Recombinant Polymerase Amplification Coupled with CRISPR/Cas12a Detection System for Rapid Visual Detection of Porcine Circovirus 3
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses, Clinical Samples, and Antibodies
2.2. The Expression and Purification of Cas12a Protein
2.3. Viral DNA or cDNA Preparation of Various Viruses and Clinical Samples
2.4. Design and Synthesization of crRNAs and ssDNA
2.5. Assay of Endonuclease Activity of the Purified Cas12a Proteins and Optimization of crRNAs and Cas12a Concentration
2.6. The Optimization of Recombinant Polymerase Amplification (RPA)
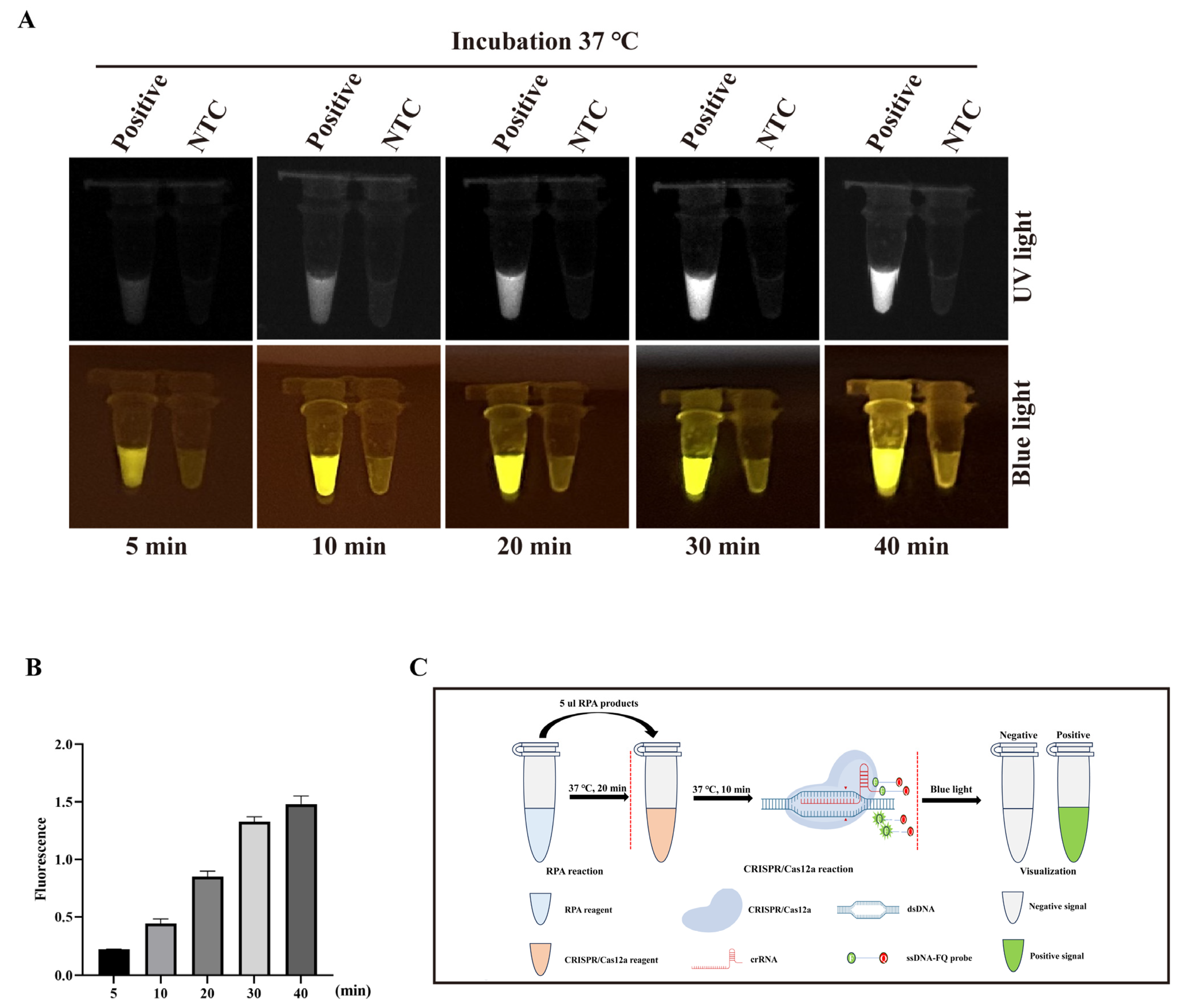
2.7. The Optimization of Sensitivity and Time in CRISPR/Cas12a-PCV3 Detection
2.8. The Analysis of Sensitivity and Specificity in Two-Pot RPA-CRISPR/Cas12a-PCV3 Detection
2.9. Establishment and Sensitivity of One-Pot RPA-CRISPR/Cas12a-PCV3 Detection Method
2.10. The Detection of Clinical Samples
2.11. Statistical Analysis
3. Results
3.1. Expression and Purification of Cas12a Protein and Assay of Endonuclease Activity
3.2. The Optimization of Cas12a and crRNA Concentration in CRISPR/Cas12a-PCV3 Detection Method
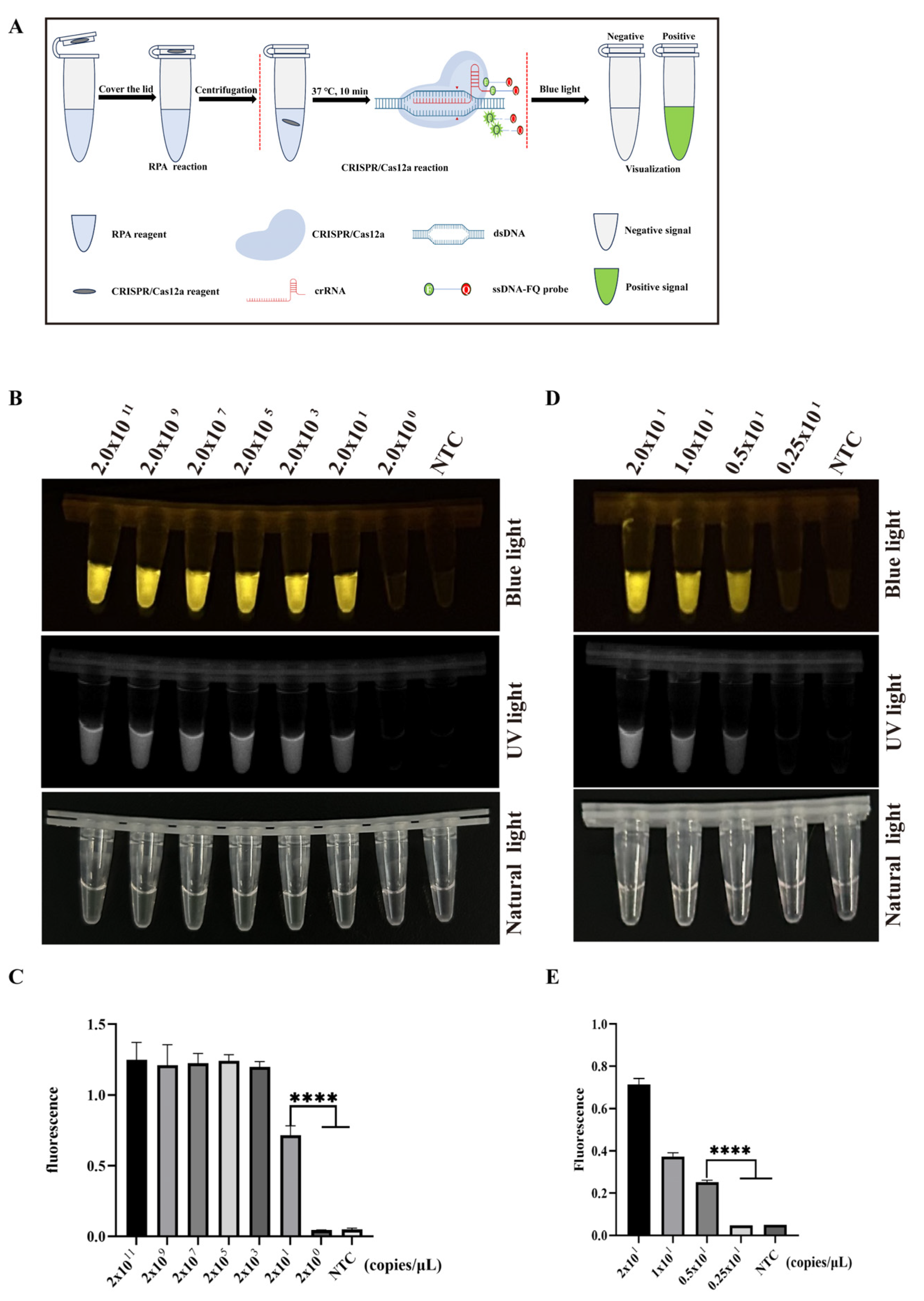
3.3. The Optimization of RPA Primers and Sensitivity of CRISPR/Cas12a-PCV3 Detection
3.4. Establishment of Two-Pot RPA-CRISPR/Cas12a-PCV3 Detection Method
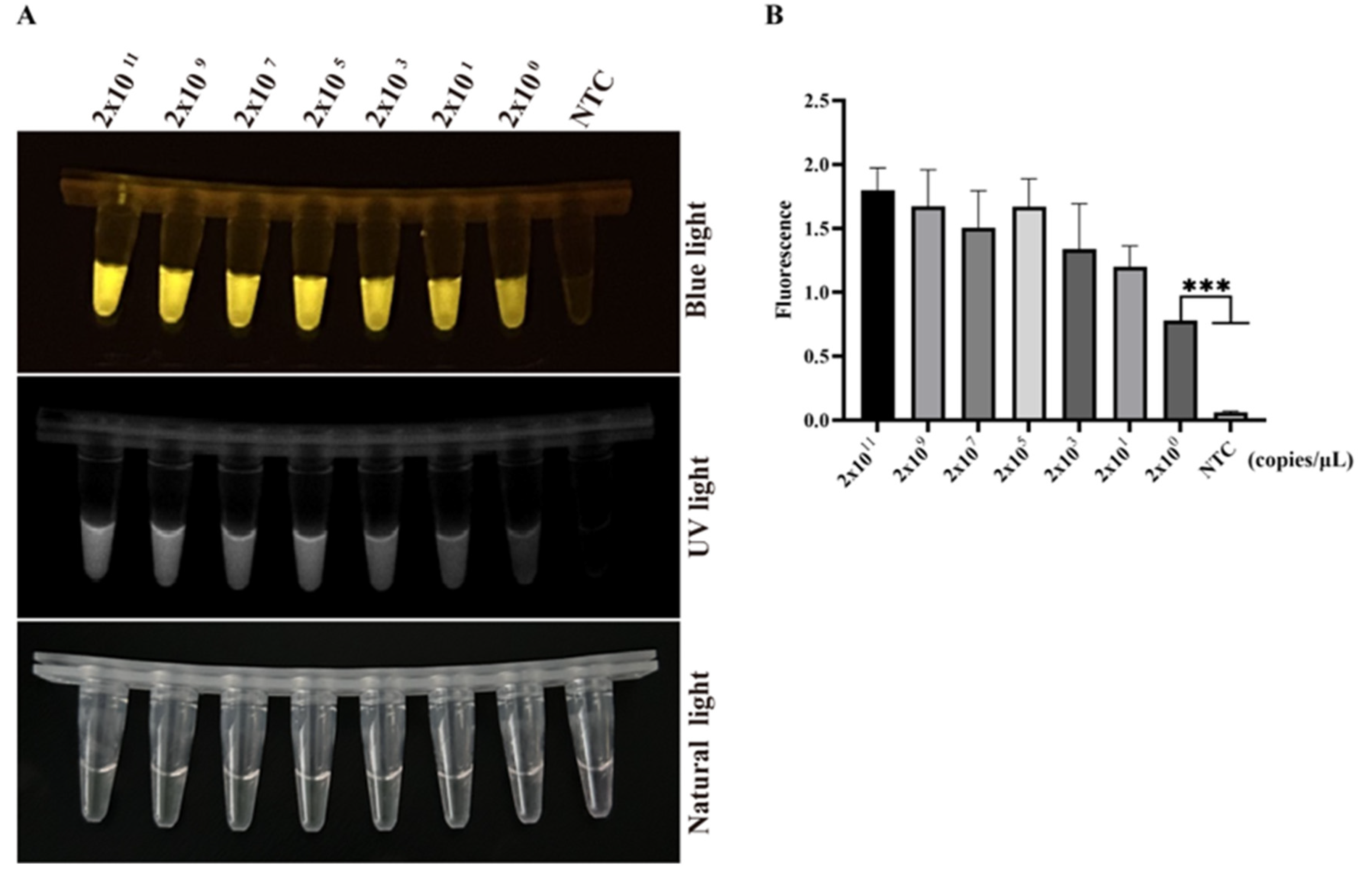
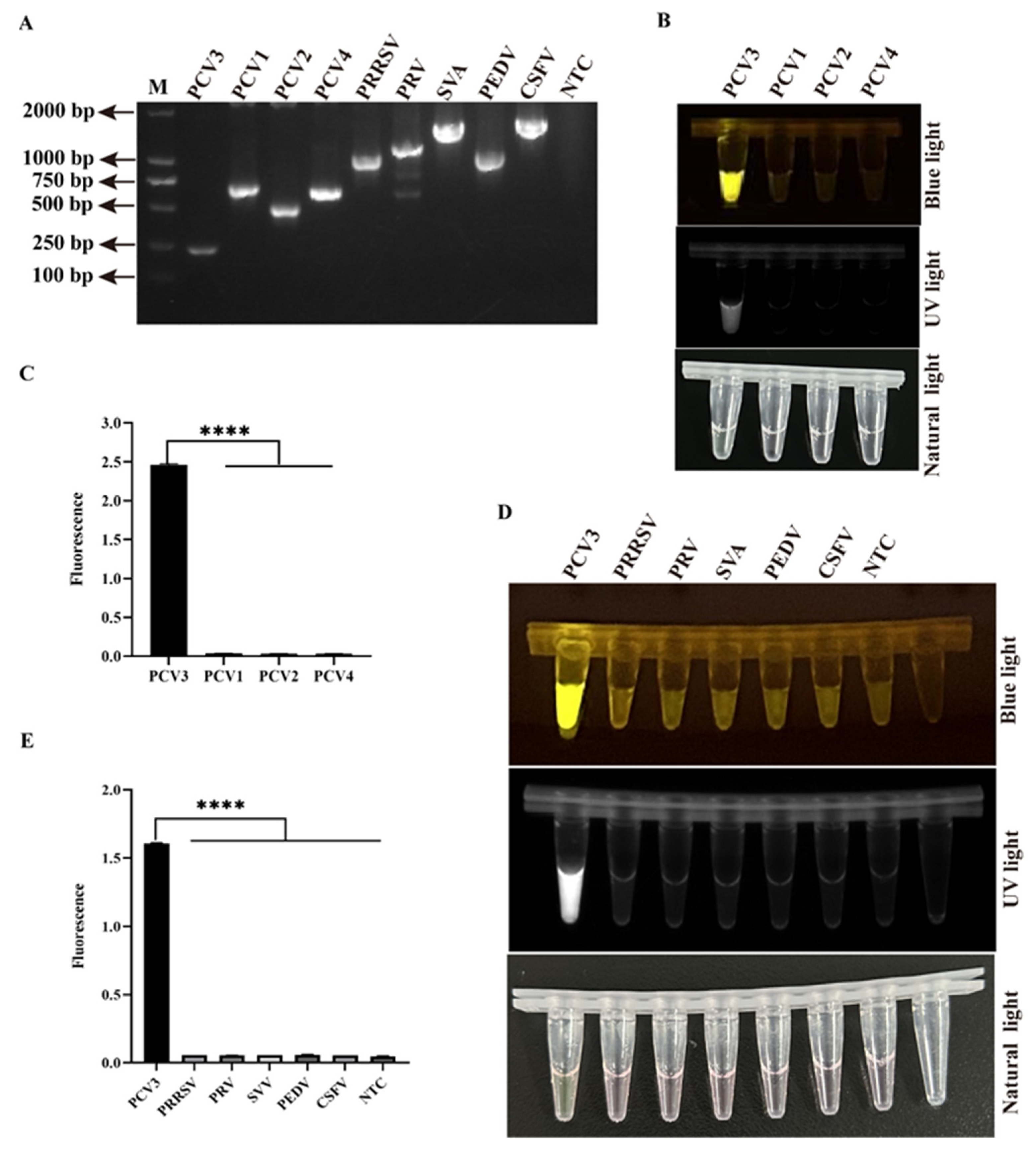
3.5. Sensitivity and Specificity of Two-Pot RPA-CRISPR/Cas12a-PCV3 Detection Method
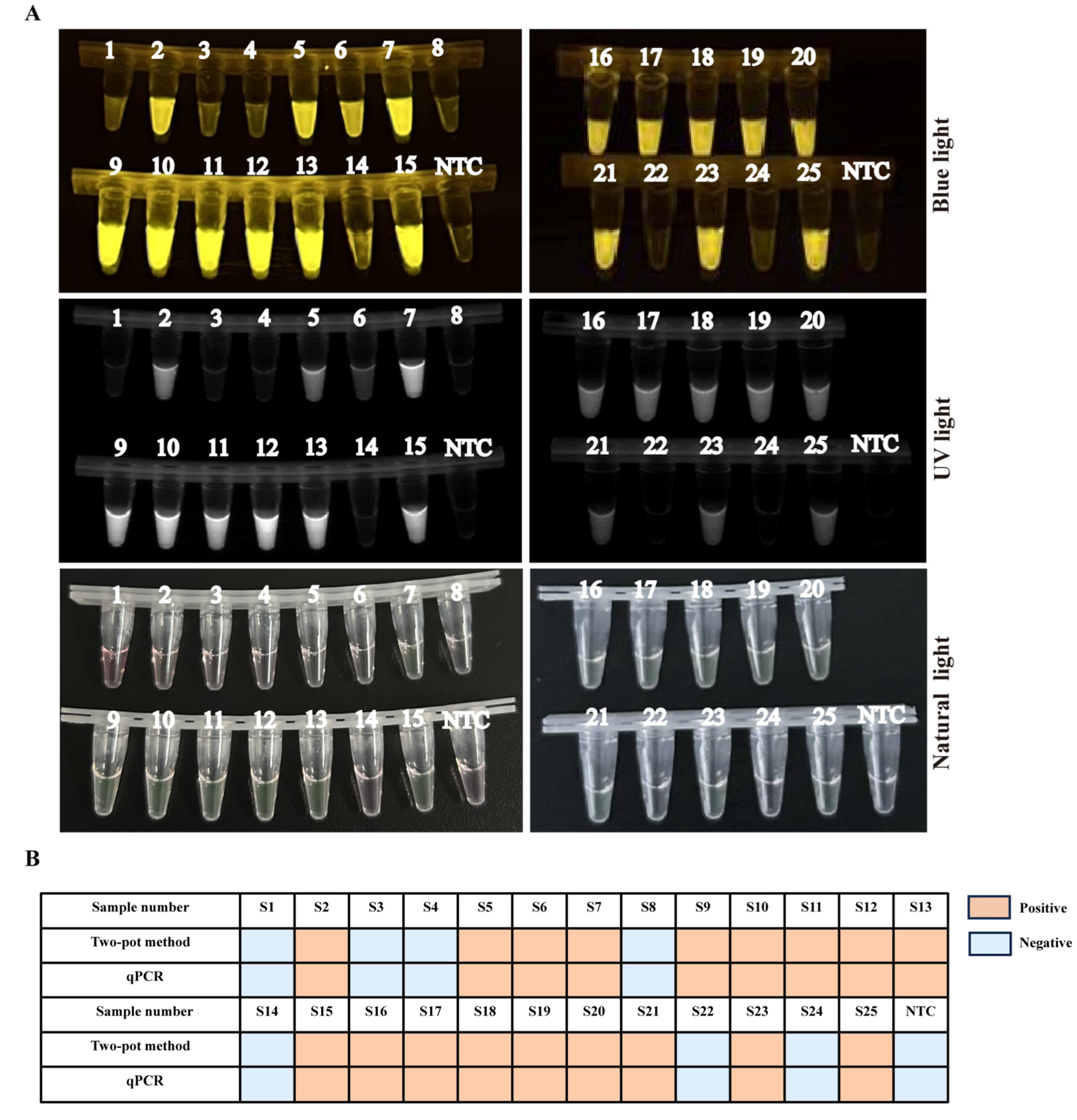
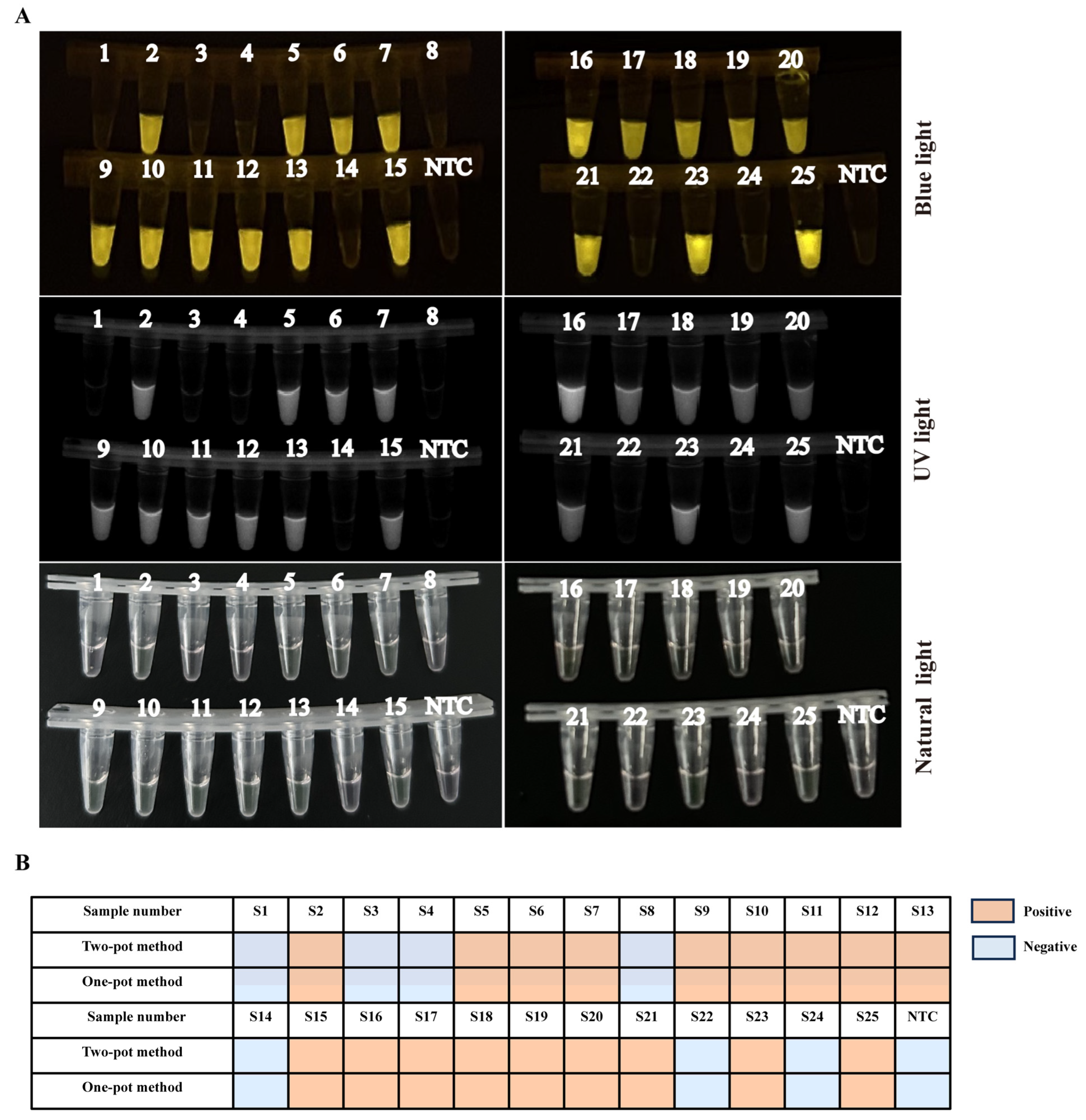
3.6. Evaluating Consistency between Two-Pot RPA-CRISPR/Cas12a-PCV3 Detection Method and Real-Time Quantitative PCR (qPCR) Method
3.7. The Clinical Samples Detection of One-Pot RPA-CRISPR/Cas12a-PCV3 Method
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Tischer, I.; Gelderblom, H.; Vettermann, W.; Koch, M.A. A very small porcine virus with circular single-stranded DNA. Nature 1982, 295, 64–66. [Google Scholar] [CrossRef]
- Opriessnig, T.; Karuppannan, A.K.; Castro, A.; Xiao, C.T. Porcine circoviruses: Current status, knowledge gaps and challenges. Virus Res. 2020, 286, 198044. [Google Scholar] [CrossRef] [PubMed]
- Tischer, I.; Rasch, R.; Tochtermann, G. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl Bakteriol. Orig. A Med. Mikrobiol. Und Parasitol. 1974, 226, 153–167. [Google Scholar]
- An, D.J.; Roh, I.S.; Song, D.S.; Park, C.K.; Park, B.K. Phylogenetic characterization of porcine circovirus type 2 in PMWS and PDNS Korean pigs between 1999 and 2006. Virus Res. 2007, 129, 115–122. [Google Scholar] [CrossRef]
- Welti, S.; Sydler, T.; Wiederkehr, D.; Pospischil, A.; Hässig, M.; Bürgi, E.; Sidler, X. Postweaning multisystemic wasting syndrome (PMWS) and porcine dermatitis and nephropathy syndrome (PDNS) in Switzerland in the years 2003–2006. Schweiz. Arch. Tierheilkd. 2012, 154, 417–427. [Google Scholar] [CrossRef]
- Palinski, R.; Piñeyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B.M. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 2017, 91, 10–1128. [Google Scholar] [CrossRef]
- Nawagitgul, P.; Morozov, I.; Bolin, S.R.; Harms, P.A.; Sorden, S.D.; Paul, P.S. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 2000, 81, 2281–2287. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Fernández-Pinero, J.; Arias, M. African swine fever (ASF) diagnosis, an essential tool in the epidemiological investigation. Virus Res. 2019, 271, 197676. [Google Scholar] [CrossRef]
- Petrovan, V.; Yuan, F.; Li, Y.; Shang, P.; Murgia, M.V.; Misra, S.; Rowland, R.R.R.; Fang, Y. Development and characterization of monoclonal antibodies against p30 protein of African swine fever virus. Virus Res. 2019, 269, 197632. [Google Scholar] [CrossRef]
- Kralik, P.; Ricchi, M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef]
- Espy, M.J.; Uhl, J.R.; Sloan, L.M.; Buckwalter, S.P.; Jones, M.F.; Vetter, E.A.; Yao, J.D.; Wengenack, N.L.; Rosenblatt, J.E.; Cockerill, F.R., 3rd; et al. Real-time PCR in clinical microbiology: Applications for routine laboratory testing. Clin. Microbiol. Rev. 2006, 19, 165–256. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sun, R.; Yang, M.; Peng, S.; Cheng, Y.; Chen, C. Conformational Dynamics and Cleavage Sites of Cas12a Are Modulated by Complementarity between crRNA and DNA. Iscience 2019, 19, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Ren, K.; Qiu, X.; Zheng, J.; Guo, M.; Guan, X.; Liu, H.; Li, N.; Zhang, B.; Yang, D.; et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 2016, 532, 522–526. [Google Scholar] [CrossRef]
- Swarts, D.C.; van der Oost, J.; Jinek, M. Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a. Mol. Cell 2017, 66, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Mei, H.; Zhou, M.; Fu, Z.F.; Han, H.; Bi, D.; Peng, F.; Zhao, L. Development of A Super-Sensitive Diagnostic Method for African Swine Fever Using CRISPR Techniques. Virol. Sin. 2021, 36, 220–230. [Google Scholar] [CrossRef]
- Huang, M.; Liu, S.; Xu, Y.; Li, A.; Wu, W.; Liang, M.; Niu, G.; Wang, Z.; Wang, T. CRISPR/Cas12a Technology Combined With RPA for Rapid and Portable SFTSV Detection. Front. Microbiol. 2022, 13, 754995. [Google Scholar] [CrossRef]
- Zhang, K.; Sun, Z.; Shi, K.; Yang, D.; Bian, Z.; Li, Y.; Gou, H.; Jiang, Z.; Yang, N.; Chu, P.; et al. RPA-CRISPR/Cas12a-Based Detection of Haemophilus parasuis. Animals 2023, 13, 3317. [Google Scholar] [CrossRef]
- Luo, M.; Pan, Y.; He, Y.; Ruhan, A.; Wu, C.; Huang, B.; Lu, R.; Zhao, L.; Peng, B.; Ye, F.; et al. Detecting SARS-CoV-2 BA.2, BA.4, and BA.5 Variants Utilizing a Robust RT-RPA-CRISPR/Cas12a-Based Method—China, 2023. China CDC Wkly. 2023, 5, 584–591. [Google Scholar] [CrossRef]
- Zhang, D.; Jiang, S.; Xia, N.; Zhang, Y.; Zhang, J.; Liu, A.; Zhang, C.; Chen, N.; Meurens, F.; Zheng, W.; et al. Rapid Visual Detection of African Swine Fever Virus with a CRISPR/Cas12a Lateral Flow Strip Based on Structural Protein Gene D117L. Animals 2023, 13, 3712. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Ding, X.; Li, Z.; Zhao, H.; Cooper, K.; Liu, C. Dynamic Aqueous Multiphase Reaction System for One-Pot CRISPR-Cas12a-Based Ultrasensitive and Quantitative Molecular Diagnosis. Anal. Chem. 2020, 92, 8561–8568. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Hu, Y.; Fan, Z.; Huang, B.; Wei, L.; Xie, Y.; Huang, Y.; Mei, S.; Wang, L.; Wang, L.; et al. Rapid and sensitive one-tube detection of mpox virus using RPA-coupled CRISPR-Cas12 assay. Cell Rep. Methods 2023, 3, 100620. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Hu, J.; Bao, Z.; Wang, M. An isothermal enzymatic recombinase amplification (ERA) assay for rapid and accurate detection of Enterocytozoon hepatopenaei infection in shrimp. J. Invertebr. Pathol. 2023, 197, 107895. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef]
- Rolando, J.C.; Jue, E.; Barlow, J.T.; Ismagilov, R.F. Real-time kinetics and high-resolution melt curves in single-molecule digital LAMP to differentiate and study specific and non-specific amplification. Nucleic Acids Res. 2020, 48, e42. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Wang, J.; Liu, G. CRISPR/Cas Systems towards Next-Generation Biosensing. Trends Biotechnol. 2019, 37, 730–743. [Google Scholar] [CrossRef]
- Chertow, D.S. Next-generation diagnostics with CRISPR. Science 2018, 360, 381–382. [Google Scholar] [CrossRef]
- Li, Y.; Liu, L.; Liu, G. CRISPR/Cas Multiplexed Biosensing: A Challenge or an Insurmountable Obstacle? Trends Biotechnol. 2019, 37, 792–795. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Wang, C.; Zhang, Y.; Du, F.; Zhang, Z.; Xiao, F.; Wang, J.; Lin, X.; Wu, S. Establishment of a sensitive TaqMan-based real-time PCR assay for porcine circovirus type 3 and its application in retrospective quarantine of imported boars to China. Vet. Med. Sci. 2019, 5, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Wu, X.; Shi, J.; Peng, Z.; Gao, M.; Xin, C.; Liu, Y.; Wang, S.; Xu, S.; Han, H.; et al. Rapid specific and visible detection of porcine circovirus type 3 using loop-mediated isothermal amplification (LAMP). Transbound. Emerg. Dis. 2018, 65, 597–601. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Zhang, R.; Han, Q.; Wang, J.; Liu, L.; Li, R.; Yuan, W. Recombinase polymerase amplification assay for rapid detection of porcine circovirus 3. Mol. Cell. Probes 2017, 36, 58–61. [Google Scholar] [CrossRef] [PubMed]
| Sample Types | Sample Numbers | RPA-CRISPR/Cas12a | qPCR |
|---|---|---|---|
| Spleen | 6 | 6/6 | 6/6 |
| Lung | 7 | 5/7 | 5/7 |
| Kidney | 2 | 1/2 | 1/2 |
| Submandibular lymph node | 3 | 3/3 | 3/3 |
| Inguinal lymph node | 7 | 3/7 | 3/7 |
| Positive rates | 72% | 72% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, G.; Yang, X.; Li, Z.; Mao, J.; Zeng, P.; Wang, D.; Wu, Z.; Liu, C.; Qiu, Y.; Cui, Y.; et al. Recombinant Polymerase Amplification Coupled with CRISPR/Cas12a Detection System for Rapid Visual Detection of Porcine Circovirus 3. Animals 2024, 14, 2527. https://doi.org/10.3390/ani14172527
Jiang G, Yang X, Li Z, Mao J, Zeng P, Wang D, Wu Z, Liu C, Qiu Y, Cui Y, et al. Recombinant Polymerase Amplification Coupled with CRISPR/Cas12a Detection System for Rapid Visual Detection of Porcine Circovirus 3. Animals. 2024; 14(17):2527. https://doi.org/10.3390/ani14172527
Chicago/Turabian StyleJiang, Genghong, Xiaoyu Yang, Zhaoyang Li, Jingyu Mao, Penghui Zeng, Dedong Wang, Zhi Wu, Changzhe Liu, Yonghui Qiu, Yongqiu Cui, and et al. 2024. "Recombinant Polymerase Amplification Coupled with CRISPR/Cas12a Detection System for Rapid Visual Detection of Porcine Circovirus 3" Animals 14, no. 17: 2527. https://doi.org/10.3390/ani14172527





