Nuclear Receptor Subfamily 4 Group A Member 1 (NR4A1) Promotes the Adipogenesis of Intramuscular Preadipocytes through PI3K/AKT Pathway in Goats
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Cell Culture
2.2. Induction of Goat Intramuscular Preadipocyte
2.3. Construction of NR4A1 Overexpression Vector
2.4. Synthesis of siRNA
2.5. Cell Transfection
2.6. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qPCR)
2.7. Oil Red O Staining
2.8. Total RNA Extraction and RNA-Seq
2.9. Differentially Expressed Genes and KEGG Analysis
2.10. Data Analysis
3. Results
3.1. The Expression Patterns of NR4A1 Gene during Differentiation of Goat Intramuscular Adipocytes
3.2. NR4A1 Overexpression Enhances Intramuscular Preadipocyte Differentiation
3.3. NR4A1 Knockdown Inhibits Goat Intramuscular Adipocyte Differentiation
3.4. KEGG Enrichment Analysis
3.5. RNA-Seq Verification Result
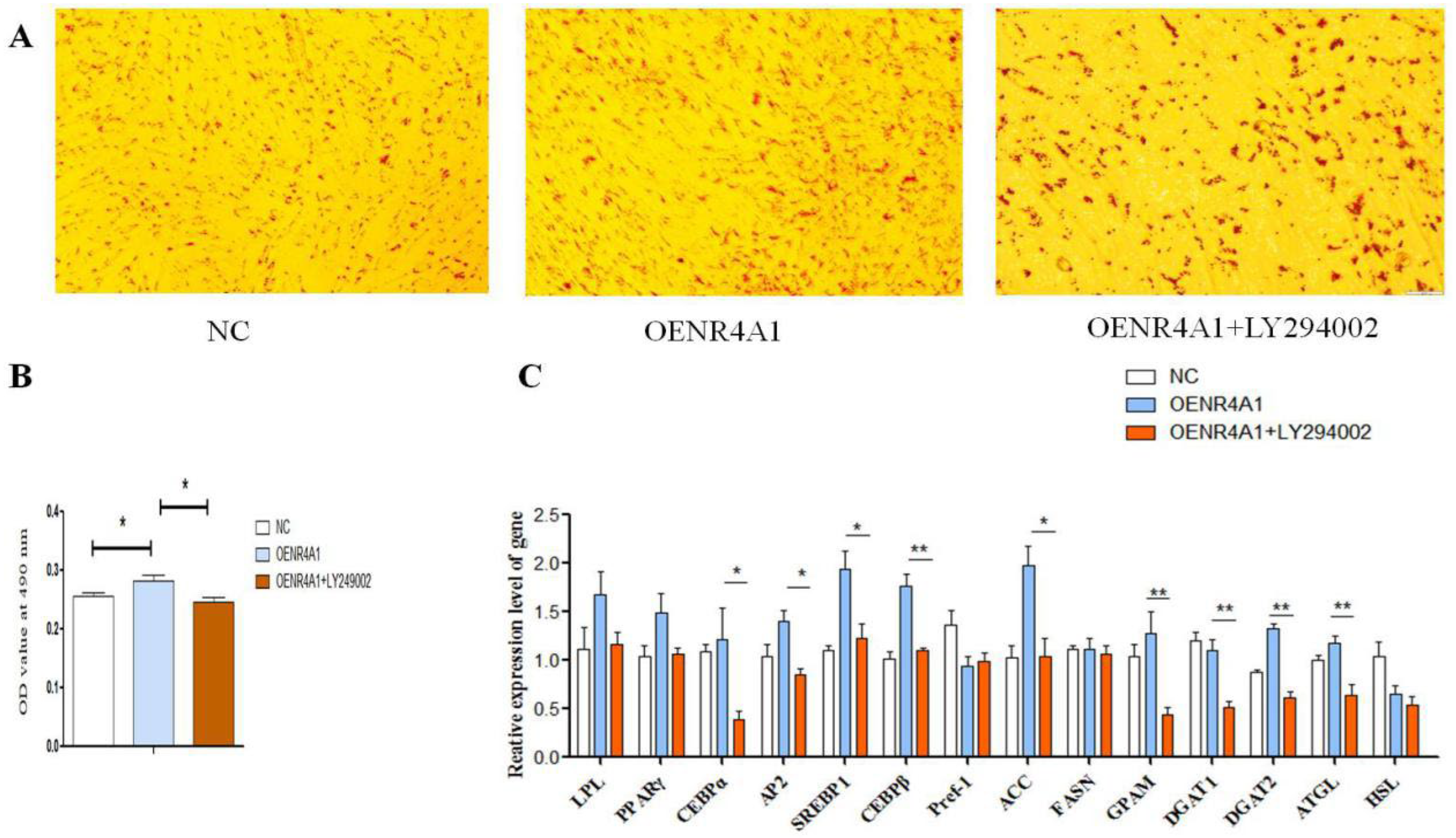
3.6. LY294002 Inhibits Overexpression of NR4A1 and Promotes Differentiation of Goat Intramuscular Adipocytes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pearen, M.A.; Muscat, G.E. Orphan nuclear receptors and the regulation of nutrient metabolism: Understanding obesity. Physiology 2012, 27, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Safe, S.; Jin, U.H.; Morpurgo, B.; Abudayyeh, A.; Singh, M.; Tjalkens, R.B. Nuclear receptor 4A (NR4A) family–orphans no more. J. Steroid Biochem. Mol. Biol. 2016, 157, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Burris, T.P.; Busby, S.A.; Griffin, P.R. Targeting orphan nuclear receptors for treatment of metabolic diseases and autoimmunity. Chem. Biol. 2012, 19, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yu, J. NR4A1 Promotes Cerebral Ischemia Reperfusion Injury by Repressing Mfn2-Mediated Mitophagy and Inactivating the MAPK-ERK-CREB Signaling Pathway. Neurochem. Res. 2018, 43, 1963–1977. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Federation, A.J.; Kim, S.; O’Keefe, J.P.; Lun, M.; Xiang, D.; Brown, J.D.; Steinhauser, M.L. Targeting nuclear receptor NR4A1-dependent adipocyte progenitor quiescence promotes metabolic adaptation to obesity. J. Clin. Investig. 2018, 128, 4898–4911. [Google Scholar] [CrossRef] [PubMed]
- Chao, L.C.; Wroblewski, K.; Zhang, Z.; Pei, L.; Vergnes, L.; Ilkayeva, O.R.; Ding, S.Y.; Reue, K.; Watt, M.J.; Newgard, C.B.; et al. Insulin resistance and altered systemic glucose metabolism in mice lacking Nur77. Diabetes 2009, 58, 2788–2796. [Google Scholar] [CrossRef] [PubMed]
- Chao, L.C.; Bensinger, S.J.; Villanueva, C.J.; Wroblewski, K.; Tontonoz, P. Inhibition of adipocyte differentiation by Nur77, Nurr1, and Nor1. Mol. Endocrinol. 2008, 22, 2596–2608. [Google Scholar] [CrossRef] [PubMed]
- Ming, Y.; Yin, Y.; Sun, Z. Interaction of Nuclear Receptor Subfamily 4 Group A Member 1 (Nr4a1) and Liver Linase B1 (LKB1) Mitigates Type 2 Diabetes Mellitus by Activating Monophosphate-Activated Protein Kinase (AMPK)/Sirtuin 1 (SIRT1) Axis and Inhibiting Nuclear Factor-kappa B (NF-kappaB) Activation. Med. Sci. Monit. 2020, 26, e920278. [Google Scholar] [CrossRef]
- Jung, Y.S.; Lee, H.S.; Cho, H.R.; Kim, K.J.; Kim, J.H.; Safe, S.; Lee, S.O. Dual targeting of Nur77 and AMPKalpha by isoalantolactone inhibits adipogenesis in vitro and decreases body fat mass in vivo. Int. J. Obes. 2019, 43, 952–962. [Google Scholar] [CrossRef]
- Shan, T.; Xiong, Y.; Zhang, P.; Li, Z.; Jiang, Q.; Bi, P.; Yue, F.; Yang, G.; Wang, Y.; Liu, X.; et al. Lkb1 controls brown adipose tissue growth and thermogenesis by regulating the intracellular localization of CRTC3. Nat. Commun. 2016, 7, 12205. [Google Scholar] [CrossRef]
- Chen, D.; Lin, Y.; Zhao, N.; Wang, Y.; Li, Y. Hoxa5 Inhibits the Proliferation and Induces Adipogenic Differentiation of Subcutaneous Preadipocytes in Goats. Animals 2022, 12, 1859. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Veum, V.L.; Dankel, S.N.; Gjerde, J.; Nielsen, H.J.; Solsvik, M.H.; Haugen, C.; Christensen, B.J.; Hoang, T.; Fadnes, D.J.; Busch, C.; et al. The nuclear receptors NUR77, NURR1 and NOR1 in obesity and during fat loss. Int. J. Obes. 2012, 36, 1195–1202. [Google Scholar] [CrossRef]
- Rosen, E.D.; Spiegelman, B.M. Molecular regulation of adipogenesis. Annu. Rev. Cell Dev. Biol. 2000, 16, 145–171. [Google Scholar] [CrossRef]
- Rosen, E.D.; MacDougald, O.A. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 2006, 7, 885–896. [Google Scholar] [CrossRef]
- Satoh, A.; Stein, L.; Imai, S. The role of mammalian sirtuins in the regulation of metabolism, aging, and longevity. Handb. Exp. Pharmacol. 2011, 206, 125–162. [Google Scholar] [CrossRef]
- Tang, Q.Q.; Otto, T.C.; Lane, M.D. CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 850–855. [Google Scholar] [CrossRef] [PubMed]
- Tontonoz, P.; Hu, E.; Spiegelman, B.M. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 1994, 79, 1147–1156. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, J.; Luo, S.; Zhan, Y.; Lu, Q. The roles of PPARgamma and its agonists in autoimmune diseases: A comprehensive review. J. Autoimmun. 2020, 113, 102510. [Google Scholar] [CrossRef] [PubMed]
- Ohlsson, E.; Hasemann, M.S.; Willer, A.; Lauridsen, F.K.; Rapin, N.; Jendholm, J.; Porse, B.T. Initiation of MLL-rearranged AML is dependent on C/EBPalpha. J. Exp. Med. 2014, 211, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zan, L.S.; Wang, H.B.; Gong, C.; Fu, C.Z. Cloning, expression analysis and sequence prediction of the CCAAT/enhancer-binding protein alpha gene of Qinchuan cattle. Genet. Mol. Res. 2012, 11, 1651–1661. [Google Scholar] [CrossRef] [PubMed]
- Yuan, E.; Duan, X.; Xiang, L.; Ren, J.; Lai, X.; Li, Q.; Sun, L.; Sun, S. Aged Oolong Tea Reduces High-Fat Diet-Induced Fat Accumulation and Dyslipidemia by Regulating the AMPK/ACC Signaling Pathway. Nutrients 2018, 10, 187. [Google Scholar] [CrossRef] [PubMed]
- Jeon, T.I.; Osborne, T.F. SREBPs: Metabolic integrators in physiology and metabolism. Trends Endocrinol. Metab. 2012, 23, 65–72. [Google Scholar] [CrossRef]
- Wang, Y.; Kim, K.A.; Kim, J.H.; Sul, H.S. Pref-1, a preadipocyte secreted factor that inhibits adipogenesis. J. Nutr. 2006, 136, 2953–2956. [Google Scholar] [CrossRef] [PubMed]
- Mead, J.R.; Irvine, S.A.; Ramji, D.P. Lipoprotein lipase: Structure, function, regulation, and role in disease. J. Mol. Med. 2002, 80, 753–769. [Google Scholar] [CrossRef]
- Chitraju, C.; Walther, T.C.; Farese, R.V., Jr. The triglyceride synthesis enzymes DGAT1 and DGAT2 have distinct and overlapping functions in adipocytes. J. Lipid Res. 2019, 60, 1112–1120. [Google Scholar] [CrossRef]
- Shimizu, Y.; Satoh, S.; Yano, H.; Minokoshi, Y.; Cushman, S.W.; Shimazu, T. Effects of noradrenaline on the cell-surface glucose transporters in cultured brown adipocytes: Novel mechanism for selective activation of GLUT1 glucose transporters. Biochem. J. 1998, 330 Pt 1, 397–403. [Google Scholar] [CrossRef]
- Hu, B.; Jin, C.; Zeng, X.; Resch, J.M.; Jedrychowski, M.P.; Yang, Z.; Desai, B.N.; Banks, A.S.; Lowell, B.B.; Mathis, D.; et al. gammadelta T cells and adipocyte IL-17RC control fat innervation and thermogenesis. Nature 2020, 578, 610–614. [Google Scholar] [CrossRef]
- Smith, U.; Kahn, B.B. Adipose tissue regulates insulin sensitivity: Role of adipogenesis, de novo lipogenesis and novel lipids. J. Intern. Med. 2016, 280, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Ardestani, A.; Maedler, K. Loss of TAZ Boosts PPARgamma to Cope with Insulin Resistance. Cell Metab. 2020, 31, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wan, X.; Pilch, P.F.; Ellisen, L.W.; Fried, S.K.; Liu, L. An AMPK-dependent, non-canonical p53 pathway plays a key role in adipocyte metabolic reprogramming. Elife 2020, 9, e63665. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Yuan, W.; Peng, X.; Wang, M.; Xiao, J.; Wu, C.; Luo, L. PPAR gamma/Nnat/NF-kappaB Axis Involved in Promoting Effects of Adiponectin on Preadipocyte Differentiation. Mediat. Inflamm. 2019, 2019, 5618023. [Google Scholar] [CrossRef] [PubMed]
- Bluthgen, N.; Legewie, S. Systems analysis of MAPK signal transduction. Essays Biochem. 2008, 45, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Gan, W.; Dai, X.; Dai, X.; Xie, J.; Yin, S.; Zhu, J.; Wang, C.; Liu, Y.; Guo, J.; Wang, M.; et al. LATS suppresses mTORC1 activity to directly coordinate Hippo and mTORC1 pathways in growth control. Nat. Cell Biol. 2020, 22, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Bai, X.Q.; Liao, L.; Zhou, M.; Peng, J.; Xiang, Q.; Ren, Z.; Wen, H.Y.; Jiang, Z.S.; Tang, Z.H.; et al. Hydrogen sulfide inhibits PCSK9 expression through the PI3K/Akt-SREBP-2 signaling pathway to influence lipid metabolism in HepG2 cells. Int. J. Mol. Med. 2019, 43, 2055–2063. [Google Scholar] [CrossRef] [PubMed]
- Song, B.Q.; Chi, Y.; Li, X.; Du, W.J.; Han, Z.B.; Tian, J.J.; Li, J.J.; Chen, F.; Wu, H.H.; Han, L.X.; et al. Inhibition of Notch Signaling Promotes the Adipogenic Differentiation of Mesenchymal Stem Cells Through Autophagy Activation and PTEN-PI3K/AKT/mTOR Pathway. Cell Physiol. Biochem. 2015, 36, 1991–2002. [Google Scholar] [CrossRef] [PubMed]
- Choe, W.K.; Kang, B.T.; Kim, S.O. Water-extracted plum (Prunus salicina L. cv. Soldam) attenuates adipogenesis in murine 3T3-L1 adipocyte cells through the PI3K/Akt signaling pathway. Exp. Ther. Med. 2018, 15, 1608–1615. [Google Scholar] [CrossRef]
- Ohashi, E.; Kohno, K.; Arai, N.; Harashima, A.; Ariyasu, T.; Ushio, S. Adenosine N1-Oxide Exerts Anti-inflammatory Effects through the PI3K/Akt/GSK-3beta Signaling Pathway and Promotes Osteogenic and Adipocyte Differentiation. Biol. Pharm. Bull. 2019, 42, 968–976. [Google Scholar] [CrossRef]
- Wang, J.; Hu, X.; Ai, W.; Zhang, F.; Yang, K.; Wang, L.; Zhu, X.; Gao, P.; Shu, G.; Jiang, Q.; et al. Phytol increases adipocyte number and glucose tolerance through activation of PI3K/Akt signaling pathway in mice fed high-fat and high-fructose diet. Biochem. Biophys. Res. Commun. 2017, 489, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Ke, C.; Cai, Z.; Wu, H.; Ye, Y.; Liang, X.; Yu, L.; Jiang, S.; Shen, J.; Wang, L.; et al. LNK deficiency decreases obesity-induced insulin resistance by regulating GLUT4 through the PI3K-Akt-AS160 pathway in adipose tissue. Aging 2020, 12, 17150–17166. [Google Scholar] [CrossRef] [PubMed]

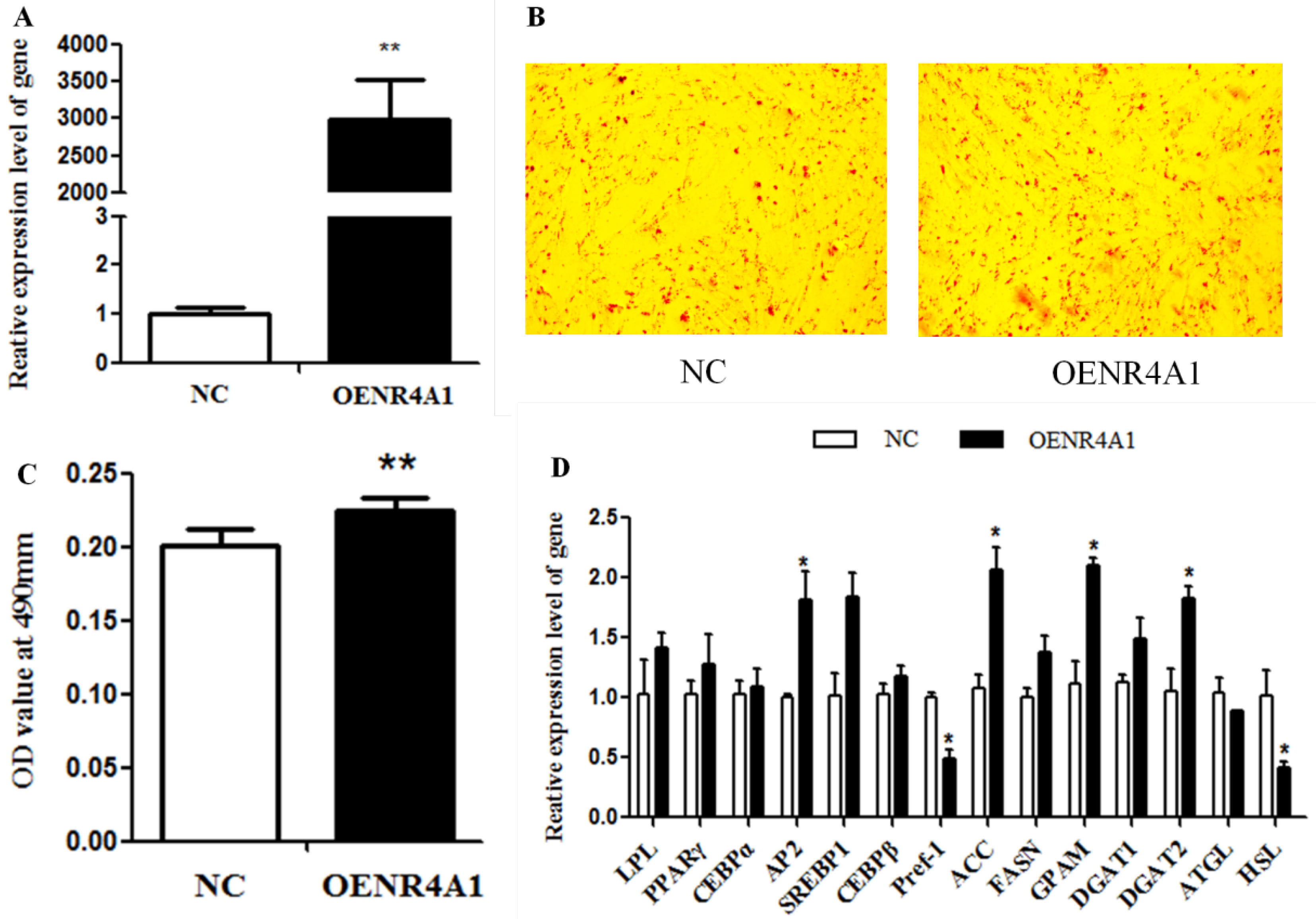

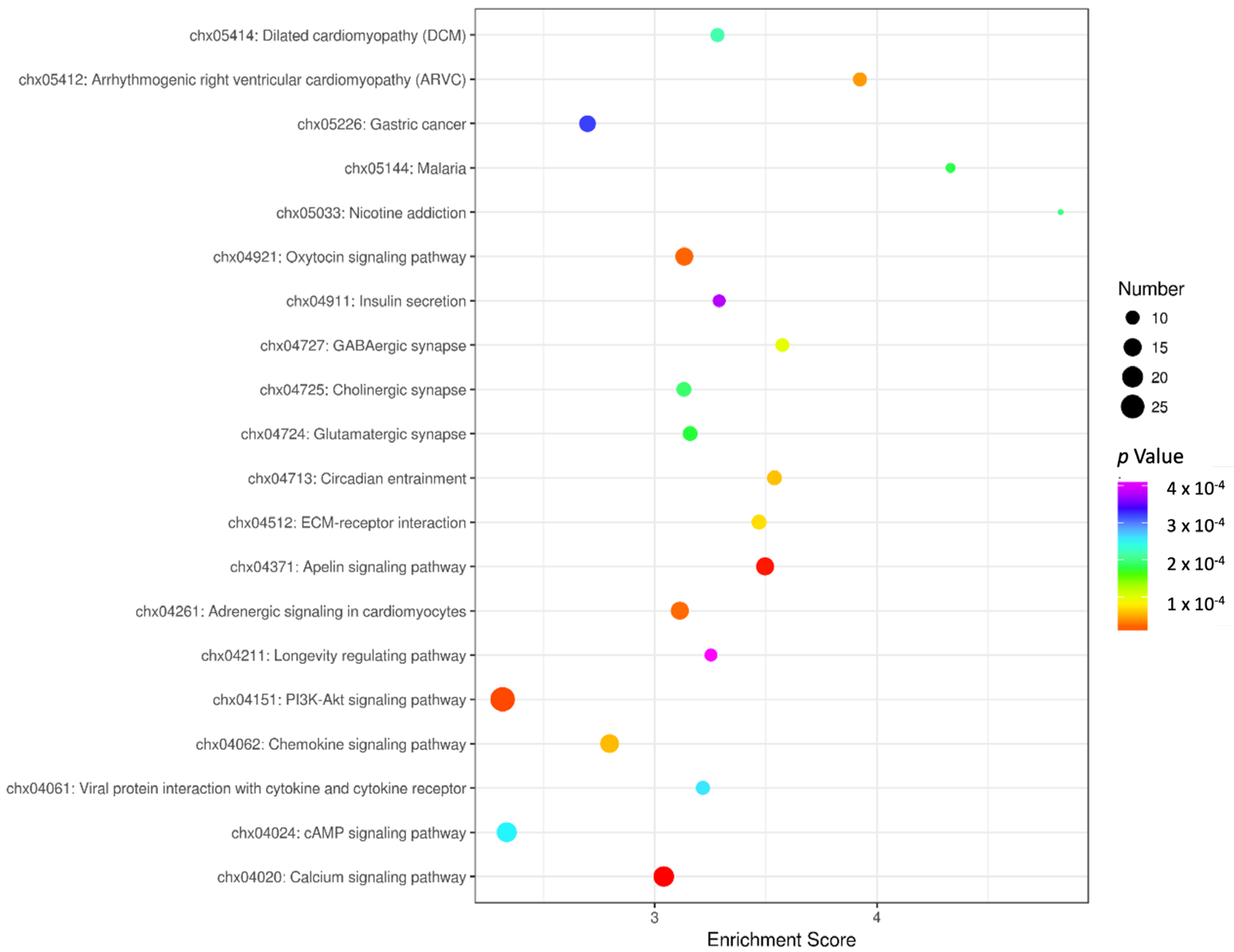
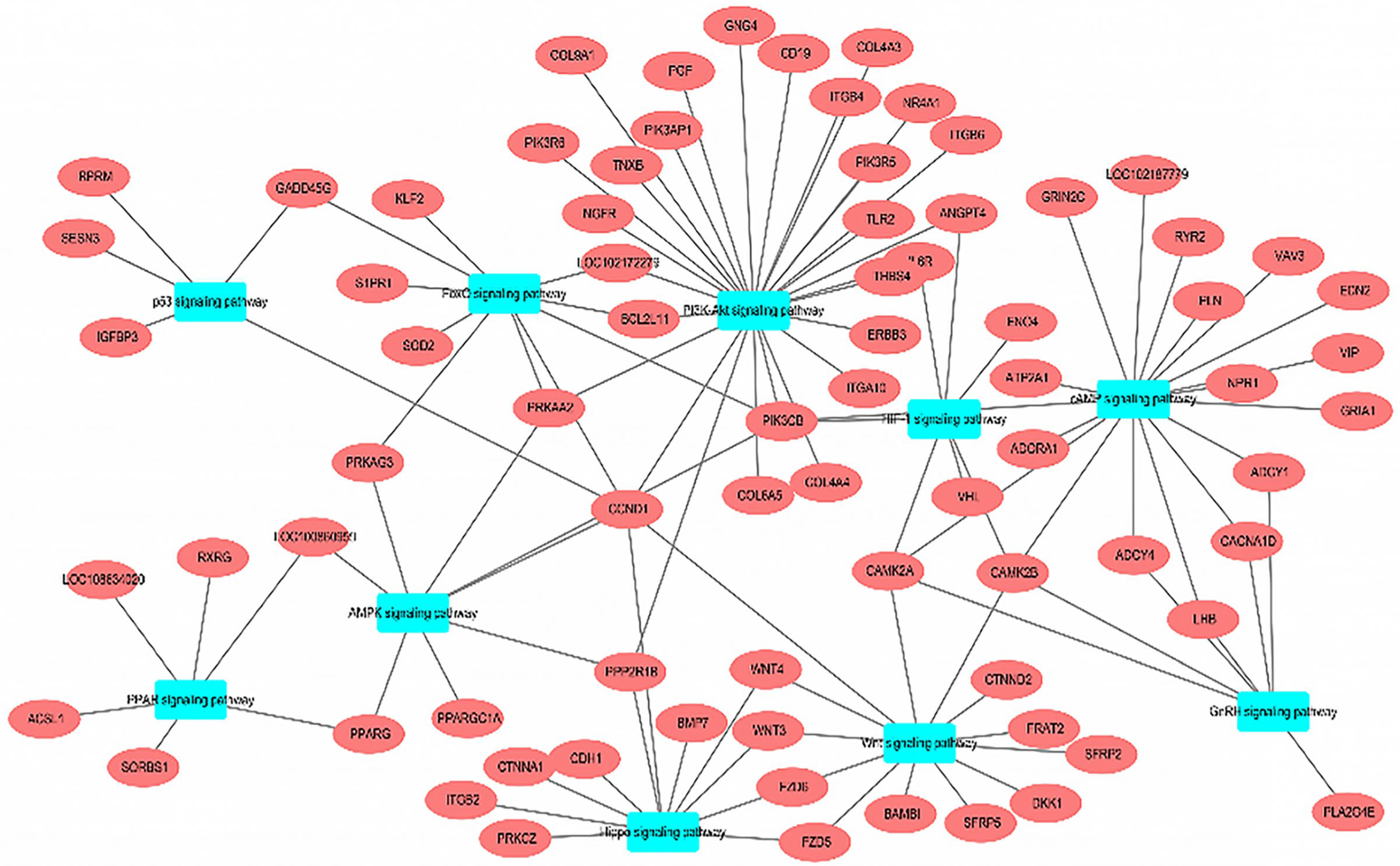
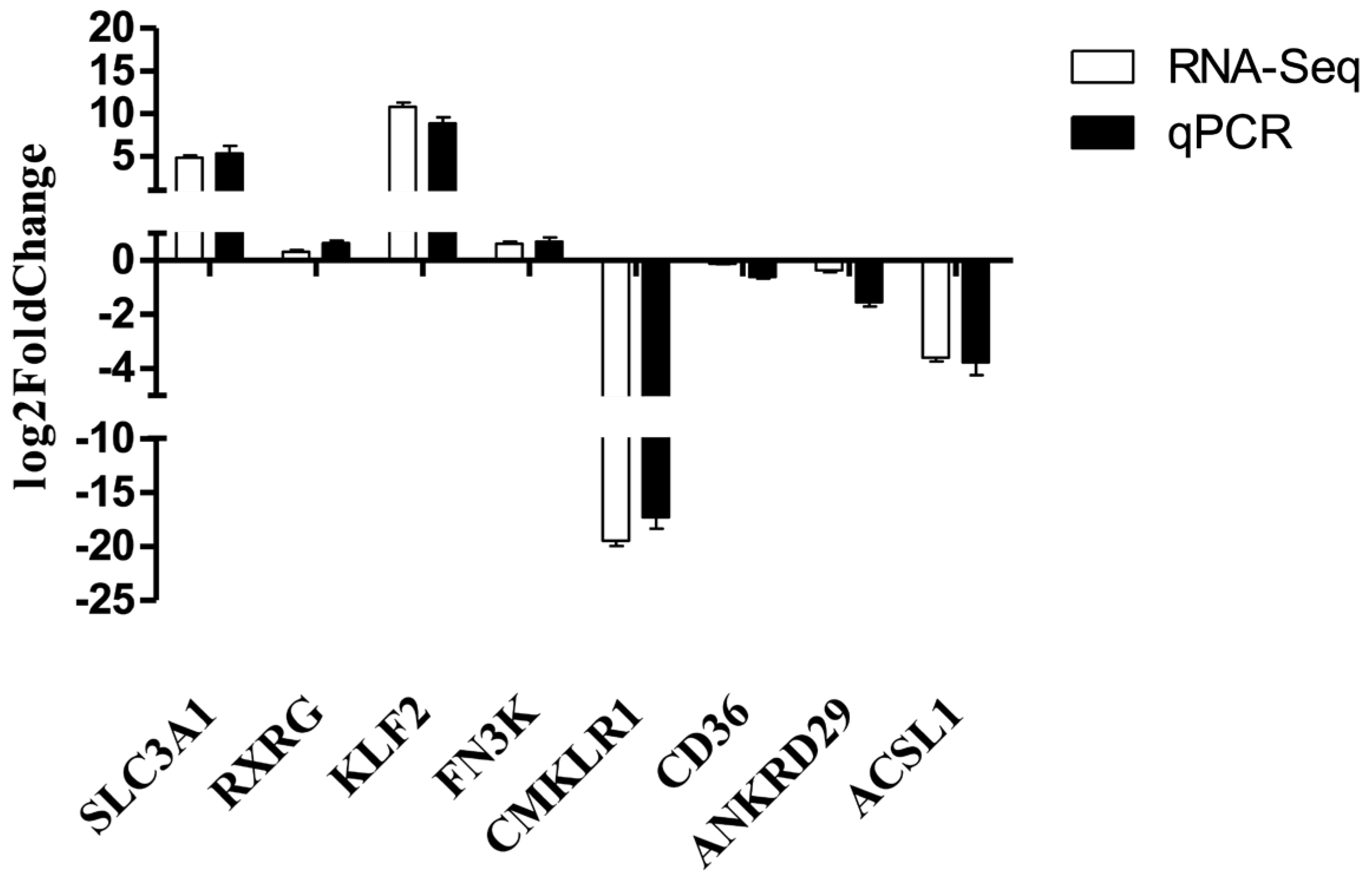
| Gene | Primer Sequence (5′~3′) | Tm/°C | Product Length/bp | GenBank Accession Number |
|---|---|---|---|---|
| LPL | S: TCCTGGAGTGACGGAATCTGT | 60 | 174 | NM_001285607.1 |
| A: GACAGCCAGTCCACCACGAT | ||||
| C/EBPα | S: CCGTGGACAAGAACAGCAAC | 58 | 142 | XM_018062278.1 |
| A: AGGCGGTCATTGTCACTGGT | ||||
| PPARγ | S: AAGCGTCAGGGTTCCACTATG | 60 | 197 | NM_001285658.1 |
| A: GAACCTGATGGCGTTATGAGAC | ||||
| SREBP1 | S: AAGTGGTGGGCCTCTCTGA | 58 | 127 | NM_001285755.1 |
| A: GCAGGGGTTTCTCGGACT | ||||
| Pref-1 | S: CCGGCTTCATGGATAAGACCT | 65 | 178 | KP686197.1 |
| A: GCCTCGCACTTGTTGAGGAA | ||||
| C/EBPβ | S: CAAGAAGACGGTGGACAAGC | 65 | 204 | XM_018058020.1 |
| A: AACAAGTTCCGCAGGGTG | ||||
| AP2 | S: TGAAGTCACTCCAGATGACAGG | 58 | 143 | NM_001285623.1 |
| A: TGACACATTCCAGCACCAGC | ||||
| ACC | S: GGAGACAAACAGGGACCATT | 60 | 146 | XM_018064169.1 |
| A: ATCAGGGACTGCCGAAAC | ||||
| FASN | S: TGTGCAACTGTGCCCTAG | 57 | 111 | NM_001285629.1 |
| A: GTCCTCTGAGCAGCGTGT | ||||
| ATGL | S: GGTGCCAATATCATCGAGGT | 64 | 133 | NM_001285739.1 |
| A: CACACCCGTGGCAGTCAG | ||||
| HSL | S: AGGGTCATTGCCGACTTCC | 60 | 161 | XM_018062484.1 |
| A: GTCTCGTTGCGTTTGTAGTGC | ||||
| GPAM | S: GCAGGTTTATCCAGTATGGCATT | 60 | 63 | XM_013975269.2 |
| A: GGACTGATATCTTCCTGATCATCTTG | ||||
| DGAT1 | S: CCACTGGGACCTGAGGTGTC | 60 | 111 | MT221183.1 |
| A: GCATCACCACACACCAATTCA | ||||
| DGAT2 | S: CAATAGGTCCAAGGTAGAGAAGC | 60 | 156 | NM_001313305.1 |
| A: ACCAGCCAGGTGAAGTAGAGC | ||||
| UXT | S: GCAAGTGGATTTGGGCTGTAAC | 60 | 180 | XM_005700842.2 |
| A: ATGGAGTCCTTGGTGAGGTTGT |
| Gene | Primers Sequence (5′-3′) | TM/°C | GenBank Accession Number |
|---|---|---|---|
| RXRG | S: TCCTCAGGAAAGCACTACGGT | 60 | XM_005677098.3 |
| A: GGCAGTATTGACAGCGGTTG | |||
| FN3K | S: CGGGAAATGTGGCAGAGGAT | 61 | XM_018065388.1 |
| A: TGGTGGTAGGCGGTGAAGAA | |||
| ANKRD29 | S: GACTCTGCTCCGCCTGCTAC | 59 | XM_018039771.1 |
| A: GAGATTGATGTCCGCTCCCT | |||
| ACSL1 | S: GCCATCACCTACATCATCAACAA | 60 | XM_005698718.3 |
| A: ACACTTCTTGCCTCGTTCCA | |||
| SLC3A1 | S: CACGGTCACTCACTACTCGCA | 56 | XM_005686551.3 |
| A: CTGTATCGCCCTGGCTCCCT | |||
| CMKLR1 | S: ACTACCCCGACGACTTGGAC | 62 | XM_005691599.3 |
| A: TCCCGAGGAGGCAGATAATG | |||
| KLF2 | S: GCGGCAAGACCTACACCAA | 60 | KU041748.1 |
| A: TGTGCTTGCGGTAGTGGC | |||
| CD36 | S: AAAGAACTATTGTGGGGCTA | 60 | JF690773.1 |
| A: TATGTGTCAATTATGGCGACT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, J.; Zheng, J.; Cui, S.; Wang, J.; Wang, Y.; Li, Y.; Zhu, J.; Lin, Y. Nuclear Receptor Subfamily 4 Group A Member 1 (NR4A1) Promotes the Adipogenesis of Intramuscular Preadipocytes through PI3K/AKT Pathway in Goats. Animals 2024, 14, 2051. https://doi.org/10.3390/ani14142051
Xing J, Zheng J, Cui S, Wang J, Wang Y, Li Y, Zhu J, Lin Y. Nuclear Receptor Subfamily 4 Group A Member 1 (NR4A1) Promotes the Adipogenesis of Intramuscular Preadipocytes through PI3K/AKT Pathway in Goats. Animals. 2024; 14(14):2051. https://doi.org/10.3390/ani14142051
Chicago/Turabian StyleXing, Jiani, Jianying Zheng, Sheng Cui, Jinling Wang, Yong Wang, Yanyan Li, Jiangjiang Zhu, and Yaqiu Lin. 2024. "Nuclear Receptor Subfamily 4 Group A Member 1 (NR4A1) Promotes the Adipogenesis of Intramuscular Preadipocytes through PI3K/AKT Pathway in Goats" Animals 14, no. 14: 2051. https://doi.org/10.3390/ani14142051
APA StyleXing, J., Zheng, J., Cui, S., Wang, J., Wang, Y., Li, Y., Zhu, J., & Lin, Y. (2024). Nuclear Receptor Subfamily 4 Group A Member 1 (NR4A1) Promotes the Adipogenesis of Intramuscular Preadipocytes through PI3K/AKT Pathway in Goats. Animals, 14(14), 2051. https://doi.org/10.3390/ani14142051




