Application of GWAS and mGWAS in Livestock and Poultry Breeding
Abstract
:Simple Summary
Abstract
1. Introduction
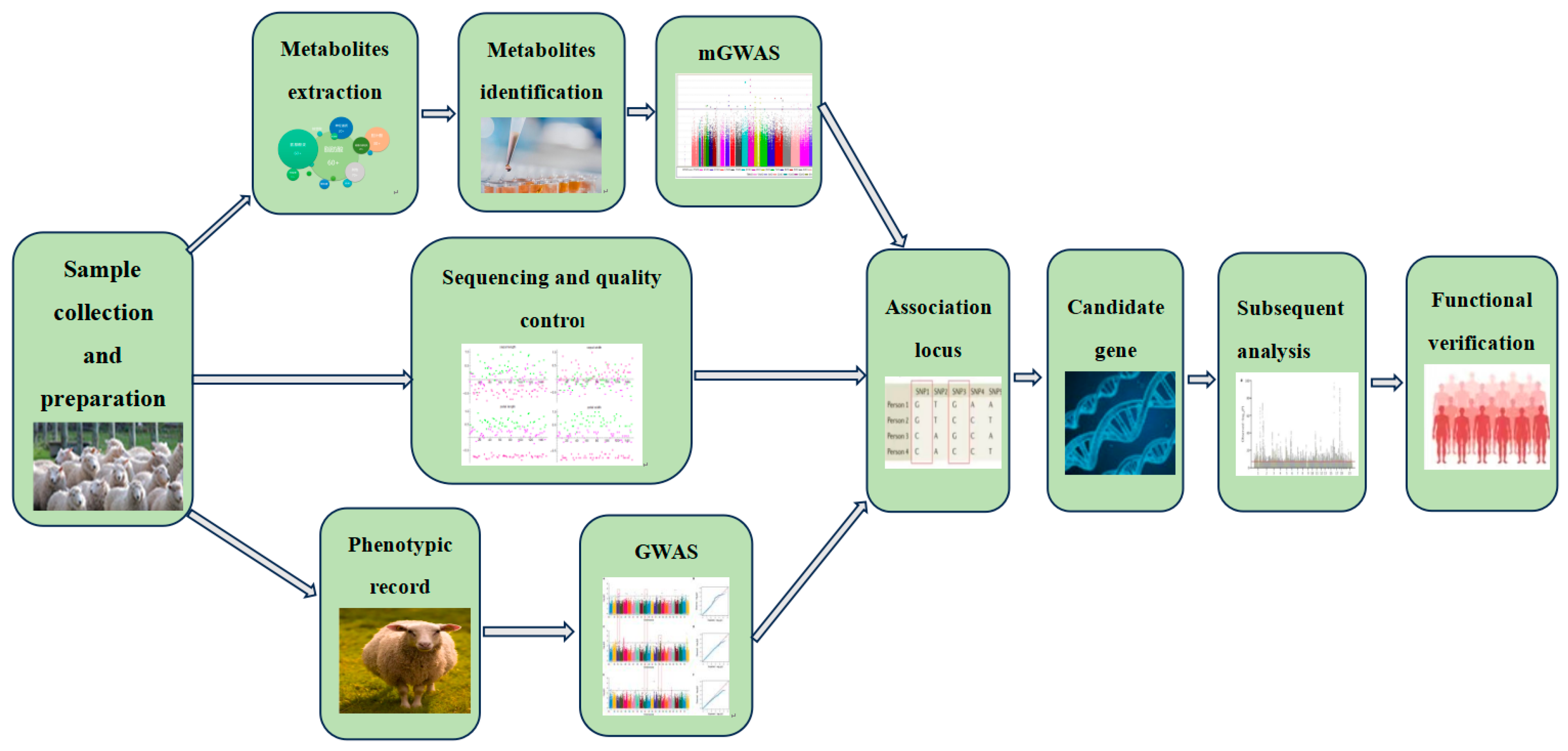
2. Research Progress of GWAS in Genetic Breeding of Livestock and Poultry
3. Research Progress of mGWAS in Genetic Breeding of Livestock and Poultry
4. The Advantages and Disadvantages of GWAS
5. The Advantages and Disadvantages of mGWAS
6. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Freebern, E.; Santos, D.J.A.; Fang, L.; Jiang, J.; Parker Gaddis, K.L.; Liu, G.E.; VanRaden, P.M.; Maltecca, C.; Cole, J.B.; Ma, L. GWAS and Fine-Mapping of Livability and Six Disease Traits in Holstein Cattle. BMC Genom. 2020, 21, 41. [Google Scholar] [CrossRef]
- Puig-Oliveras, A.; Revilla, M.; Castelló, A.; Fernández, A.I.; Folch, J.M.; Ballester, M. Expression-Based GWAS Identifies Variants, Gene Interactions and Key Regulators Affecting Intramuscular Fatty Acid Content and Composition in Porcine Meat. Sci. Rep. 2016, 6, 31803. [Google Scholar] [CrossRef]
- Baron, C.; Cherkaoui, S.; Therrien-Laperriere, S.; Ilboudo, Y.; Poujol, R.; Mehanna, P.; Garrett, M.E.; Telen, M.J.; Ashley-Koch, A.E.; Bartolucci, P.; et al. Gene-Metabolite Annotation with Shortest Reactional Distance Enhances Metabolite Genome-Wide Association Studies Results. bioRxiv 2023. [Google Scholar] [CrossRef]
- Zhang, Y.; Lyu, Y.; Chen, L.; Cao, K.; Chen, J.; He, C.; Lyu, X.; Jiang, Y.; Xiang, J.; Liu, B.; et al. Exploring the Prognosis-Related Genetic Variation in Gastric Cancer Based on mGWAS. Int. J. Mol. Sci. 2023, 24, 15259. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, Y.; Yin, H.; Liu, W.; Hu, X.; Li, D.; Lan, C.; Gao, L.; He, Z.; Cui, F.; et al. The Pathway of Melatonin Biosynthesis in Common Wheat (Triticum aestivum). J. Pineal Res. 2023, 74, e12841. [Google Scholar] [CrossRef]
- Zhou, S.; Kremling, K.A.; Bandillo, N.; Richter, A.; Zhang, Y.K.; Ahern, K.R.; Artyukhin, A.B.; Hui, J.X.; Younkin, G.C.; Schroeder, F.C.; et al. Metabolome-Scale Genome-Wide Association Studies Reveal Chemical Diversity and Genetic Control of Maize Specialized Metabolites. Plant Cell 2019, 31, 937–955. [Google Scholar] [CrossRef]
- Malik, P.; Kumar, J.; Sharma, S.; Sharma, R.; Sharma, S. Multi-Locus Genome-Wide Association Mapping for Spike-Related Traits in Bread Wheat (Triticum aestivum L.). BMC Genom. 2021, 22, 597. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.; Su, M.; Wu, Y.; Zhou, L.; Fu, R.; Li, Y.; Guo, H.; Luo, J.; Wang, S.; Zhang, Y. Trichome Regulator SlMIXTA-like Directly Manipulates Primary Metabolism in Tomato Fruit. Plant Biotechnol. J. 2020, 18, 354–363. [Google Scholar] [CrossRef]
- Lin, Q.; Chen, J.; Liu, X.; Wang, B.; Zhao, Y.; Liao, L.; Allan, A.C.; Sun, C.; Duan, Y.; Li, X.; et al. A Metabolic Perspective of Selection for Fruit Quality Related to Apple Domestication and Improvement. Genome Biol. 2023, 24, 95. [Google Scholar] [CrossRef]
- Nica, A.C.; Dermitzakis, E.T. Using Gene Expression to Investigate the Genetic Basis of Complex Disorders. Hum. Mol. Genet. 2008, 17, R129–R134. [Google Scholar] [CrossRef]
- Slavov, G.; Allison, G.; Bosch, M. Advances in the Genetic Dissection of Plant Cell Walls: Tools and Resources Available in Miscanthus. Front. Plant Sci. 2013, 4, 217. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, S.P.; Sindt, J.J.; Greenquist, M.A.; Miller, W.F.; Pike, J.N.; Loe, E.R.; Sulpizio, M.J.; Drouillard, J.S. Plasma Metabolites of Receiving Heifers and the Relationship between Apparent Bovine Respiratory Disease, Body Weight Gain, and Carcass Characteristics. J. Anim. Sci. 2009, 87, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Ossipov, V.; Ossipova, S.; Bykov, V.; Oksanen, E.; Koricheva, J.; Haukioja, E. Application of Metabolomics to Genotype and Phenotype Discrimination of Birch Trees Grown in a Long-Term Open-Field Experiment|Metabolomics. Metabolomics 2008, 4, 39–51. Available online: https://link.springer.com/article/10.1007/s11306-007-0097-8 (accessed on 28 July 2024). [CrossRef]
- Chan, E.K.F.; Rowe, H.C.; Kliebenstein, D.J. Understanding the Evolution of Defense Metabolites in Arabidopsis Thaliana Using Genome-Wide Association Mapping. Genetics 2010, 185, 991–1007. [Google Scholar] [CrossRef]
- Illig, T.; Gieger, C.; Zhai, G.; Römisch-Margl, W.; Wang-Sattler, R.; Prehn, C.; Altmaier, E.; Kastenmüller, G.; Kato, B.S.; Mewes, H.-W.; et al. A Genome-Wide Perspective of Genetic Variation in Human Metabolism. Nat. Genet. 2010, 42, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Akanno, E.C.; Valente, T.S.; Abo-Ismail, M.; Karisa, B.K.; Wang, Z.; Plastow, G.S. Genomic Heritability and Genome-Wide Association Studies of Plasma Metabolites in Crossbred Beef Cattle. Front. Genet. 2020, 11, 538600. [Google Scholar] [CrossRef]
- Putra, W.P.B.; Hartati, H.; Mariyono, M.; Noor, R.R.; Sumantri, C.; Margawati, E.T. Early Detection of Candidate Genes for Body Weight in Indonesian Cattle Breeds with Genome-Wide Association Study (GWAS). Acta Vet. 2024, 74, 246–260. [Google Scholar] [CrossRef]
- Oliveira, H.R.; Chud, T.C.S.; Oliveira, G.A.; Hermisdorff, I.C.; Narayana, S.G.; Rochus, C.M.; Butty, A.M.; Malchiodi, F.; Stothard, P.; Miglior, F.; et al. Genome-Wide Association Analyses Reveals Copy Number Variant Regions Associated with Reproduction and Disease Traits in Canadian Holstein Cattle. J. Dairy Sci. 2024, 107, 7052–7063. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Yu, S.; Guo, J.; Cheng, G.; Mei, C.; Zan, L. Genome-Wide Association Study Reveals Novel Loci Associated with Body Conformation Traits in Qinchuan Cattle. Animals 2023, 13, 3628. [Google Scholar] [CrossRef]
- Gao, Y.; Marceau, A.; Iqbal, V.; Torres-Vázquez, J.A.; Neupane, M.; Jiang, J.; Liu, G.E.; Ma, L. Genome-Wide Association Analysis of Heifer Livability and Early First Calving in Holstein Cattle. BMC Genom. 2023, 24, 628. [Google Scholar] [CrossRef]
- Adhikari, M.; Kantar, M.B.; Longman, R.J.; Lee, C.N.; Oshiro, M.; Caires, K.; He, Y. Genome-Wide Association Study for Carcass Weight in Pasture-Finished Beef Cattle in Hawai’i. Front. Genet. 2023, 14, 1168150. [Google Scholar] [CrossRef]
- Arikawa, L.M.; Mota, L.F.M.; Schmidt, P.I.; Frezarim, G.B.; Fonseca, L.F.S.; Magalhães, A.F.B.; Silva, D.A.; Carvalheiro, R.; Chardulo, L.A.L.; de Albuquerque, L.G. Genome-Wide Scans Identify Biological and Metabolic Pathways Regulating Carcass and Meat Quality Traits in Beef Cattle. Meat Sci. 2024, 209, 109402. [Google Scholar] [CrossRef]
- Wang, J.; Fan, T.; Du, Z.; Xu, L.; Chen, Y.; Zhang, L.; Gao, H.; Li, J.; Ma, Y.; Gao, X. Genome-Wide Association Analysis Identifies the PMEL Gene Affecting Coat Color and Birth Weight in Simmental × Holstein. Animals 2023, 13, 3821. [Google Scholar] [CrossRef]
- Anaya, G.; Laseca, N.; Granero, A.; Ziadi, C.; Arrebola, F.; Domingo, A.; Molina, A. Genomic Characterization of Quality Wool Traits in Spanish Merino Sheep. Genes 2024, 15, 795. [Google Scholar] [CrossRef]
- Smitchger, J.A.; Taylor, J.B.; Mousel, M.R.; Schaub, D.; Thorne, J.W.; Becker, G.M.; Murdoch, B.M. Genome-Wide Associations with Longevity and Reproductive Traits in U.S. Rangeland Ewes. Front. Genet. 2024, 15, 1398123. [Google Scholar] [CrossRef]
- Li, C.; Li, J.; Wang, H.; Zhang, R.; An, X.; Yuan, C.; Guo, T.; Yue, Y. Genomic Selection for Live Weight in the 14th Month in Alpine Merino Sheep Combining GWAS Information. Animals 2023, 13, 3516. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Cao, Y.; Shan, H.; Wu, J.; Song, X.; Jiang, Y. The GWAS Analysis of Body Size and Population Verification of Related SNPs in Hu Sheep. Front. Genet. 2021, 12, 642552. [Google Scholar] [CrossRef]
- Bolormaa, S.; Swan, A.A.; Stothard, P.; Khansefid, M.; Moghaddar, N.; Duijvesteijn, N.; van der Werf, J.H.J.; Daetwyler, H.D.; MacLeod, I.M. A Conditional Multi-Trait Sequence GWAS Discovers Pleiotropic Candidate Genes and Variants for Sheep Wool, Skin Wrinkle and Breech Cover Traits. Genet. Sel. Evol. 2021, 53, 58. [Google Scholar] [CrossRef]
- Gholizadeh, M.; Esmaeili-Fard, S.M. Meta-Analysis of Genome-Wide Association Studies for Litter Size in Sheep. Theriogenology 2022, 180, 103–112. [Google Scholar] [CrossRef]
- Revelo, H.A.; López-Alvarez, D.; Palacios, Y.A.; Vergara, O.D.; Yánez, M.B.; Ariza, M.F.; Molina, S.L.C.; Sanchez, Y.O.; Alvarez, L.Á. Genome-Wide Association Study Reveals Candidate Genes for Traits Related to Meat Quality in Colombian Creole Hair Sheep. Trop. Anim. Health Prod. 2023, 55, 357. [Google Scholar] [CrossRef]
- Liu, C.; Chen, Z.; Zhang, Z.; Wang, Z.; Guo, X.; Pan, Y.; Wang, Q. Unveiling the Genetic Mechanism of Meat Color in Pigs through GWAS, Multi-Tissue, and Single-Cell Transcriptome Signatures Exploration. Int. J. Mol. Sci. 2024, 25, 3682. [Google Scholar] [CrossRef]
- Shen, Y.; Chen, Y.; Zhang, S.; Wu, Z.; Lu, X.; Liu, W.; Liu, B.; Zhou, X. Smartphone-Based Digital Phenotyping for Genome-Wide Association Study of Intramuscular Fat Traits in Longissimus Dorsi Muscle of Pigs. Anim. Genet. 2024, 55, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhou, S.; Liu, Z.; Ruan, D.; Wu, J.; Quan, J.; Zheng, E.; Yang, J.; Cai, G.; Wu, Z.; et al. Genome-Wide Association Study for Somatic Skeletal Traits in Duroc × (Landrace × Yorkshire) Pigs. Animals 2023, 14, 37. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, S.; Yuan, M.; Zhan, F.; Song, M.; Shang, P.; Yang, F.; Li, X.; Qiao, R.; Han, X.; et al. Genome-Wide Association Studies and Runs of Homozygosity to Identify Reproduction-Related Genes in Yorkshire Pig Population. Genes 2023, 14, 2133. [Google Scholar] [CrossRef]
- Zeng, M.; Wang, B.; Liu, L.; Yang, Y.; Tang, Z. Genome-Wide Association Study Identifies 12 New Genetic Loci Associated with Growth Traits in Pigs. J. Integr. Agric. 2024, 23, 217–227. [Google Scholar] [CrossRef]
- Zhou, F.; Quan, J.; Ruan, D.; Qiu, Y.; Ding, R.; Xu, C.; Ye, Y.; Cai, G.; Liu, L.; Zhang, Z.; et al. Identification of Candidate Genes for Economically Important Carcass Cutting in Commercial Pigs through GWAS. Animals 2023, 13, 3243. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Li, M.; Sun, H.; Chen, Q.; Yan, D.; Dong, X.; Pan, Y.; Lu, S. Genome-Wide Association Study Reveals Genetic Loci and Candidate Genes for Meat Quality Traits in a Four-Way Crossbred Pig Population. Front. Genet. 2023, 14, 1001352. [Google Scholar] [CrossRef]
- Zeng, H.; Zhong, Z.; Xu, Z.; Teng, J.; Wei, C.; Chen, Z.; Zhang, W.; Ding, X.; Li, J.; Zhang, Z. Meta-Analysis of Genome-Wide Association Studies Uncovers Shared Candidate Genes across Breeds for Pig Fatness Trait. BMC Genom. 2022, 23, 786. [Google Scholar] [CrossRef]
- Cai, K.; Liu, R.; Wei, L.; Wang, X.; Cui, H.; Luo, N.; Wen, J.; Chang, Y.; Zhao, G. Genome-Wide Association Analysis Identify Candidate Genes for Feed Efficiency and Growth Traits in Wenchang Chickens. BMC Genom. 2024, 25, 645. [Google Scholar] [CrossRef]
- Perini, F.; Cendron, F.; Lasagna, E.; Cassandro, M.; Penasa, M. Genomic Insights into Shank and Eggshell Color in Italian Local Chickens. Poult. Sci. 2024, 103, 103677. [Google Scholar] [CrossRef]
- Wang, J.; Liu, J.; Lei, Q.; Liu, Z.; Han, H.; Zhang, S.; Qi, C.; Liu, W.; Li, D.; Li, F.; et al. Elucidation of the Genetic Determination of Body Weight and Size in Chinese Local Chicken Breeds by Large-Scale Genomic Analyses. BMC Genom. 2024, 25, 296. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, X.; Zhong, C.; Jiang, X.; Wu, G.; Li, G.; Yan, Y.; Yang, N.; Sun, C. Genetic Patterns and Genome-Wide Association Analysis of Eggshell Quality Traits of Egg-Type Chicken across an Extended Laying Period. Poult. Sci. 2024, 103, 103458. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.; Jin, D.; Kim, J.-H.; Kim, S.-C.; Lim, J.A.; Chai, H.-H.; Jung, S.A.; Lee, J.-H.; Lee, S.-H. Genome-Wide Association Study Revealed the Genomic Regions Associated with Skin Pigmentation in an Ogye x White Leghorn F2 Chicken Population. Poult. Sci. 2023, 102, 102720. [Google Scholar] [CrossRef]
- Zhu, X.-N.; Wang, Y.-Z.; Li, C.; Wu, H.-Y.; Zhang, R.; Hu, X.-X. Chicken Chromatin Accessibility Atlas Accelerates Epigenetic Annotation of Birds and Gene Fine-Mapping Associated with Growth Traits. Zool. Res. 2023, 44, 53–62. [Google Scholar] [CrossRef]
- Li, Y.; Bai, X.; Liu, X.; Wang, W.; Li, Z.; Wang, N.; Xiao, F.; Gao, H.; Guo, H.; Li, H.; et al. Integration of Genome-Wide Association Study and Selection Signatures Reveals Genetic Determinants for Skeletal Muscle Production Traits in an F2 Chicken Population. J. Integr. Agric. 2022, 21, 2065–2075. [Google Scholar] [CrossRef]
- Fan, S.; Yuan, P.; Li, S.; Li, H.; Zhai, B.; Li, Y.; Zhang, H.; Gu, J.; Li, H.; Tian, Y.; et al. Genetic Architecture and Key Regulatory Genes of Fatty Acid Composition in Gushi Chicken Breast Muscle Determined by GWAS and WGCNA. BMC Genom. 2023, 24, 434. [Google Scholar] [CrossRef]
- Dou, D.; Shen, L.; Zhou, J.; Cao, Z.; Luan, P.; Li, Y.; Xiao, F.; Guo, H.; Li, H.; Zhang, H. Genome-Wide Association Studies for Growth Traits in Broilers. BMC Genom. Data 2022, 23, 1. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Gong, H.; Cui, L.; Zhang, W.; Ma, J.; Chen, C.; Ai, H.; Xiao, S.; Huang, L.; et al. Genetic Correlation of Fatty Acid Composition with Growth, Carcass, Fat Deposition and Meat Quality Traits Based on GWAS Data in Six Pig Populations. Meat Sci. 2019, 150, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yang, B.; Zhang, J.; Cui, L.; Ma, J.; Chen, C.; Ai, H.; Xiao, S.; Ren, J.; Huang, L. Genome-Wide Association Studies for Fatty Acid Metabolic Traits in Five Divergent Pig Populations. Sci. Rep. 2016, 6, 24718. [Google Scholar] [CrossRef] [PubMed]
- Tetens, J.; Heuer, C.; Heyer, I.; Klein, M.S.; Gronwald, W.; Junge, W.; Oefner, P.J.; Thaller, G.; Krattenmacher, N. Polymorphisms within the APOBR Gene Are Highly Associated with Milk Levels of Prognostic Ketosis Biomarkers in Dairy Cows. Physiol. Genom. 2015, 47, 129–137. [Google Scholar] [CrossRef]
- Park, J. Genome-Wide Association Study to Reveal New Candidate Genes Using Single-Step Approaches for Productive Traits of Yorkshire Pig in Korea. Anim. Biosci. 2024, 37, 451–460. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Li, M.; Sun, H.; Chen, Q.; Yan, D.; Dong, X.; Pan, Y.; Lu, S. Genome-Wide Association Study of Growth Traits in a Four-Way Crossbred Pig Population. Genes 2022, 13, 1990. [Google Scholar] [CrossRef]
- Reis, H.B.D.; Carvalho, M.E.; Espigolan, R.; Poleti, M.D.; Ambrizi, D.R.; Berton, M.P.; Ferraz, J.B.S.; de Mattos Oliveira, E.C.; Eler, J.P. Genome-Wide Association (GWAS) Applied to Carcass and Meat Traits of Nellore Cattle. Metabolites 2023, 14, 6. [Google Scholar] [CrossRef]
- Velayudhan, S.M.; Yin, T.; Alam, S.; Brügemann, K.; Sejian, V.; Bhatta, R.; Schlecht, E.; König, S. Unraveling the Genomic Association for Milk Production Traits and Signatures of Selection of Cattle in a Harsh Tropical Environment. Biology 2023, 12, 1483. [Google Scholar] [CrossRef]
- Massender, E.; Oliveira, H.R.; Brito, L.F.; Maignel, L.; Jafarikia, M.; Baes, C.F.; Sullivan, B.; Schenkel, F.S. Genome-Wide Association Study for Milk Production and Conformation Traits in Canadian Alpine and Saanen Dairy Goats. J. Dairy Sci. 2023, 106, 1168–1189. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Wu, Y.; Shen, J.; Pan, A.; Zhang, H.; Sun, J.; Liang, Z.; Huang, T.; Du, J.; Pi, J. Genome-Wide Association Study of Egg Production Traits in Shuanglian Chickens Using Whole Genome Sequencing. Genes 2023, 14, 2129. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-W.; Lee, C.-C.; Liu, H.; Wu, C.-S.; Pickering, C.R.; Huang, P.-J.; Wang, J.; Chang, I.Y.-F.; Yeh, Y.-M.; Chen, C.-D.; et al. APOBEC3A Is an Oral Cancer Prognostic Biomarker in Taiwanese Carriers of an APOBEC Deletion Polymorphism. Nat. Commun. 2017, 8, 465. [Google Scholar] [CrossRef]
- Zhao, H.; He, Y.; Zhang, K.; Li, S.; Chen, Y.; He, M.; He, F.; Gao, B.; Yang, D.; Fan, Y.; et al. Rewiring of the Seed Metabolome during Tartary Buckwheat Domestication. Plant Biotechnol. J. 2023, 21, 150–164. [Google Scholar] [CrossRef]
- Li, J.; Mukiibi, R.; Wang, Y.; Plastow, G.S.; Li, C. Identification of Candidate Genes and Enriched Biological Functions for Feed Efficiency Traits by Integrating Plasma Metabolites and Imputed Whole Genome Sequence Variants in Beef Cattle. BMC Genomics 2021, 22, 823. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.; Mukiibi, R.; Karisa, B.; Plastow, G.S.; Li, C. Integrative Analyses of Genomic and Metabolomic Data Reveal Genetic Mechanisms Associated with Carcass Merit Traits in Beef Cattle. Sci. Rep. 2022, 12, 3389. [Google Scholar] [CrossRef]
- Deng, T.; Ma, X.; Duan, A.; Lu, X.; Abdel-Shafy, H. Multi-Omics Analysis Provides Insight into the Genetic Basis of Proline-Derived Milk Microbiota in Buffalo. Food Biosci. 2024, 59, 103942. [Google Scholar] [CrossRef]
- Wang, X.; Kadarmideen, H.N. Metabolite Genome-Wide Association Study (mGWAS) and Gene-Metabolite Interaction Network Analysis Reveal Potential Biomarkers for Feed Efficiency in Pigs. Metabolites 2020, 10, 201. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, H.; Yang, Y.; Liu, T.; Guo, Z.; Fan, W.; Wang, Z.; Yang, X.; Zhang, B.; Liu, H.; et al. Metabolome-Based Genome-Wide Association Study of Duck Meat Leads to Novel Genetic and Biochemical Insights. Adv. Sci. 2023, 10, e2300148. [Google Scholar] [CrossRef]
- Tian, J.; Zhu, X.; Wu, H.; Wang, Y.; Hu, X. Serum Metabolic Profile and Metabolome Genome-Wide Association Study in Chicken. J. Anim. Sci. Biotechnol. 2023, 14, 69. [Google Scholar] [CrossRef]
- Xu, N.; Chen, B.; Cheng, Y.; Su, Y.; Song, M.; Guo, R.; Wang, M.; Deng, K.; Lan, T.; Bao, S.; et al. Integration of GWAS and RNA-Seq Analysis to Identify SNPs and Candidate Genes Associated with Alkali Stress Tolerance at the Germination Stage in Mung Bean. Genes 2023, 14, 1294. [Google Scholar] [CrossRef]
- Ho, S.S.; Urban, A.E.; Mills, R.E. Structural Variation in the Sequencing Era. Nat. Rev. Genet. 2020, 21, 171–189. [Google Scholar] [CrossRef]
- He, J.; Gai, J. Genome-Wide Association Studies (GWAS). In Plant Genotyping: Methods and Protocols; Shavrukov, Y., Ed.; Springer: New York, NY, USA, 2023; pp. 123–146. ISBN 978-1-07-163024-2. [Google Scholar]
- Ma, S.; Li, P.; Liu, H.; Xi, Y.; Xu, Q.; Qi, J.; Wang, J.; Li, L.; Wang, J.; Hu, J.; et al. Genome-Wide Association Analysis of the Primary Feather Growth Traits of Duck: Identification of Potential Loci for Growth Regulation. Poult. Sci. 2023, 102, 102243. [Google Scholar] [CrossRef]
- Shriner, D.; Vaughan, L.K.; Padilla, M.A.; Tiwari, H.K. Problems with Genome-Wide Association Studies. Science 2007, 316, 1840–1842. [Google Scholar] [CrossRef]
- Mkize, N.; Maiwashe, A.; Dzama, K.; Dube, B.; Mapholi, N. Suitability of GWAS as a Tool to Discover SNPs Associated with Tick Resistance in Cattle: A Review. Pathogens 2021, 10, 1604. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, Y.; Wei, J.; Hu, X.; Yin, H.; Liu, W.; Li, D.; Tian, W.; Hao, Y.; He, Z.; et al. Beyond Pathways: Accelerated Flavonoids Candidate Identification and Novel Exploration of Enzymatic Properties Using Combined Mapping Populations of Wheat. Plant Biotechnol. J. 2024, 22, 2033–2050. [Google Scholar] [CrossRef]
- Adamski, J.; Suhre, K. Metabolomics Platforms for Genome Wide Association Studies--Linking the Genome to the Metabolome. Curr. Opin. Biotechnol. 2013, 24, 39–47. [Google Scholar] [CrossRef] [PubMed]
| Species | Variety | Research Traits | Candidate Gene | References |
|---|---|---|---|---|
| Cattle | Indonesian cattle | Body weight | SUGT1, SF3A3, DSCAM | [17] |
| Canadian Holstein cattle | Reproduction | CSh, FSTCc, NRRh | [18] | |
| Qinchuan cattle | Body conformation | ADAMTS17, ALDH1A3, CHSY1, MAGEL2, MEF2A, SYNM, CNTNAP5, CTNNA3 | [19] | |
| Holstein cattle | Heifer livability | MOG, OR12D2E, OR12D3, OR2H1, OR5V1, OR5V1C, OR5V2, TRIM10, TRIM15 | [20] | |
| Hawaiian beef cattle | Carcass weight | RGS20, TCEA1, LYPLA1, MRPL15, EIF5 | [21] | |
| Brazilian beef cattle | Carcass and meat quality traits | CAST, PLAG1, XKR4, PLAGL2, AQP3/AQP7, MYLK2, WWOX, CARTPT, PLA2G, etc. | [22] | |
| Simmental Holstein cattle | Hair color and birth weight | RNF41, ZC3H10, ERBB3, PMEL, OR10A7 | [23] | |
| Sheep | Spanish Merino sheep | Quality wool | EDN2, COL18A1, LRP1B, FGF12, ADAM17 | [24] |
| U.S. rangeland ewes | Longevity and reproduction | LPL, ANOS1, ARHGEF26, ASIC2, ASTN2, ATP8A2, CAMK2D, etc. | [25] | |
| High mountain Merino sheep | 14 months live weight | FAM184B, NCAPG, MACF1, ANKRD44, DCAF16, FUK, LCORL, SYN3 | [26] | |
| Hu sheep | Shape | KITLG, CADM2, MCTP1, COL4A6 | [27] | |
| Merino sheep | Fiber and skin wrinkles | ALX4, EIF2AK2, ESRP1, HAS2, MC5R, MX2 | [28] | |
| 5 varieties including Wadi, Icelandic, Finnsheep, etc. | Litter size | CASK, PLCB4, RPTOR, GRIA2, PLCB1 | [29] | |
| Colombian Creole hair sheep | Meat quality | ELOVL2, ARAP2, IBN2, TPM2, etc. | [30] | |
| Pig | Jinhua × Piétrain | Meat color | ZBTB17, FAM131C, KIFC3, NTPCR, etc. | [31] |
| Large White × Tongcheng pigs | Intramuscular fat traits in longissimus dorsi muscle | NR2F2, MCTP2, MTLN, ST3GAL5, NDUFAB1, etc. | [32] | |
| Duroc × (Landrace × Yorkshire) pigs | Somatic skeletal traits | OPRM1, SLC44A5, WASHC4, NOPCHAP1, RHOT1, etc. | [33] | |
| Yorkshire pigs | Reproductive traits | ELMO1, AOAH, INSIG2, NUP205, LYPLAL1, etc. | [34] | |
| Duroc, Changbai, Dabai | Growth traits | SKAP2, SATB1, PDE7B, PPP1R16B, WNT3, WNT9B | [35] | |
| Duroc × Landrace × Yorkshire | Economic characteristics of carcass | TIMP2, EML1, SMN1 | [36] | |
| Duroc × Saba, Yorkshire × (Landrace × Saba) | Meat quality traits | GRM8, ANKRD6, MACROD2, CDYL2, CHL1, etc. | [37] | |
| Duroc, Yorkshire, Landrace | Pig fatness trait | MC4R, PPARD, SLC27A1, PHLPP1, NUDT3, ILRUN, RELCH, KCNQ5, ITPR3, and U3 | [38] | |
| Birds | Wenchang chickens | Feed efficiency and growth traits | PLCE1, LAP3, MED28, QDPR, LDB2 and SEL1L3 | [39] |
| Italian local chickens | Shank and eggshell color | MTAP, CDKN2A, CDKN2B, SLC7A11 and MITF | [40] | |
| Wenshang Barred, Recessive White, Luxi Mini | Body weight and size | LCORL, LDB2, and PPARGC1A | [41] | |
| Rhode Island Red chickens | Eggshell strength | FRY and PCNX2 | [42] | |
| Ogye x White Leghorn | Skin color | MTAP, FEM1C, GNAS and EDN3 | [43] | |
| Lingnan Yellow chicken×Chinese Huiyang Bearded | Body weight | CAB39L, RCBTB1 | [44] | |
| Arbor Acres broiler× Baier layer | Skeletal muscle production traits | LRCH1, CDADC1, CAB39L, FOXO1, NBEA, GPALPP1, etc. | [45] | |
| Tibetan chicken, Wenchang chicken, etc. | Chest muscle fatty acid composition | ENO1, ADH1, ASAH1, ADH1C, PIK3CD, WISP1, AKT1, PANK3, C1QTNF2 | [46] | |
| NEAUHLF | Growth traits | ACTA1, IGF2BP1, TAPT1, LDB2, PRKCA, TGFBR2, GLI3, SLC16A7, INHBA, BAMBI, GATA4, etc. | [47] |
| Species | Variety | Research Contents | Candidate Gene | References |
|---|---|---|---|---|
| Cattle | Canadian hybrid beef cattle | Heritability of plasma metabolites | CTNNA2 | [16] |
| Charolais, Hereford–Angus crosses, and a Beefbooster composite breed | Feed efficiency traits | PLSCR1, AQP9, NEDD4, PRTG, PYGO1, CUX2, NOS1, etc. | [59] | |
| Charolais, Hereford–Angus crosses, and a Beefbooster composite breed | Carcass merit traits | CDH13, KMT5B, NDUFS8, ALDH3B1, CHKA, TCIRG1 | [60] | |
| Guangxi Buffalo | Buffalo milk traits | ATG16L1 | [61] | |
| Pigs | Duroc pig, Landrace pig | Genetic variation in feed efficiency | LRRC4C, SH2D4A, MBOAT1 | [62] |
| Birds | Beijing Duck × Liancheng Duck | Skeletal muscle metabolism | AOX1, ACBD5, GADL1, CARNMT2 | [63] |
| High-quality chicken strain A03 × Huiyang Bearded Chicken | Chicken blood metabolites from the Qing Dynasty | TDH, AASS, ABCB1, CD36 | [64] |
| mGWAS | GWAS | |
|---|---|---|
| Principle | Metabolite genome association analysis is conducted, utilizing sample metabolomic data as the phenotype [72]. | Phenotypic trait information was gathered from samples, followed by an association analysis between the phenotype and the genome [1]. |
| Characteristic | Among different varieties or individuals, the types and contents of metabolites exhibit significant variation, characterized by rich data and precise identification. | Traditional phenotypes exhibit fewer types, are challenging to quantify, and are heavily influenced by environmental factors, leading to elevated rates of false positives. |
| Summary | The increased availability of phenotypic data enables the identification of a broader spectrum of gene phenotypes that can be quantified. Enhanced quantification accuracy corresponds to more precise SNP localization. Moreover, ample data facilitate the identification of rare SNP loci. | The localization of genes is relatively limited in terms of quantity, and their localization effect is often poor. Additionally, there can be simultaneous associations with multiple genes, making it challenging to distinguish the main gene responsible. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, J.; Gao, Z.; Lu, Y.; Li, M.; Hong, J.; Wu, J.; Wu, D.; Deng, W.; Xi, D.; Chong, Y. Application of GWAS and mGWAS in Livestock and Poultry Breeding. Animals 2024, 14, 2382. https://doi.org/10.3390/ani14162382
Ren J, Gao Z, Lu Y, Li M, Hong J, Wu J, Wu D, Deng W, Xi D, Chong Y. Application of GWAS and mGWAS in Livestock and Poultry Breeding. Animals. 2024; 14(16):2382. https://doi.org/10.3390/ani14162382
Chicago/Turabian StyleRen, Jing, Zhendong Gao, Ying Lu, Mengfei Li, Jieyun Hong, Jiao Wu, Dongwang Wu, Weidong Deng, Dongmei Xi, and Yuqing Chong. 2024. "Application of GWAS and mGWAS in Livestock and Poultry Breeding" Animals 14, no. 16: 2382. https://doi.org/10.3390/ani14162382
APA StyleRen, J., Gao, Z., Lu, Y., Li, M., Hong, J., Wu, J., Wu, D., Deng, W., Xi, D., & Chong, Y. (2024). Application of GWAS and mGWAS in Livestock and Poultry Breeding. Animals, 14(16), 2382. https://doi.org/10.3390/ani14162382





