. 2015, 7, 223–242. [Google Scholar] [CrossRef]
- Yasumaru, F.; Lemos, D. Species Specific in vitro Protein Digestion (PH-Stat) for Fish: Method Development and Application for Juvenile Rainbow Trout (Oncorhynchus mykiss), Cobia (Rachycentron canadum), and Nile Tilapia (Oreochromis niloticus). Aquaculture 2014, 426–427, 74–84. [Google Scholar] [CrossRef]
- Han, S.; Lu, Y.; Xie, J.; Fei, Y.; Zheng, G.; Wang, Z.; Liu, J.; Lv, L.; Ling, Z.; Berglund, B.; et al. Probiotic Gastrointestinal Transit and Colonization after Oral Administration: A Long Journey. Front. Cell. Infect. Microbiol. 2021, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Guan, N.; Liu, L. Microbial Response to Acid Stress: Mechanisms and Applications. Appl. Microbiol. Biotechnol. 2020, 104, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Fazilah, N.F.; Hamidon, N.H.; Ariff, A.B.; Khayat, M.E.; Wasoh, H.; Halim, M. Microencapsulation of Lactococcus lactis Gh1 with Gum Arabic and Synsepalum Dulcificum via Spray Drying for Potential Inclusion in Functional Yogurt. Molecules 2019, 24, 1422. [Google Scholar] [CrossRef] [PubMed]
- Bustos, A.Y.; Taranto, M.P.; Gerez, C.L.; Agriopoulou, S.; Smaoui, S.; Varzakas, T.; Enshasy, H.A. El Recent Advances in the Understanding of Stress Resistance Mechanisms in Probiotics: Relevance for the Design of Functional Food Systems. Probiotics Antimicrob. Proteins 2024. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.B.; Ki, K.S.; Khan, M.A.; Lee, W.S.; Lee, H.J.; Ahn, B.S.; Kim, H.S. Peptic and Tryptic Hydrolysis of Native and Heated Whey Protein to Reduce Its Antigenicity. J. Dairy Sci. 2007, 90, 4043–4050. [Google Scholar] [CrossRef] [PubMed]
- Galvão, C.M.A.; Silva, A.F.S.; Custódio, M.F.; Monti, R.; Giordano, R.D.L.C. Controlled Hydrolysis of Cheese Whey Proteins Using Trypsin and α-Chymotrypsin. Appl. Biochem. Biotechnol. 2001, 91–93, 761–776. [Google Scholar] [CrossRef] [PubMed]
- Hai, N. Van Research Findings from the Use of Probiotics in Tilapia Aquaculture: A Review. Fish Shellfish Immunol. 2015, 45, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Akhter, N.; Wu, B.; Memon, A.M.; Mohsin, M. Probiotics and Prebiotics Associated with Aquaculture: A Review. Fish Shellfish Immunol. 2015, 45, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.; Singh, R. Probiotics in Aquaculture: A Promising Emerging Alternative Approach. Symbiosis 2019, 77, 99–113. [Google Scholar] [CrossRef]
- Kiousi, D.E.; Efstathiou, C.; Tzampazlis, V.; Plessas, S.; Panopoulou, M.; Koffa, M.; Galanis, A. Genetic and Phenotypic Assessment of the Antimicrobial Activity of Three Potential Probiotic Lactobacilli against Human Enteropathogenic Bacteria. Front. Cell. Infect. Microbiol. 2023, 13, 1127256. [Google Scholar] [CrossRef] [PubMed]
- Nami, Y.; Kahieshesfandiari, M.; Lornezhad, G.; Kiani, A.; Elieh-Ali-Komi, D.; Jafari, M.; Jaymand, M.; Haghshenas, B. Administration of Microencapsulated Enterococcus faecium ABRIINW.N7 with Fructo-Oligosaccharides and Fenugreek on the Mortality of Tilapia Challenged with Streptococcus agalactiae. Front. Vet. Sci. 2022, 9, 938380. [Google Scholar] [CrossRef] [PubMed]
- Aragón-Rojas, S.; Quintanilla-Carvajal, M.X.; Hernández-Sánchez, H. Multifunctional Role of the Whey Culture Medium in the Spray-Drying Microencapsulation of Lactic Acid Bacteria. Food Technol. Biotechnol. 2018, 56, 381–397. [Google Scholar] [CrossRef] [PubMed]
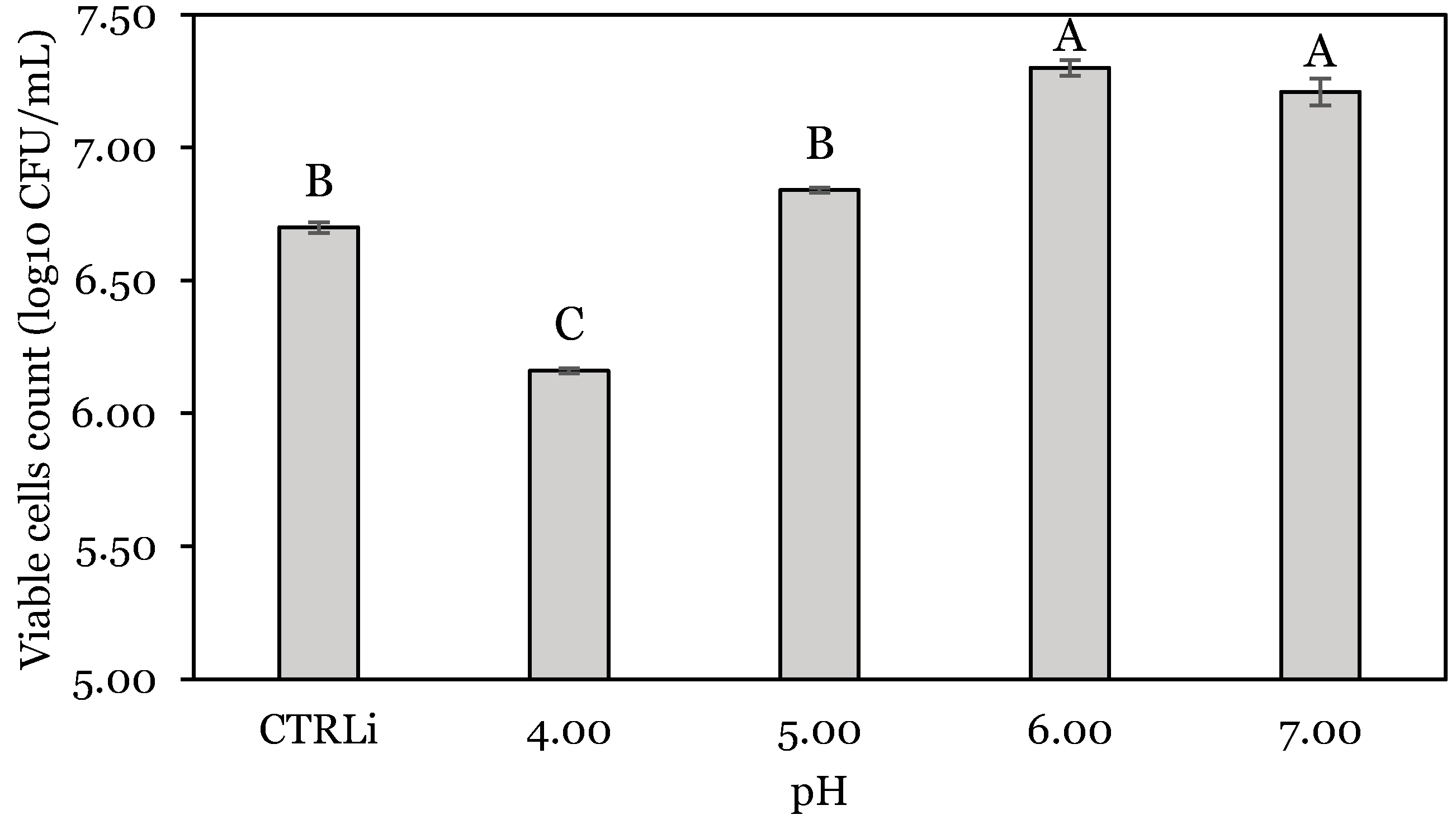
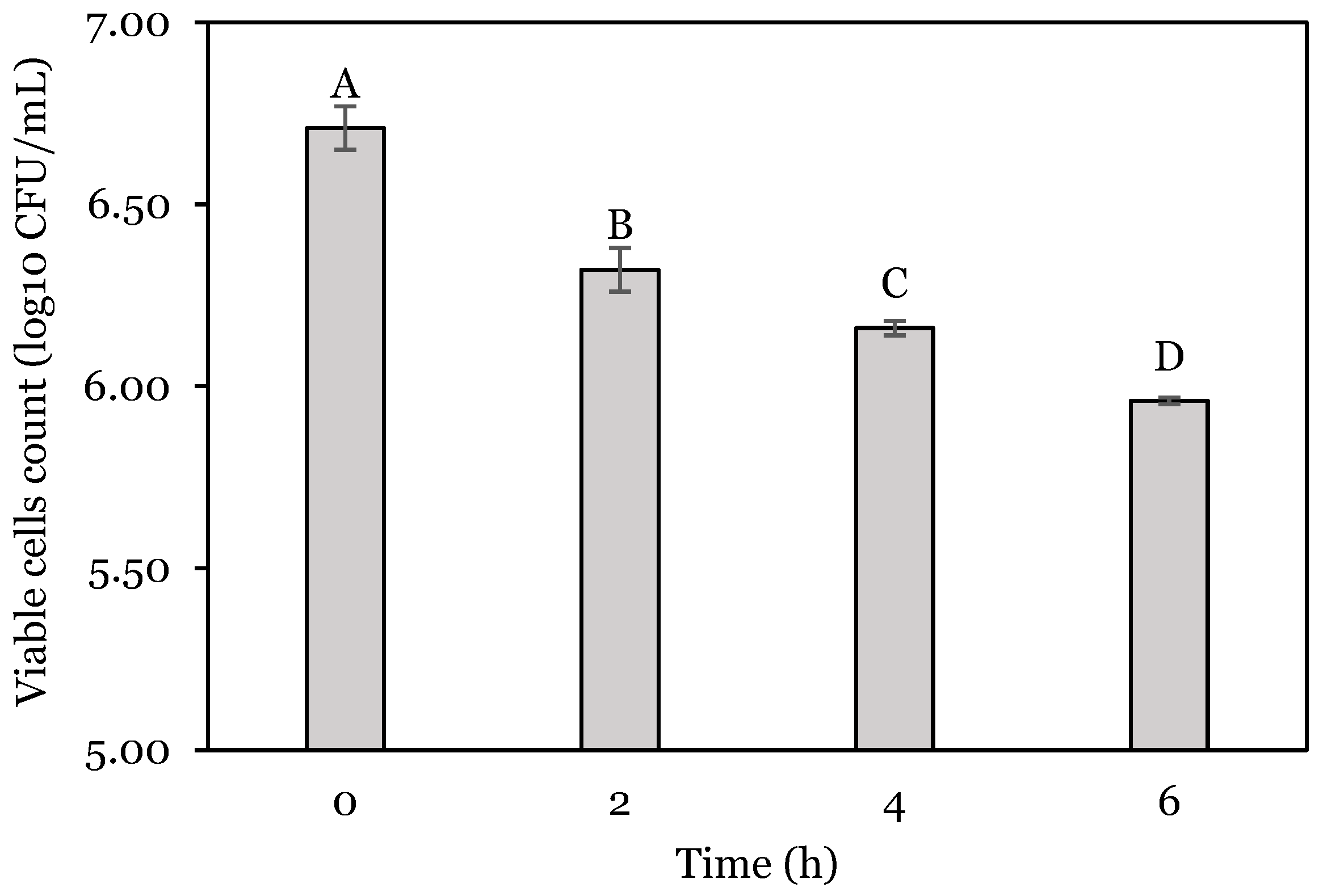
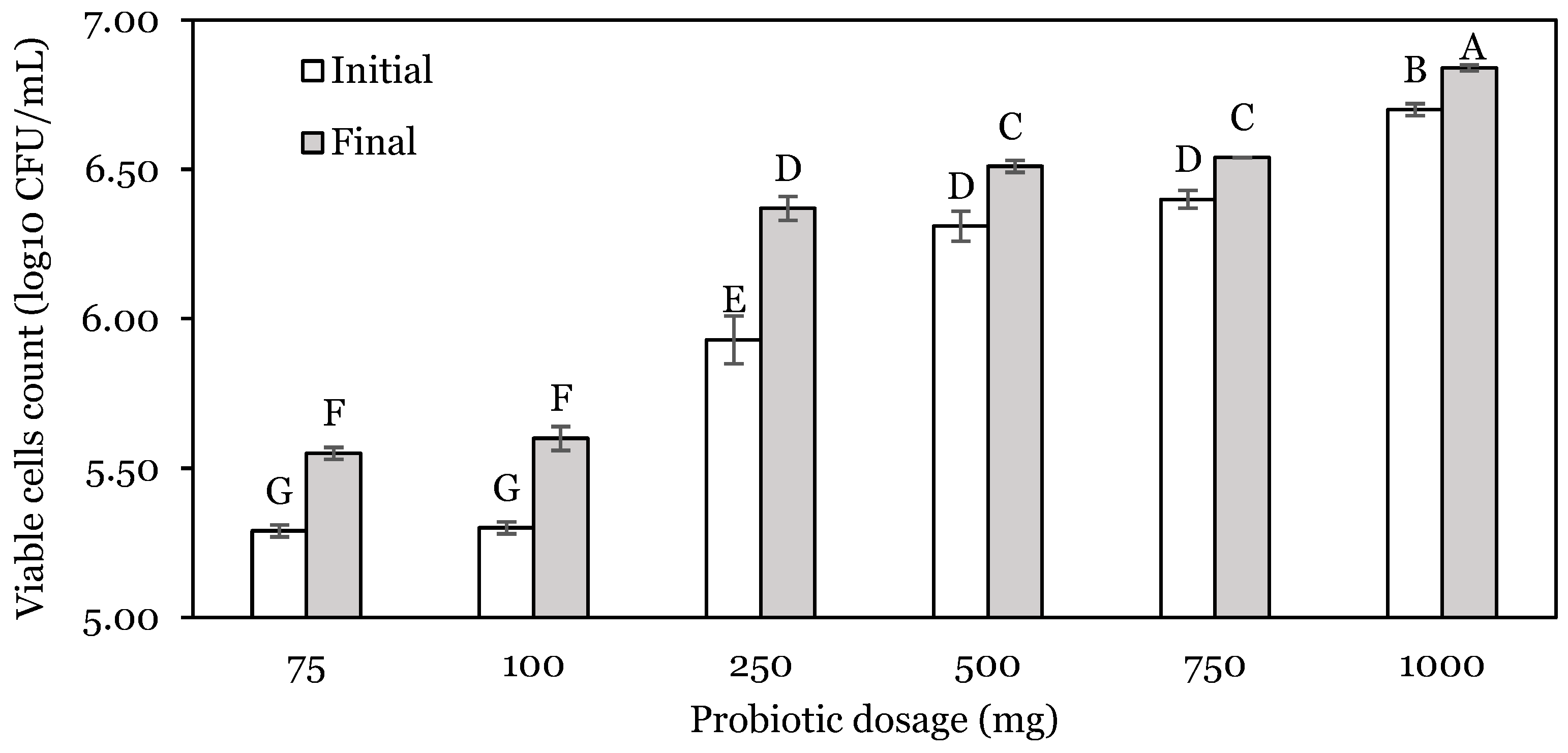
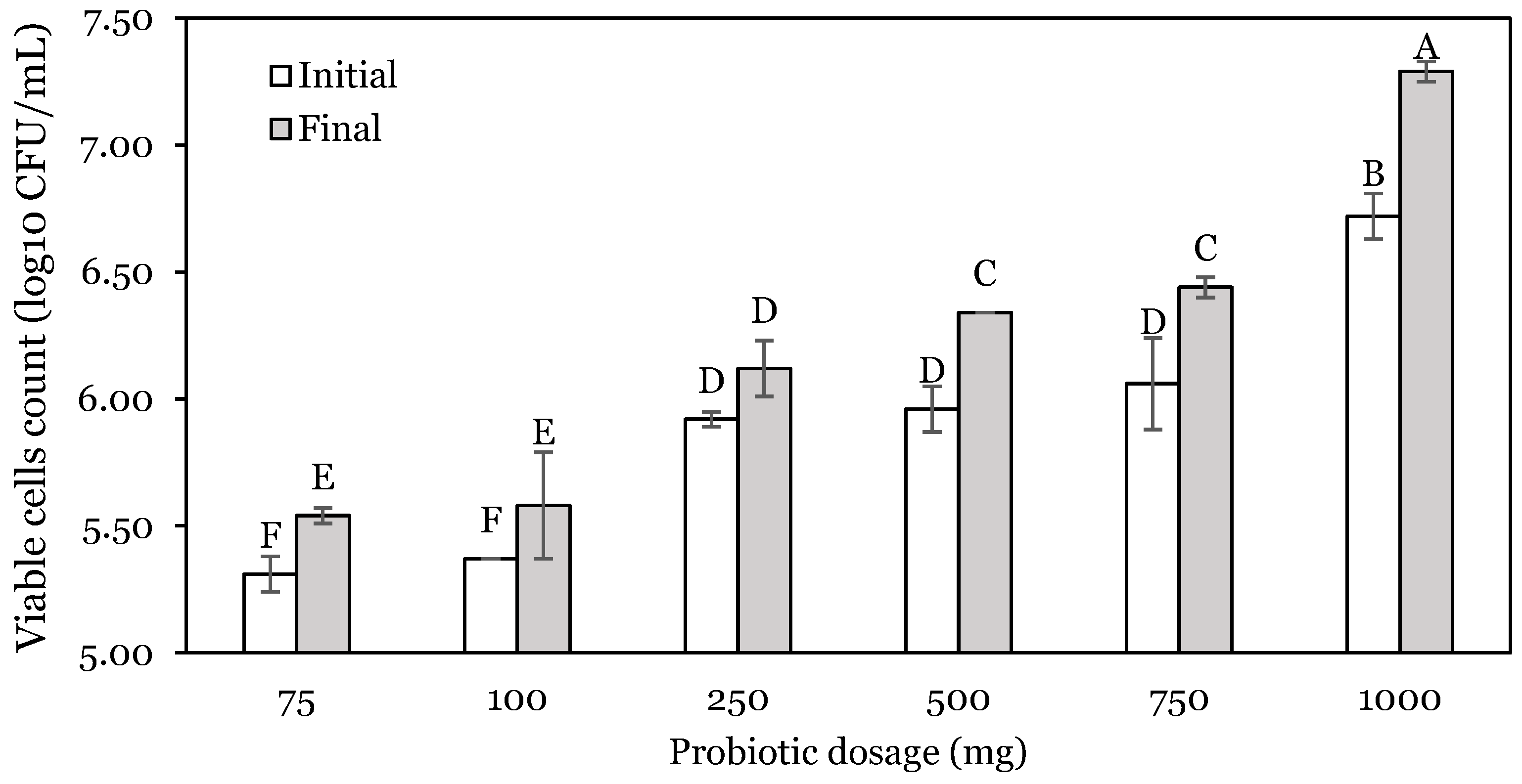
| Run | Stomach: pH | Stomach Time: h | Enzyme: U | Stomach Change (SC): log10 CFU | Intestine Change (IC): log10 CFU | Total Change (TC): log10 CFU |
|---|---|---|---|---|---|---|
| 1 | 5.00 | 1.0 | 2.68 | −0.21 | 0.14 | −0.06 |
| 2 | 6.00 | 2.5 | 1.58 | −0.11 | 1.76 | 1.82 |
| 3 | 4.00 | 1.0 | 2.13 | −1.51 | −0.14 | −1.52 |
| 4 | 5.00 | 4.0 | 2.68 | −1.20 | 1.30 | 0.52 |
| 5 | 4.00 | 4.0 | 2.13 | −1.60 | 1.06 | −0.47 |
| 6 | 6.00 | 4.0 | 2.13 | 0.15 | 1.27 | 1.36 |
| 7 | 5.00 | 2.5 | 2.13 | −0.72 | 1.74 | 1.01 |
| 8 | 5.00 | 1.0 | 1.58 | −0.30 | 0.82 | 0.54 |
| 9 | 5.00 | 2.5 | 2.13 | −0.75 | 1.75 | 1.11 |
| 10 | 5.00 | 4.0 | 1.58 | −0.75 | 1.68 | 1.26 |
| 11 | 5.00 | 2.5 | 2.13 | −0.78 | 1.84 | 1.05 |
| 12 | 6.00 | 2.5 | 2.68 | −0.19 | 1.48 | 1.45 |
| 13 | 4.00 | 2.5 | 2.68 | −1.07 | 0.19 | −0.69 |
| 14 | 6.00 | 1.0 | 2.13 | 0.35 | 0.60 | 1.01 |
| 15 | 4.00 | 2.5 | 1.58 | −1.17 | 1.07 | 0.14 |
| p Value | |||
|---|---|---|---|
| Term | Stomach Change | Intestine Change | Total Change |
| Model | <0.0001 | <0.0001 | <0.0001 |
| A: Stomach pH | <0.0001 | 0.0012 | <0.0001 |
| B: Stomach time | 0.0289 | 0.0002 | <0.0001 |
| C: Enzyme | 0.6314 | 0.0065 | <0.0001 |
| AB | - | - | 0.0179 |
| A2 | - | 0.0034 | 0.0001 |
| B2 | - | 0.0006 | <0.0001 |
| Lack of fit | 0.0123 | 0.0466 | 0.1307 |
| R2 | 0.8665 | 0.9251 | 0.9907 |
| R2 adjusted | 0.8301 | 0.8836 | 0.9838 |
| R2 predicted | 0.7198 | 0.7705 | 0.9584 |
| Adeq precision | 14.4673 | 14.8264 | 42.4524 |
| Factors | Predicted | Observed | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Run | pH | Time | Enzyme | SC | IC | TC | SC | IC | TC |
| 1 | 4.00 | 4.0 h | 2.68 U | −1.61 | −0.46 | −0.71 | −1.55 (3.72%) | 0.50 (8.69%) | −0.76 (7.04%) |
| 2 | 5.50 | 4.0 h | 2.68 U | −0.57 | 1.35 | 0.88 | −0.54 (5.26%) | 1.44 (6.66%) | 0.89 (1.13%) |
| Final Cell Count (log10 CFU/mL) | AA (mm) | |
|---|---|---|
| N-ENCP_pH 4.00 | 7.40 ± 0.08 b | 5.16 ± 0.90 b |
| N-ENCP_pH 5.50 | 7.49 ± 0.23 b | 4.99 ± 0.28 b |
| ENCP_pH 4.00 | 7.91 ± 0.03 a | 5.34 ± 0.92 b |
| ENCP_pH 5.50 | 7.92 ± 0.01 a | 5.95 ± 0.94 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valle Vargas, M.F.; Quintanilla-Carvajal, M.X.; Villamil-Diaz, L.; Ruiz Pardo, R.Y.; Moyano, F.J. Assessment of Encapsulated Probiotic Lactococcus lactis A12 Viability Using an In Vitro Digestion Model for Tilapia. Animals 2024, 14, 1981. https://doi.org/10.3390/ani14131981
Valle Vargas MF, Quintanilla-Carvajal MX, Villamil-Diaz L, Ruiz Pardo RY, Moyano FJ. Assessment of Encapsulated Probiotic Lactococcus lactis A12 Viability Using an In Vitro Digestion Model for Tilapia. Animals. 2024; 14(13):1981. https://doi.org/10.3390/ani14131981
Chicago/Turabian StyleValle Vargas, Marcelo Fernando, María Ximena Quintanilla-Carvajal, Luisa Villamil-Diaz, Ruth Yolanda Ruiz Pardo, and Francisco Javier Moyano. 2024. "Assessment of Encapsulated Probiotic Lactococcus lactis A12 Viability Using an In Vitro Digestion Model for Tilapia" Animals 14, no. 13: 1981. https://doi.org/10.3390/ani14131981






