Transcriptomic Profiles of Pectoralis major Muscles Affected by Spaghetti Meat and Woody Breast in Broiler Chickens
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Procurement and Classification
2.2. RNA Isolation, Quantification, and Quality Assessment
2.3. Library Preparation and Sequencing
2.4. Sequence Quality Assessment, Alignment, and DEG Analysis
2.5. Pathway Enrichment Analysis
3. Results
3.1. Sequence Quality Assessment and Alignment
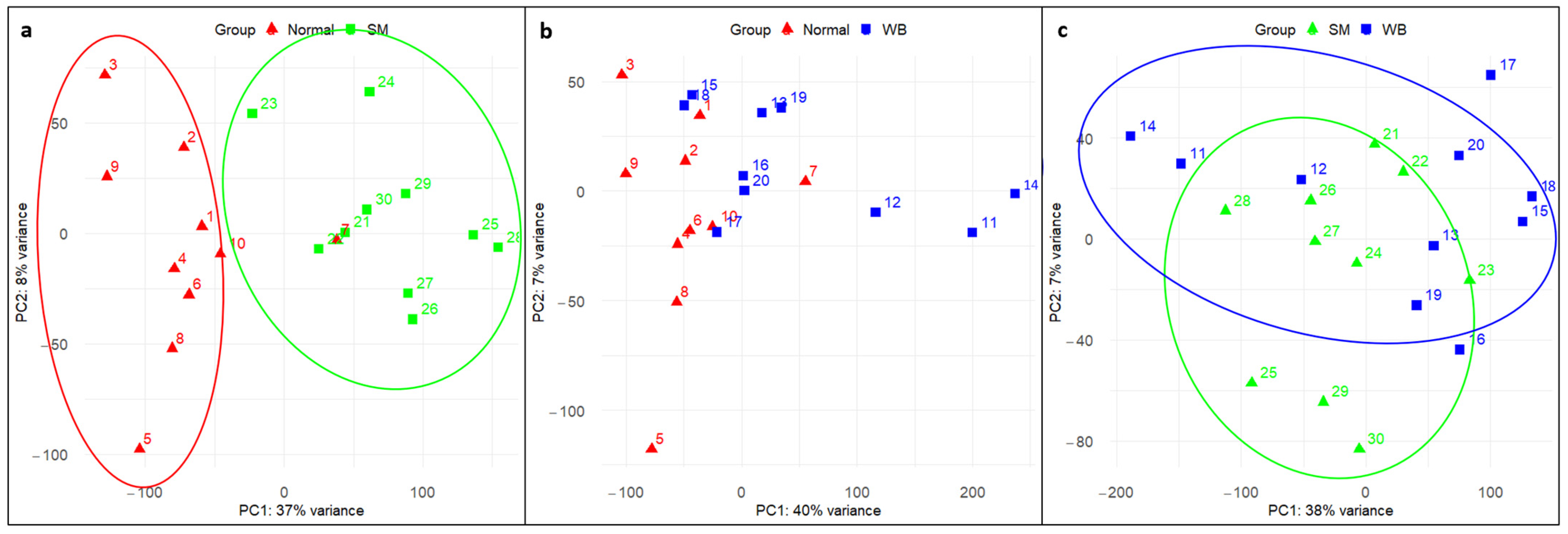
3.2. Principal Component Analysis (PCA)
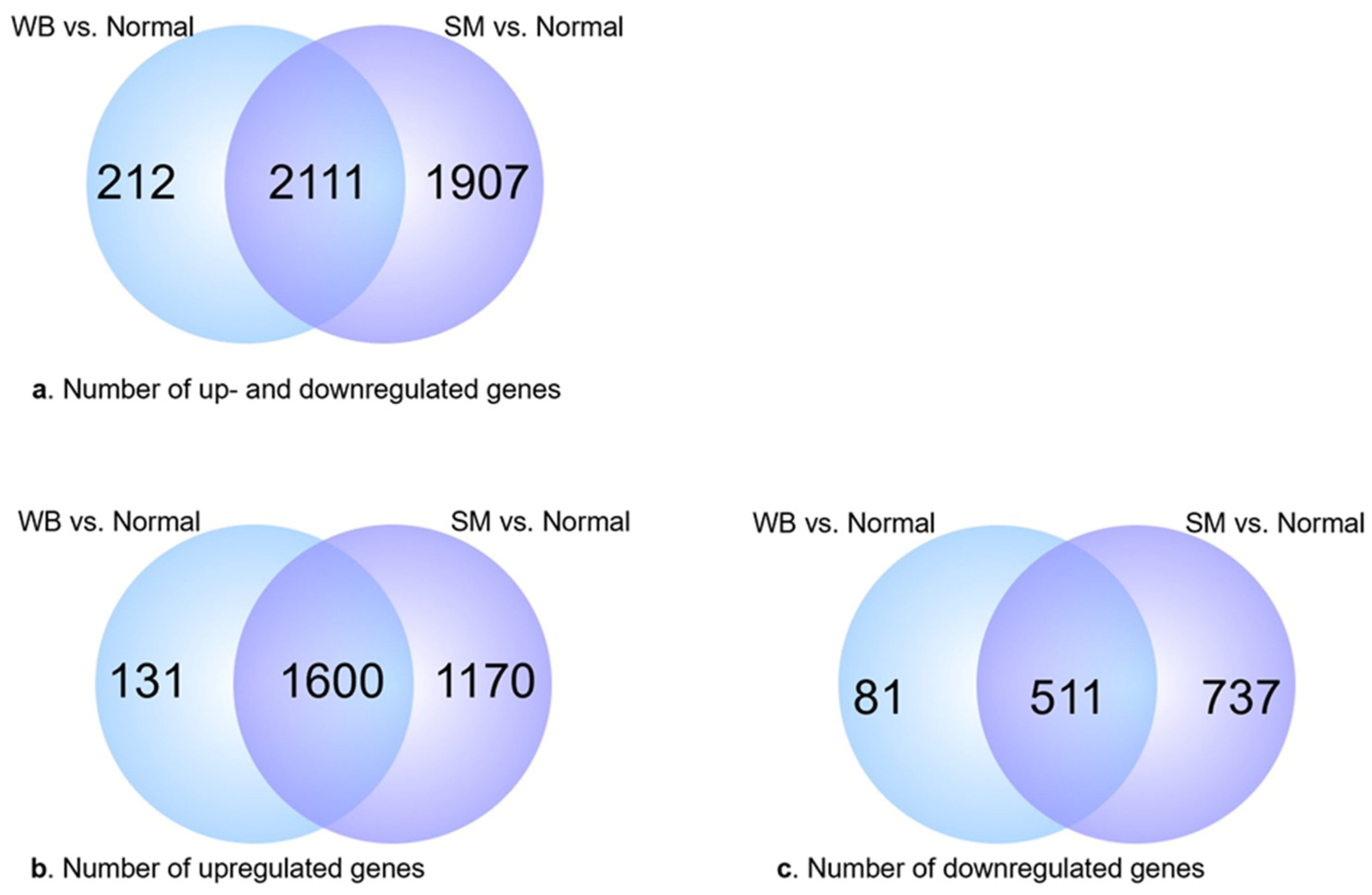
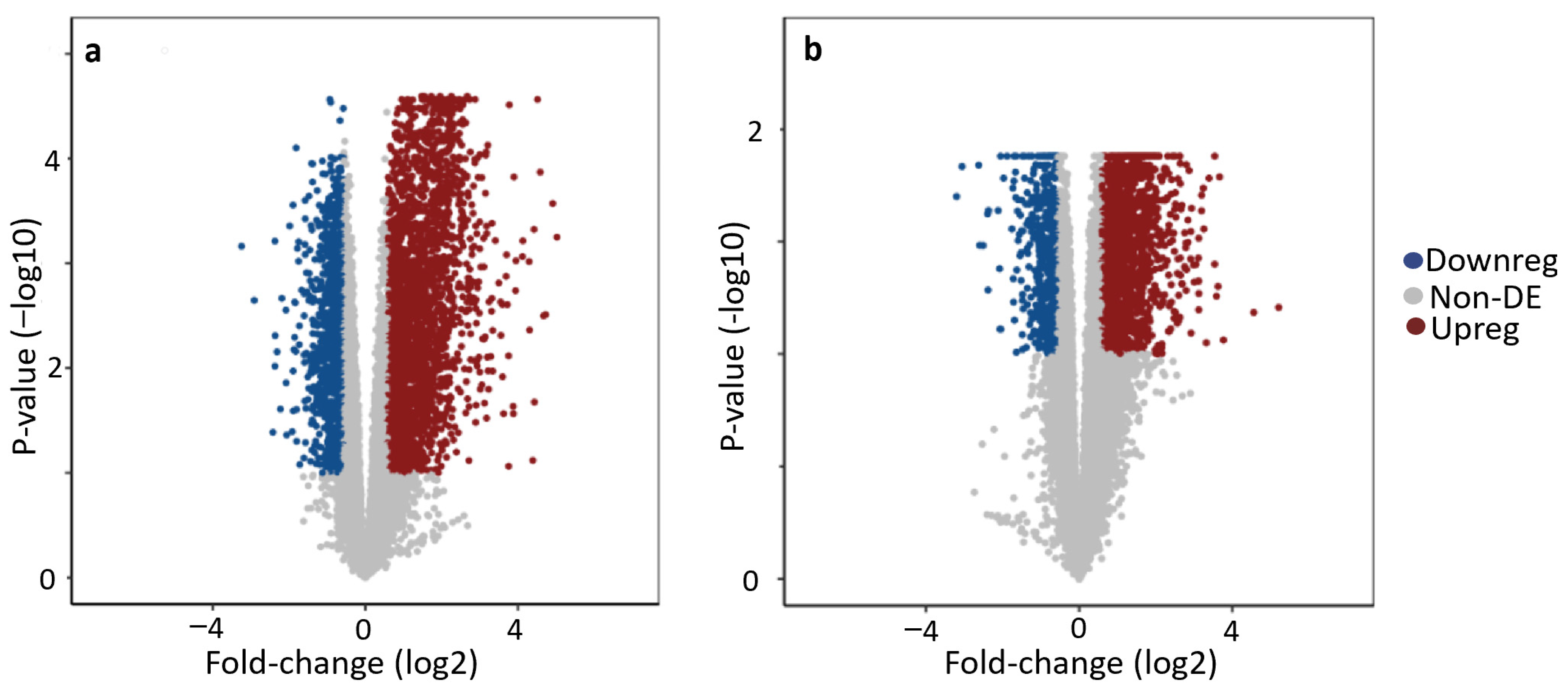
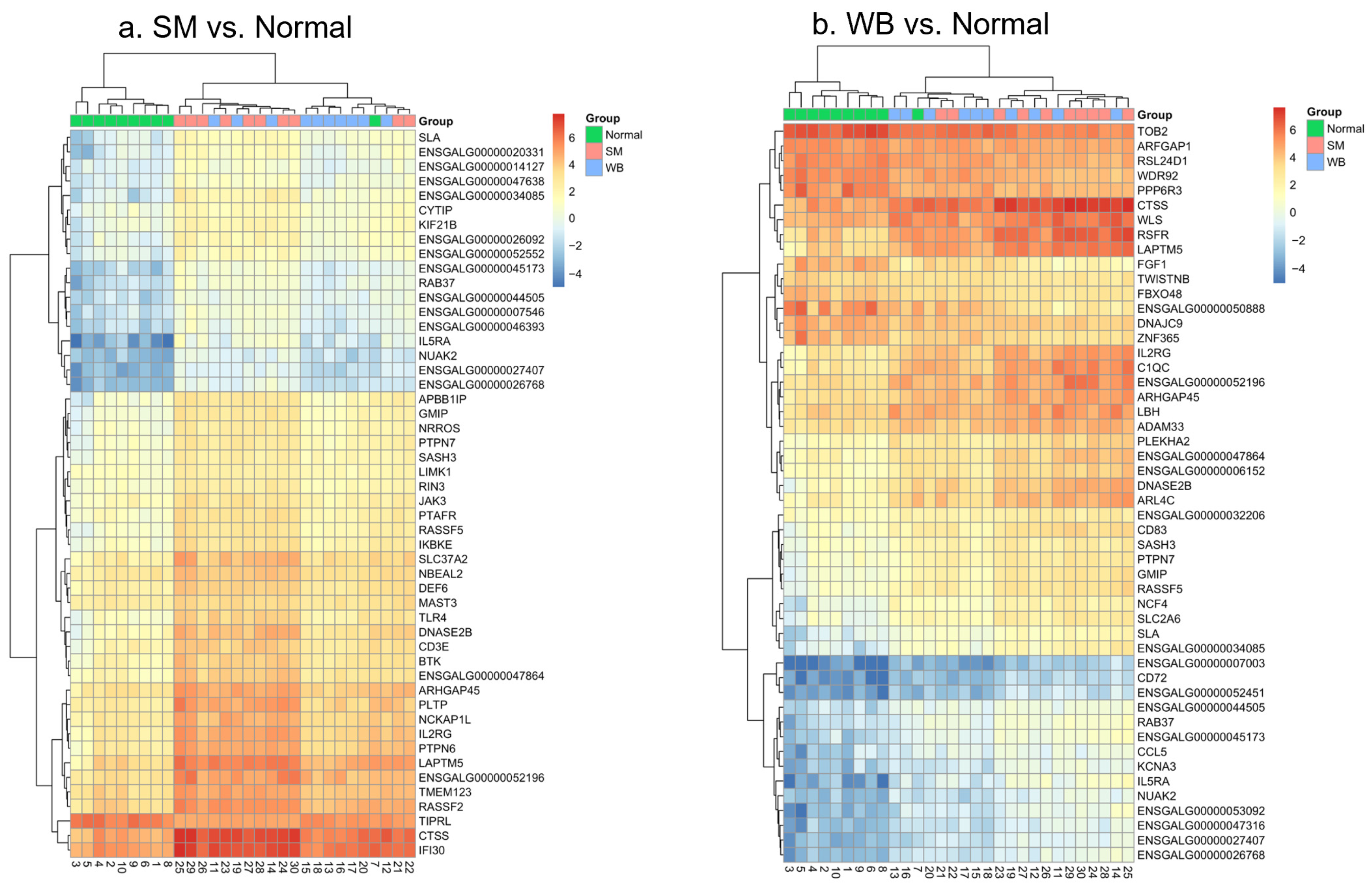
3.3. Analysis of Differentially Expressed Genes (DEGs)
3.4. Collagen-Related DEGs
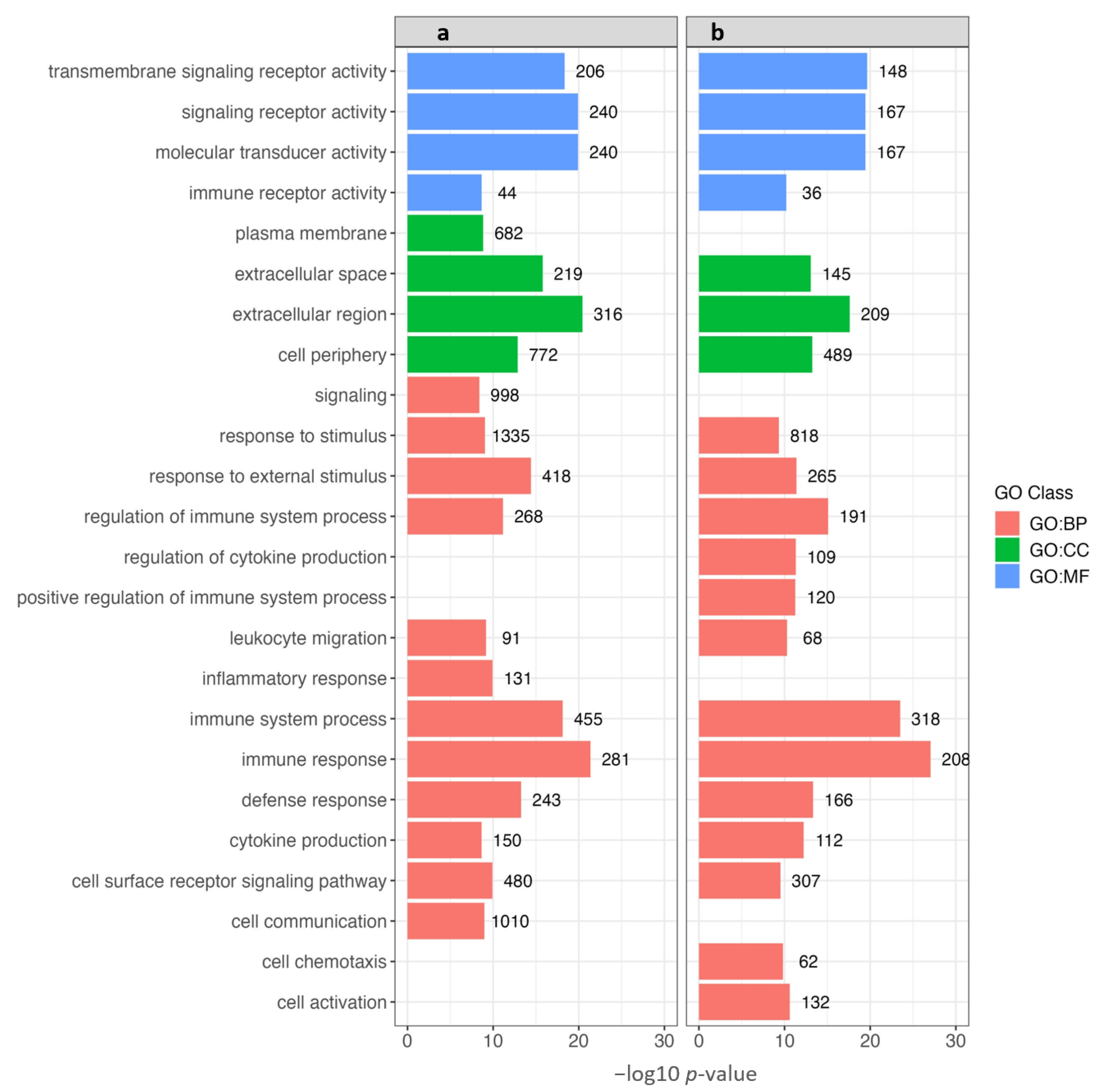
3.5. Gene Ontology (GO) Enrichment Analysis
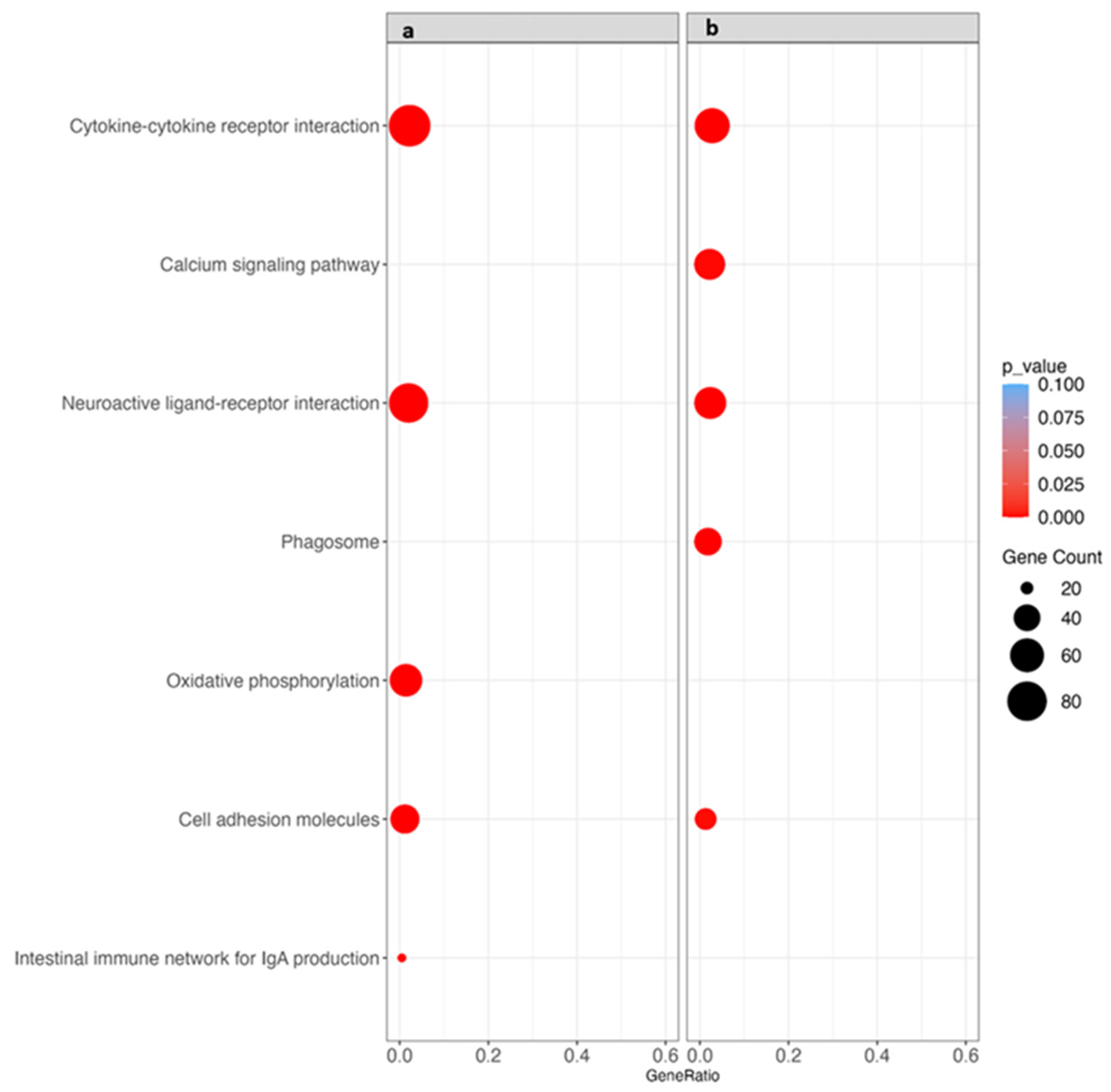
3.6. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis
4. Discussion
4.1. Inflammation and Cytokine Signaling
4.2. Extracellular Space and Collagen
4.3. Tissue Morphogenesis and Proliferation
4.4. Hypoxia and Oxidative Stress
4.5. Calcium Signaling and Phagosome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baldi, G.; Soglia, F.; Laghi, L.; Tappi, S.; Rocculi, P.; Tavaniello, S.; Prioriello, D.; Mucci, R.; Maiorano, G.; Petracci, M. Comparison of Quality Traits among Breast Meat Affected by Current Muscle Abnormalities. Food Res. Int. 2019, 115, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Sihvo, H.-K.; Lindén, J.; Airas, N.; Immonen, K.; Valaja, J.; Puolanne, E. Wooden Breast Myodegeneration of Pectoralis major Muscle Over the Growth Period in Broilers. Vet. Pathol. 2017, 54, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Che, S.; Wang, C.; Varga, C.; Barbut, S.; Susta, L. Prevalence of Breast Muscle Myopathies (Spaghetti Meat, Woody Breast, White Striping) and Associated Risk Factors in Broiler Chickens from Ontario Canada. PLoS ONE 2022, 17, e0267019. [Google Scholar] [CrossRef] [PubMed]
- Baldi, G.; Soglia, F.; Petracci, M. Current Status of Poultry Meat Abnormalities. Meat Muscle Biol. 2020, 4, 1–7. [Google Scholar] [CrossRef]
- Dalgaard, L.B.; Rasmussen, M.K.; Bertram, H.C.; Jensen, J.A.; Møller, H.S.; Aaslyng, M.D.; Hejbøl, E.K.; Pedersen, J.R.; Elsser-Gravesen, D.; Young, J.F. Classification of Wooden Breast Myopathy in Chicken Pectoralis major by a Standardised Method and Association with Conventional Quality Assessments. Int. J. Food Sci. Tech. 2018, 53, 1744–1752. [Google Scholar] [CrossRef]
- Kuttappan, V.A.; Lee, Y.S.; Erf, G.F.; Meullenet, J.-F.C.; McKee, S.R.; Owens, C.M. Consumer Acceptance of Visual Appearance of Broiler Breast Meat with Varying Degrees of White Striping. Poult. Sci. 2012, 91, 1240–1247. [Google Scholar] [CrossRef] [PubMed]
- Barbut, S. Recent Myopathies in Broiler’s Breast Meat Fillets. World’s Poult. Sci. J. 2019, 75, 559–582. [Google Scholar] [CrossRef]
- Petracci, M.; Soglia, F.; Madruga, M.; Carvalho, L.; Ida, E.; Estévez, M. Wooden-Breast, White Striping, and Spaghetti Meat: Causes, Consequences and Consumer Perception of Emerging Broiler Meat Abnormalities. Comp. Rev. Food Sci. Food Safe 2019, 18, 565–583. [Google Scholar] [CrossRef]
- Baldi, G.; Soglia, F.; Mazzoni, M.; Sirri, F.; Canonico, L.; Babini, E.; Laghi, L.; Cavani, C.; Petracci, M. Implications of White Striping and Spaghetti Meat Abnormalities on Meat Quality and Histological Features in Broilers. Animal 2018, 12, 164–173. [Google Scholar] [CrossRef]
- Che, S.; Wang, C.; Iverson, M.; Varga, C.; Barbut, S.; Bienzle, D.; Susta, L. Characteristics of Broiler Chicken Breast Myopathies (Spaghetti Meat, Woody Breast, White Striping) in Ontario, Canada. Poult. Sci. 2022, 101, 101747. [Google Scholar] [CrossRef]
- Chen, L.R.; Suyemoto, M.M.; Sarsour, A.H.; Cordova, H.A.; Oviedo-Rondón, E.O.; Wineland, M.; Barnes, H.J.; Borst, L.B. Temporal Characterization of Wooden Breast Myopathy (“Woody Breast”) Severity and Correlation with Growth Rate and Lymphocytic Phlebitis in Three Commercial Broiler Strains and a Random-Bred Broiler Strain. Avian Pathol. 2019, 48, 319–328. [Google Scholar] [CrossRef]
- Pampouille, E.; Hennequet-Antier, C.; Praud, C.; Juanchich, A.; Brionne, A.; Godet, E.; Bordeau, T.; Fagnoul, F.; Le Bihan-Duval, E.; Berri, C. Differential Expression and Co-Expression Gene Network Analyses Reveal Molecular Mechanisms and Candidate Biomarkers Involved in Breast Muscle Myopathies in Chicken. Sci. Rep. 2019, 9, 14905. [Google Scholar] [CrossRef] [PubMed]
- Lorenzi, M.; Mudalal, S.; Cavani, C.; Petracci, M. Incidence of White Striping under Commercial Conditions in Medium and Heavy Broiler Chickens in Italy. J. Appl. Poult. Res. 2014, 23, 754–758. [Google Scholar] [CrossRef]
- Kuttappan, V.A.; Brewer, V.B.; Mauromoustakos, A.; McKee, S.R.; Emmert, J.L.; Meullenet, J.F.; Owens, C.M. Estimation of Factors Associated with the Occurrence of White Striping in Broiler Breast Fillets. Poult. Sci. 2013, 92, 811–819. [Google Scholar] [CrossRef]
- Caldas-Cueva, J.P.; Owens, C.M. A Review on the Woody Breast Condition, Detection Methods, and Product Utilization in the Contemporary Poultry Industry. J. Anim. Sci. 2020, 98, skaa207. [Google Scholar] [CrossRef] [PubMed]
- Bailey, R.A.; Watson, K.A.; Bilgili, S.F.; Avendano, S. The Genetic Basis of Pectoralis major Myopathies in Modern Broiler Chicken Lines. Poult. Sci. 2015, 94, 2870–2879. [Google Scholar] [CrossRef] [PubMed]
- Bailey, R.A.; Souza, E.; Avendano, S. Characterising the Influence of Genetics on Breast Muscle Myopathies in Broiler Chickens. Front. Physiol. 2020, 11, 1041. [Google Scholar] [CrossRef]
- Santos, M.N.; Rothschild, D.; Widowski, T.M.; Barbut, S.; Kiarie, E.G.; Mandell, I.; Guerin, M.T.; Edwards, A.M.; Torrey, S. In Pursuit of a Better Broiler: Carcass Traits and Muscle Myopathies in Conventional and Slower-Growing Strains of Broiler Chickens. Poult. Sci. 2021, 100, 101309. [Google Scholar] [CrossRef]
- Lake, J.A.; Papah, M.B.; Abasht, B. Increased Expression of Lipid Metabolism Genes in Early Stages of Wooden Breast Links Myopathy of Broilers to Metabolic Syndrome in Humans. Genes 2019, 10, 746. [Google Scholar] [CrossRef]
- Papah, M.B.; Brannick, E.M.; Schmidt, C.J.; Abasht, B. Gene Expression Profiling of the Early Pathogenesis of Wooden Breast Disease in Commercial Broiler Chickens Using RNA-Sequencing. PLoS ONE 2018, 13, e0207346. [Google Scholar] [CrossRef]
- Mutryn, M.F.; Brannick, E.M.; Fu, W.; Lee, W.R.; Abasht, B. Characterization of a Novel Chicken Muscle Disorder through Differential Gene Expression and Pathway Analysis Using RNA-Sequencing. BMC Genom. 2015, 16, 399. [Google Scholar] [CrossRef] [PubMed]
- Soglia, F.; Mazzoni, M.; Zappaterra, M.; Di Nunzio, M.; Babini, E.; Bordini, M.; Sirri, F.; Clavenzani, P.; Davoli, R.; Petracci, M. Distribution and Expression of Vimentin and Desmin in Broiler Pectoralis major Affected by the Growth-Related Muscular Abnormalities. Front. Physiol. 2020, 10, 1581. [Google Scholar] [CrossRef] [PubMed]
- Baldi, G.; Soglia, F.; Petracci, M. Spaghetti Meat Abnormality in Broilers: Current Understanding and Future Research Directions. Front. Physiol. 2021, 12, 684497. [Google Scholar] [CrossRef] [PubMed]
- Pham, P.H.; Tockovska, T.; Leacy, A.; Iverson, M.; Ricker, N.; Susta, L. Transcriptome Analysis of Duck and Chicken Brains Infected with Aquatic Bird Bornavirus-1 (ABBV-1). Viruses 2022, 14, 2211. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. G:Profiler—A Web Server for Functional Interpretation of Gene Lists (2016 Update). Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef]
- Tonniges, J.R.; Clark, D.L.; Velleman, S.G. The Effect of the Wooden Breast Fibrotic Myopathy in Broilers on Fibrillar Collagen Organization and Decorin-Collagen Binding. Avian Dis. 2018, 63, 48–60. [Google Scholar] [CrossRef]
- Clark, D.L.; Velleman, S.G. Spatial Influence on Breast Muscle Morphological Structure, Myofiber Size, and Gene Expression Associated with the Wooden Breast Myopathy in Broilers. Poult. Sci. 2017, 95, 2930–2945. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T. Do Results Obtained with RNA-Sequencing Require Independent Verification? Biofilm 2021, 3, 100043. [Google Scholar] [CrossRef] [PubMed]
- SEQC/MAQC-III Consortium. A Comprehensive Assessment of RNA-Seq Accuracy, Reproducibility and Information Content by the Sequencing Quality Control Consortium. Nat. Biotechnol. 2014, 32, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Sihvo, H.-K.; Immonen, K.; Puolanne, E. Myodegeneration With Fibrosis and Regeneration in the Pectoralis major Muscle of Broilers. Vet. Pathol. 2014, 51, 619–623. [Google Scholar] [CrossRef] [PubMed]
- Papah, M.B.; Brannick, E.M.; Schmidt, C.J.; Abasht, B. Evidence and Role of Phlebitis and Lipid Infiltration in the Onset and Pathogenesis of Wooden Breast Disease in Modern Broiler Chickens. Avian Pathol. 2017, 46, 623–643. [Google Scholar] [CrossRef] [PubMed]
- Kong, B.W.; Hudson, N.; Seo, D.; Lee, S.; Khatri, B.; Lassiter, K.; Cook, D.; Piekarski, A.; Dridi, S.; Anthony, N.; et al. RNA Sequencing for Global Gene Expression Associated with Muscle Growth in a Single Male Modern Broiler Line Compared to a Foundational Barred Plymouth Rock Chicken Line. BMC Genom. 2017, 18, 82. [Google Scholar] [CrossRef] [PubMed]
- Mann, C.J.; Perdiguero, E.; Kharraz, Y.; Aguilar, S.; Pessina, P.; Serrano, A.L.; Muñoz-Cánoves, P. Aberrant Repair and Fibrosis Development in Skeletal Muscle. Skelet. Muscle 2011, 1, 21. [Google Scholar] [CrossRef] [PubMed]
- Sugg, K.B.; Korn, M.A.; Sarver, D.C.; Markworth, J.F.; Mendias, C.L. Inhibition of Platelet-Derived Growth Factor Signaling Prevents Muscle Fiber Growth during Skeletal Muscle Hypertrophy. FEBS Lett. 2017, 591, 801–809. [Google Scholar] [CrossRef]
- Burkhardt, N.B.; Röll, S.; Staudt, A.; Elleder, D.; Härtle, S.; Costa, T.; Alber, A.; Stevens, M.P.; Vervelde, L.; Schusser, B.; et al. The Long Pentraxin PTX3 Is of Major Importance among Acute Phase Proteins in Chickens. Front. Immunol. 2019, 10, 124. [Google Scholar] [CrossRef]
- Deban, L.; Russo, R.C.; Sironi, M.; Moalli, F.; Scanziani, M.; Zambelli, V.; Cuccovillo, I.; Bastone, A.; Gobbi, M.; Valentino, S.; et al. Regulation of Leukocyte Recruitment by the Long Pentraxin PTX3. Nat. Immunol. 2010, 11, 328–334. [Google Scholar] [CrossRef]
- Jing, J.; Yang, I.V.; Hui, L.; Patel, J.A.; Evans, C.M.; Prikeris, R.; Kobzik, L.; O’Connor, B.P.; Schwartz, D.A. Role of Macrophage Receptor with Collagenous Structure in Innate Immune Tolerance. J. Immunol. 2013, 190, 6360–6367. [Google Scholar] [CrossRef] [PubMed]
- Van Anken, E.; Pena, F.; Hafkemeijer, N.; Christis, C.; Romijn, E.P.; Grauschopf, U.; Oorschot, V.M.J.; Pertel, T.; Engels, S.; Ora, A.; et al. Efficient IgM Assembly and Secretion Require the Plasma Cell Induced Endoplasmic Reticulum Protein pERp1. Proc. Natl. Acad. Sci. USA 2009, 106, 17019–17024. [Google Scholar] [CrossRef] [PubMed]
- Maharjan, P.; Owens, C.M.; Coon, C. In-Vivo Intramuscular Collagen Synthesis, Muscle Fiber Growth and Histomorphology of Pectoralis major of a Fast-Growing Broiler Strain Gallus Gallus Domesticus. Front. Vet. Sci. 2020, 6, 470. [Google Scholar] [CrossRef] [PubMed]
- Velleman, S.G.; Clark, D.L.; Tonniges, J.R. Fibrillar Collagen Organization Associated with Broiler Wooden Breast Fibrotic Myopathy. Avian Dis. 2017, 61, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Velleman, S.G.; Clark, D.L.; Tonniges, J.R. The Effect of the Wooden Breast Myopathy on Sarcomere Structure and Organization. Avian Dis. 2018, 62, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Soglia, F.; Laghi, L.; Canonico, L.; Cavani, C.; Petracci, M. Functional Property Issues in Broiler Breast Meat Related to Emerging Muscle Abnormalities. Food Res. Int. 2016, 89, 1071–1076. [Google Scholar] [CrossRef]
- Velleman, S.G. Pectoralis major (Breast) Muscle Extracellular Matrix Fibrillar Collagen Modifications Associated with the Wooden Breast Fibrotic Myopathy in Broilers. Front. Physiol. 2020, 11, 461. [Google Scholar] [CrossRef]
- Sanden, K.W.; Böcker, U.; Ofstad, R.; Pedersen, M.E.; Høst, V.; Afseth, N.K.; Rønning, S.B.; Pleshko, N. Characterization of Collagen Structure in Normal, Wooden Breast and Spaghetti Meat Chicken Fillets by FTIR Microspectroscopy and Histology. Foods 2021, 10, 548. [Google Scholar] [CrossRef]
- Lim, T.S.; Goh, J.K.H.; Mortellaro, A.; Lim, C.T.; Hämmerling, G.J.; Ricciardi-Castagnoli, P. CD80 and CD86 Differentially Regulate Mechanical Interactions of T-Cells with Antigen-Presenting Dendritic Cells and B-Cells. PLoS ONE 2012, 7, e45185. [Google Scholar] [CrossRef]
- Marckmann, S.; Wiesemann, E.; Hilse, R.; Trebst, C.; Stangel, M.; Windhagen, A. Interferon-β up-Regulates the Expression of Co-Stimulatory Molecules CD80, CD86 and CD40 on Monocytes: Significance for Treatment of Multiple Sclerosis. Clin. Exp. Immunol. 2004, 138, 499–506. [Google Scholar] [CrossRef]
- Marchesi, J.A.P.; Ibelli, A.M.G.; Peixoto, J.O.; Cantão, M.E.; Pandolfi, J.R.C.; Marciano, C.M.M.; Zanella, R.; Settles, M.L.; Coutinho, L.L.; Ledur, M.C. Whole Transcriptome Analysis of the Pectoralis major Muscle Reveals Molecular Mechanisms Involved with White Striping in Broiler Chickens. Poult. Sci. 2019, 98, 590–601. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Balasubramanian, B.; Guo, Y.; Qiu, S.J.; Jha, R.; Liu, W.C. Dietary Enteromorpha Polysaccharides Supplementation Improves Breast Muscle Yield and Is Associated with Modification of mRNA Transcriptome in Broiler Chickens. Front. Vet. Sci. 2021, 8, 663988. [Google Scholar] [CrossRef] [PubMed]
- Petri, B.; Bixel, M.G. Molecular Events during Leukocyte Diapedesis. FEBS J. 2006, 273, 4399–4407. [Google Scholar] [CrossRef] [PubMed]
- Lau, D.; Elezagic, D.; Hermes, G.; Mörgelin, M.; Wohl, A.P.; Koch, M.; Hartmann, U.; Höllriegl, S.; Wagener, R.; Paulsson, M.; et al. The Cartilage-Specific Lectin C-Type Lectin Domain Family 3 Member A (CLEC3A) Enhances Tissue Plasminogen Activator-Mediated Plasminogen Activation. J. Biol. Chem. 2018, 293, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Miki, M.; Oono, T.; Fujimori, N.; Takaoka, T.; Kawabe, K.; Miyasaka, Y.; Ohtsuka, T.; Saito, D.; Nakamura, M.; Ohkawa, Y.; et al. CLEC3A, MMP7, and LCN2 as Novel Markers for Predicting Recurrence in Resected G1 and G2 Pancreatic Neuroendocrine Tumors. Cancer Med. 2019, 8, 3748–3760. [Google Scholar] [CrossRef]
- Chen, X.; Ji, Y.; Feng, F.; Liu, Z.; Qian, L.; Shen, H.; Lao, L. C-Type Lectin Domain-Containing Protein CLEC3A Regulates Proliferation, Regeneration and Maintenance of Nucleus Pulposus Cells. Cell. Mol. Life Sci. 2022, 79, 435. [Google Scholar] [CrossRef]
- Guerin, C.; Holland, P.C. Synthesis and Secretion of Matrix-Degrading Metalloproteases by Human Skeletal Muscle Satellite Cells. Dev. Dyn. 1995, 202, 91–99. [Google Scholar] [CrossRef]
- Ohtake, Y.; Tojo, H.; Seiki, M. Multifunctional Roles of MT1-MMP in Myofiber Formation and Morphostatic Maintenance of Skeletal Muscle. J. Cell Sci. 2006, 119, 3822–3832. [Google Scholar] [CrossRef]
- Xiaoping, C.; Yong, L. Role of Matrix Metalloproteinases in Skeletal Muscle: Migration, Differentiation, Regeneration and Fibrosis. Cell Adhes. Migr. 2009, 3, 337–341. [Google Scholar] [CrossRef]
- Orbe, J.; Rodríguez, J.A.; Calvayrac, O.; Rodríguez-Calvo, R.; Rodríguez, C.; Roncal, C.; De Lizarrondo, S.M.; Barrenetxe, J.; Reverter, J.C.; Martínez-González, J.; et al. Matrix Metalloproteinase-10 Is Upregulated by Thrombin in Endothelial Cells and Increased in Patients with Enhanced Thrombin Generation. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 2109–2116. [Google Scholar] [CrossRef]
- Bobadilla, M.; Sáinz, N.; Rodriguez, J.; Abizanda, G.; Orbe, J.; De Martino, A.; Verdugo, J.G.; Páramo, J.A.; Prósper, F.; Pérez-Ruiz, A. MMP-10 Is Required for Efficient Muscle Regeneration in Mouse Models of Injury and Muscular Dystrophy. Stem Cells 2014, 32, 447–461. [Google Scholar] [CrossRef] [PubMed]
- Reinecke, H.; Robey, T.E.; Mignone, J.L.; Muskheli, V.; Bornstein, P.; Murry, C.E. Lack of Thrombospondin-2 Reduces Fibrosis and Increases Vascularity around Cardiac Cell Grafts. Cardiovasc. Pathol. 2013, 22, 91–95. [Google Scholar] [CrossRef]
- Wang, Y.; Zheng, K.; Chen, X.; Chen, R.; Zou, Y. Bioinformatics Analysis Identifies COL1A1, THBS2 and SPP1 as Potential Predictors of Patient Prognosis and Immunotherapy Response in Gastric Cancer. Biosci. Rep. 2021, 41, BSR20202564. [Google Scholar] [CrossRef]
- Brothers, B.; Zhuo, Z.; Papah, M.B.; Abasht, B. RNA-Seq Analysis Reveals Spatial and Sex Differences in Pectoralis major Muscle of Broiler Chickens Contributing to Difference in Susceptibility to Wooden Breast Disease. Front. Physiol. 2019, 10, 764. [Google Scholar] [CrossRef]
- Keever, M.R.; Zhang, P.; Bolt, C.R.; Antonson, A.M.; Rymut, H.E.; Caputo, M.P.; Houser, A.K.; Hernandez, A.G.; Southey, B.R.; Rund, L.A.; et al. Lasting and Sex-Dependent Impact of Maternal Immune Activation on Molecular Pathways of the Amygdala. Front. Neurosci. 2020, 14, 774. [Google Scholar] [CrossRef]
- Calhabeu, F.; Lafont, J.; Le Dreau, G.; Laurent, M.; Kazazian, C.; Schaeffer, L.; Martinerie, C.; Dubois, C. NOV/CCN3 Impairs Muscle Cell Commitment and Differentiation. Exp. Cell Res. 2006, 312, 1876–1889. [Google Scholar] [CrossRef] [PubMed]
- Wiriyasermkul, P.; Moriyama, S.; Nagamori, S. Membrane Transport Proteins in Melanosomes: Regulation of Ions for Pigmentation. Biochim. Biophys. Acta-Biomembr. 2020, 1862, 183318. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, R.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Iny Stein, T.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The Genecards Suite: From Gene Data Mining to Disease Genome Sequence Analysis. Available online: https://www.genecards.org/ (accessed on 18 December 2022).
- Dou, T.; Yan, S.; Liu, L.; Wang, K.; Jian, Z.; Xu, Z.; Zhao, J.; Wang, Q.; Sun, S.; Talpur, M.Z.; et al. Integrative Analysis of Transcriptomics and Metabolomics to Reveal the Melanogenesis Pathway of Muscle and Related Meat Characters in Wuliangshan Black-Boned Chickens. BMC Genom. 2022, 23, 173. [Google Scholar] [CrossRef]
- Rossi, A.C.; Mammucari, C.; Argentini, C.; Reggiani, C.; Schiaffino, S. Two Novel/Ancient Myosins in Mammalian Skeletal Muscles: MYH14/7b and MYH15 Are Expressed in Extraocular Muscles and Muscle Spindles. J. Physiol. 2010, 588, 353–364. [Google Scholar] [CrossRef]
- Praud, C.; Jimenez, J.; Pampouille, E.; Couroussé, N.; Godet, E.; Le Bihan-Duval, E.; Berri, C. Molecular Phenotyping of White Striping and Wooden Breast Myopathies in Chicken. Front. Physiol. 2020, 11, 633. [Google Scholar] [CrossRef]
- Kimura, S.; Kato, E. TAS2R Expression Profile in Brown Adipose, White Adipose, Skeletal Muscle, Small Intestine, Liver and Common Cell Lines Derived from Mice. Gene Rep. 2020, 20, 100763. [Google Scholar] [CrossRef]
- Dotson, C.D.; Zhang, L.; Xu, H.; Shin, Y.K.; Vigues, S.; Ott, S.H.; Elson, A.E.T.; Choi, H.J.; Shaw, H.; Egan, J.M.; et al. Bitter Taste Receptors Influence Glucose Homeostasis. PLoS ONE 2008, 3, e3974. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.A.; Dotson, C.D.; Elson, A.E.T.; Voigt, A.; Boehm, U.; Meyerhof, W.; Steinle, N.I.; Munger, S.D. TAS2R Bitter Taste Receptors Regulate Thyroid Function. FASEB J. 2015, 29, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Avau, B.; Bauters, D.; Steensels, S.; Vancleef, L.; Laermans, J.; Lesuisse, J.; Buyse, J.; Roger Lijnen, H.; Tack, J.; Depoortere, I. The Gustatory Signaling Pathway and Bitter Taste Receptors Affect the Development of Obesity and Adipocyte Metabolism in Mice. PLoS ONE 2015, 10, e0145538. [Google Scholar] [CrossRef] [PubMed]
- Di Pizio, A.; Shy, N.; Behrens, M.; Meyerhof, W.; Niv, M.Y. Molecular Features Underlying Selectivity in Chicken Bitter Taste Receptors. Front. Mol. Biosci. 2018, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Hall, A. Rho GTpases and the Actin Cytoskeleton. Science 1998, 279, 509–514. [Google Scholar] [CrossRef]
- Spang, A.; Schekman, R. Reconstitution of Retrograde Transport from the Golgi to the ER in Vitro. J. Cell Biol. 1998, 143, 589–599. [Google Scholar] [CrossRef]
- Julian, L.; Olson, M.F. Rho-Associated Coiled-Coil Containing Kinases (ROCK), Structure, Regulation, and Functions. Small GTPases 2014, 5, e29846. [Google Scholar] [CrossRef]
- Tojkander, S.; Gateva, G.; Lappalainen, P. Actin Stress Fibers—Assembly, Dynamics and Biological Roles. J. Cell Sci. 2012, 125, 1855–1864. [Google Scholar] [CrossRef]
- Wigge, P.; Kö, K.; Vallis, Y.; Doyle, C.A.; Owen, D.; Hunt, S.P.; Mcmahon, H.T. Amphiphysin Heterodimers: Potential Role in Clathrin-Mediated Endocytosis. Mol. Biol. Cell 2003, 8, 2003–2015. [Google Scholar] [CrossRef]
- Yamada, H.; Ohashi, E.; Abe, T.; Kusumi, N.; Li, S.-A.; Yoshida, Y.; Watanabe, M.; Tomizawa, K.; Kashiwakura, Y.; Kumon, H.; et al. Amphiphysin 1 Is Important for Actin Polymerization during Phagocytosis. Mol. Biol. Cell 2007, 18, 4669–4680. [Google Scholar] [CrossRef] [PubMed]
- Cowling, B.S.; Prokic, I.; Tasfaout, H.; Rabai, A.; Humbert, F.; Rinaldi, B.; Nicot, A.S.; Kretz, C.; Friant, S.; Roux, A.; et al. Amphiphysin (BIN1) Negatively Regulates Dynamin 2 for Normal Muscle Maturation. J. Clin. Investig. 2017, 127, 4477–4487. [Google Scholar] [CrossRef] [PubMed]
- Lionello, V.M.; Nicot, A.S.; Sartori, M.; Kretz, C.; Kessler, P.; Buono, S.; Djerroud, S.; Messaddeq, N.; Koebel, P.; Prokic, I.; et al. Amphiphysin 2 Modulation Rescues Myotubular Myopathy and Prevents Focal Adhesion Defects in Mice. Sci. Transl. Med. 2019, 11, eaav1866. [Google Scholar] [CrossRef] [PubMed]
- Crawford, R.R.; Prescott, E.T.; Sylvester, C.F.; Higdon, A.N.; Shan, J.; Kilberg, M.S.; Mungrue, I.N. Human CHAC1 Protein Degrades Glutathione, and mRNA Induction Is Regulated by the Transcription Factors ATF4 and ATF3 and a Bipartite ATF/CRE Regulatory Element. J. Biol. Chem. 2015, 290, 15878–15891. [Google Scholar] [CrossRef] [PubMed]
- Napoli, J.L. Cellular Retinoid Binding-Proteins, CRBP, CRABP, FABP5: Effects on Retinoid Metabolism, Function and Related Diseases. Pharmacol. Ther. 2017, 173, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Henning, T.; Kraus, M.; Brischwein, M.; Otto, A.M.; Wolf, B. Relevance of Tumor Microenvironment for Progression, Therapy and Drug Development. Anti-Cancer Drugs 2004, 15, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Antonelo, D.; Hendrix, J.; To, V.; Campbell, Y.; Von Staden, M.; Li, S.; Suman, S.P.; Zhai, W.; Chen, J.; et al. Proteomic Characterization of Normal and Woody Breast Meat from Broilers of Five Genetic Strains. Meat Muscle Biol. 2020, 4, 1–17. [Google Scholar] [CrossRef]
- Malila, Y.; Thanatsang, K.; Arayamethakorn, S.; Uengwetwanit, T.; Srimarut, Y.; Petracci, M.; Strasburg, G.M.; Rungrassamee, W.; Visessanguan, W. Absolute Expressions of Hypoxia-Inducible Factor-1 Alpha (HIF1A) Transcript and the Associated Genes in Chicken Skeletal Muscle with White Striping and Wooden Breast Myopathies. PLoS ONE 2019, 14, e0220904. [Google Scholar] [CrossRef]
- Haguenauer, A.; Raimbault, S.; Masscheleyn, S.; Gonzalez-Barroso, M.D.M.; Criscuolo, F.; Plamondon, J.; Miroux, B.; Ricquier, D.; Richard, D.; Bouillaud, F.; et al. A New Renal Mitochondrial Carrier, KMCP1, Is up-Regulated during Tubular Cell Regeneration and Induction of Antioxidant Enzymes. J. Biol. Chem. 2005, 280, 22036–22043. [Google Scholar] [CrossRef]
- Rizzuto, R.; Pinton, P.; Ferrari, D.; Chami, M.; Szabadkai, G.; Magalhães, P.J.; Di Virgilio, F.; Pozzan, T. Calcium and Apoptosis: Facts and Hypotheses. Oncogene 2003, 22, 8619–8627. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, R.; Xu, L.; Wan, Q.; Zhu, J.; Gu, J.; Huang, Z.; Ma, W.; Shen, M.; Ding, F.; et al. Microarray Analysis of Gene Expression Provides New Insights into Denervation-Induced Skeletal Muscle Atrophy. Front. Physiol. 2019, 10, 1298. [Google Scholar] [CrossRef] [PubMed]
| Symbol | Description | Log2 Fold Change | Adjusted p-Value |
|---|---|---|---|
| PTX3 | Pentraxin 3 | 4.91 | 2.69^−04 |
| MLANA | Melan-A | 4.72 | 3.10^−03 |
| CLEC3A | C-type lectin domain family 3 member A | 4.67 | 3.18^−03 |
| GABRA5 | Gamma-aminobutyric acid type A receptor alpha5 subunit | 4.58 | 1.35^−04 |
| IL5RA | Interleukin 5 receptor subunit alpha | 4.51 | 2.73^−05 |
| CCN3 | Cellular Communication Network Factor 3 | 4.43 | 2.12^−02 |
| ASB18 | Ankyrin repeat and SOCS box containing 18 | 4.39 | 7.61^−02 |
| MARCO | Macrophage receptor with collagenous structure | 4.13 | 6.11^−04 |
| PERP1 | PERP1, TP53 apoptosis effector | 3.95 | 9.48^−04 |
| KRT24 | Keratin 24 | 3.93 | 1.83^−03 |
| PGR2/3 | Tandem PRG2/PRG3 gene pair | 3.89 | 1.50^−04 |
| THBS2 | Thrombospondin 2 | 3.87 | 2.32^−02 |
| GPR141 | G protein-coupled receptor 141 | 3.77 | 3.07^−05 |
| DSG2 | Desmoglein 2 | 3.75 | 7.67^−03 |
| MDK | Midkine (neurite growth-promoting factor 2) | 3.70 | 8.31^−04 |
| RSPO4 | R-spondin 4 | 3.40 | 6.08^−04 |
| MMP10 | Matrix metallopeptidase 10 | 3.33 | 1.07^−02 |
| CYP1C1 | Cytochrome P450 family 1 subfamily C polypeptide 1 | 3.31 | 2.16^−03 |
| MYH15 | Myosin, heavy chain 15 | 3.28 | 7.79^−03 |
| CTHRC1 | Collagen triple helix repeat containing 1 | 3.23 | 1.60^−02 |
| AMPH | Amphiphysin | −3.25 | 6.88^−04 |
| CHAC1 | ChaC glutathione specific gamma-glutamylcyclotransferase 1 | −2.37 | 9.63^−03 |
| PGPEP1L | Pyroglutamyl-peptidase I like | −2.37 | 4.93^−03 |
| SLC25A30 | Solute carrier family 25 member 30 | −2.19 | 2.16^−03 |
| 7SK | 7SK RNA | −2.08 | 2.81^−03 |
| FGF1 | Fibroblast growth factor 1 | −1.90 | 2.78^−04 |
| GPR160 | G protein-coupled receptor 160 | −1.89 | 1.07^−02 |
| FRMD5 | FERM domain containing 5 | −1.89 | 2.57^−02 |
| AMBP | Alpha-1-microglobulin/bikunin precursor | −1.86 | 5.11^−03 |
| ESR2 | Estrogen receptor 2 | −1.83 | 6.96^−03 |
| HIBADH | 3-hydroxyisobutyrate dehydrogenase | −1.82 | 7.91^−05 |
| TAS2R7 | Taste receptor, type 2, member 7 | −1.80 | 5.01^−02 |
| TENT5B | Terminal nucleotidyltransferase 5B | −1.78 | 7.22^−04 |
| GLUL | Glutamate-ammonia ligase | −1.75 | 9.58^−04 |
| KCTD20 | Potassium channel tetramerization domain containing 20 | −1.75 | 6.25^−04 |
| RHOBTB3 | Rho related BTB domain containing 3 | −1.73 | 5.99^−03 |
| RBP1 | Retinol binding protein 1 | −1.73 | 3.96^−03 |
| FSTL4 | Follistatin like 4 | −1.72 | 8.36^−02 |
| GJB6 | Gap junction protein beta 6 | −1.61 | 7.24^−02 |
| ARRDC2 | Arrestin domain containing 2 | −1.61 | 7.49^−03 |
| Symbol | Description | Log2 Fold Change | Adjusted p-Value |
|---|---|---|---|
| ASB18 | Ankyrin repeat and SOCS box containing 18 | 5.21 | 6.21^−02 |
| MYL3 | Myosin, light chain 3, alkali; ventricular, skeletal, slow | 4.56 | 6.53^−02 |
| CCN3 | Cellular Communication Network Factor 3 | 3.77 | 8.66^−02 |
| PTX3 | Pentraxin 3 | 3.66 | 1.63^−02 |
| CLEC3A | C-type lectin domain family 3 member A | 3.62 | 5.00^−02 |
| MLANA | Melan-A | 3.58 | 5.54^−02 |
| IL5RA | Interleukin 5 receptor subunit alpha | 3.54 | 1.32^−02 |
| MARCO | Macrophage receptor with collagenous structure | 3.39 | 1.65^−02 |
| THBS2 | Thrombospondin 2 | 3.32 | 8.92^−02 |
| KRT24 | Keratin 24 | 3.25 | 2.78^−02 |
| GABRA5 | Gamma-aminobutyric acid type A receptor alpha5 subunit | 3.24 | 1.83^−02 |
| CSRP3 | Cysteine and glycine rich protein 3 | 3.20 | 1.96^−02 |
| PERP1 | PERP1, TP53 apoptosis effector | 3.20 | 2.31^−02 |
| DSG2 | Desmoglein 2 | 3.12 | 5.38^−02 |
| CALB2 | Calbindin 2 | 3.12 | 6.36^−02 |
| MYH15 | Myosin, heavy chain 15 | 3.12 | 3.05^−02 |
| MYL10 | Myosin, light chain 10, regulatory | 3.12 | 4.74^−02 |
| MMP7 | Matrix metallopeptidase 7 | 3.11 | 3.74^−02 |
| IL8L1 | Interleukin 8-like 1 | 3.05 | 4.00^−02 |
| MMP10 | Matrix metallopeptidase 10 | 3.00 | 4.79^−02 |
| TAS2R7 | Taste receptor, type 2, member 7 | −3.20 | 1.99^−02 |
| AMPH | Amphiphysin | −2.62 | 1.44^−02 |
| FRMD5 | FERM domain containing 5 | −2.39 | 2.38^−02 |
| GPR160 | G protein-coupled receptor 160 | −2.12 | 2.30^−02 |
| CHAC1 | ChaC glutathione specific gamma-glutamylcyclotransfease 1 | −2.08 | 4.17^−02 |
| FSTL4 | Follistatin like 4 | −2.07 | 7.75^−02 |
| RBP1 | Retinol binding protein 1 | −2.05 | 1.32^−02 |
| KY | Kyphoscoliosis peptidase | −2.05 | 7.76^−02 |
| RHOBTB3 | Rho related BTB domain containing 3 | −1.97 | 1.65^−02 |
| PGPEP1L | Pyroglutamyl-peptidase I like | −1.72 | 4.62^−02 |
| VAMP1 | Vesicle-associated membrane protein 1 (synaptobrevin 1) | −1.70 | 1.70^−02 |
| FGF1 | Fibroblast growth factor 1 | −1.70 | 1.32^−02 |
| GADL1 | Glutamate decarboxylase like 1 | −1.69 | 7.07^−02 |
| gga-mir-1754 | Gga-mir-1754 | −1.67 | 1.55^−02 |
| USH2A | Usherin | −1.63 | 4.23^−02 |
| SBK2 | SH3 domain binding kinase family member 2 | −1.60 | 2.06^−02 |
| NRGN | Neurogranin (protein kinase C substrate, RC3) | −1.59 | 3.13^−02 |
| THSD7B | Thrombospondin type 1 domain containing 7B | −1.56 | 2.94^−02 |
| NMNAT3 | Nicotinamide nucleotide adenylyltransferase 3 | −1.53 | 2.41^−02 |
| SLC25A30 | Solute carrier family 25 member 30 | −1.52 | 4.71^−02 |
| Symbol | Description | SM vs. Normal | WB vs. Normal | ||
|---|---|---|---|---|---|
| Log2 Fold Change | Adjusted p-Value | Log2 Fold Change | Adjusted p-Value | ||
| PTX3 | Pentraxin 3 | 4.91 | 2.69^−04 | 3.66 | 1.63^−02 |
| MLANA | Melan-A | 4.72 | 3.10^−03 | 3.58 | 5.54^−02 |
| CLEC3A | C-type lectin domain family 3 member A | 4.67 | 3.18^−03 | 3.62 | 5.00^−02 |
| GABRA5 | Gamma-aminobutyric acid type A receptor alpha5 subunit | 4.58 | 1.35^−04 | 3.24 | 1.83^−02 |
| IL5RA | Interleukin 5 receptor subunit alpha | 4.51 | 2.73^−05 | 3.54 | 1.32^−02 |
| CCN3 | Cellular Communication Network Factor 3 | 4.43 | 2.12^−02 | 3.77 | 8.66^−02 |
| ASB18 | Ankyrin repeat and SOCS box containing 18 | 4.39 | 7.61^−02 | 5.21 | 6.21^−02 |
| MARCO | Macrophage receptor with collagenous structure | 4.13 | 6.11^−04 | 3.39 | 1.65^−02 |
| PERP1 | P53 apoptosis effector related to PMP-22 | 3.95 | 9.48^−04 | 3.20 | 2.31^−02 |
| KRT24 | Keratin 24 | 3.93 | 1.83^−03 | 3.25 | 2.78^−02 |
| THBS2 | Thrombospondin 2 | 3.87 | 2.32^−02 | 3.32 | 8.92^−02 |
| DSG2 | Desmoglein 2 | 3.75 | 7.67^−03 | 3.12 | 5.38^−02 |
| MMP10 | Matrix metallopeptidase 10 | 3.33 | 1.07^−02 | 3.00 | 4.79^−02 |
| MYH15 | Myosin, heavy chain 15 | 3.28 | 7.79^−03 | 3.12 | 3.05^−02 |
| FSTL4 | Follistatin like 4 | −1.72 | 8.36^−02 | −2.07 | 7.75^−02 |
| RBP1 | Retinol binding protein 1 | −1.73 | 3.96^−03 | −2.05 | 1.32^−02 |
| RHOBTB3 | Rho related BTB domain containing 3 | −1.73 | 5.99^−03 | −1.97 | 1.65^−02 |
| TAS2R7 | Taste receptor, type 2, member 7 | −1.80 | 5.01^−02 | −3.20 | 1.99^−02 |
| FRMD5 | FERM domain containing 5 | −1.89 | 2.57^−02 | −2.39 | 2.38^−02 |
| GPR160 | G protein-coupled receptor 160 | −1.89 | 1.07^−02 | −2.12 | 2.30^−02 |
| FGF1 | Fibroblast growth factor 1 | −1.90 | 2.78^−04 | −1.70 | 1.32^−02 |
| SLC25A30 | Solute carrier family 25 member 30 | −2.19 | 2.16^−03 | −1.52 | 4.71^−02 |
| CHAC1 | ChaC glutathione specific gamma-glutamylcyclotransferase 1 | −2.37 | 9.63^−03 | −2.08 | 4.17^−02 |
| PGPEP1L | Pyroglutamyl-peptidase I like | −2.37 | 4.93^−03 | −1.72 | 4.62^−02 |
| AMPH | Amphiphysin | −3.25 | 6.88^−04 | −2.62 | 4.62^−02 |
| Symbol | Description | SM vs. Normal | WB vs. Normal | ||
|---|---|---|---|---|---|
| Log2 Fold Change | Adjusted p-Value | Log2 Fold Change | Adjusted p-Value | ||
| Collagen genes upregulated in both SM and WB | |||||
| COL1A1 | Collagen type I alpha 1 chain | 1.40 | 1.14^−02 | 1.35 | 3.40^−02 |
| COL1A2 | Collagen type I alpha 2 chain | 1.36 | 1.21^−02 | 1.22 | 4.47^−02 |
| COL22A1 | Collagen type XXII alpha 1 chain | 1.23 | 4.14^−03 | 1.07 | 3.13^−02 |
| COL3A1 | Collagen type III alpha 1 chain | 1.36 | 6.70^−03 | 1.00 | 6.14^−02 |
| COL5A2 | Collagen type V alpha 2 chain | 1.04 | 8.30^−03 | 0.81 | 5.91^−02 |
| COL6A1 | Collagen type VI alpha 1 chain | 1.21 | 2.74^−03 | 0.97 | 3.06^−02 |
| COL6A2 | Collagen type VI alpha 2 chain | 1.14 | 2.36^−03 | 0.93 | 2.70^−02 |
| COL6A3 | Collagen type VI alpha 3 chain | 1.33 | 7.15^−04 | 0.89 | 3.10^−02 |
| COL8A1 | Collagen type VIII alpha 1 chain | 1.41 | 3.10^−03 | 1.13 | 3.52^−02 |
| COL8A2 | Collagen type VIII alpha 2 chain | 1.67 | 2.58^−02 | 1.64 | 5.87^−02 |
| COL9A2 | Collagen type IX alpha 2 chain | 1.43 | 1.45^−03 | 1.18 | 2.46^−02 |
| COL12A1 | Collagen type XII alpha 1 chain | 2.12 | 1.50^−02 | 1.91 | 5.26^−02 |
| COL13A1 | Collagen type XIII alpha 1 chain | 1.34 | 1.39^−03 | 0.91 | 4.84^−02 |
| Collagen genes upregulated in SM but not in WB | |||||
| COL14A1 | Collagen type XIV alpha 1 chain | 1.17 | 6.57^−03 | NA | NA |
| COL5A1 | Collagen type V alpha 1 chain | 0.83 | 2.84^−02 | NA | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Che, S.; Pham, P.H.; Barbut, S.; Bienzle, D.; Susta, L. Transcriptomic Profiles of Pectoralis major Muscles Affected by Spaghetti Meat and Woody Breast in Broiler Chickens. Animals 2024, 14, 176. https://doi.org/10.3390/ani14020176
Che S, Pham PH, Barbut S, Bienzle D, Susta L. Transcriptomic Profiles of Pectoralis major Muscles Affected by Spaghetti Meat and Woody Breast in Broiler Chickens. Animals. 2024; 14(2):176. https://doi.org/10.3390/ani14020176
Chicago/Turabian StyleChe, Sunoh, Phuc H. Pham, Shai Barbut, Dorothee Bienzle, and Leonardo Susta. 2024. "Transcriptomic Profiles of Pectoralis major Muscles Affected by Spaghetti Meat and Woody Breast in Broiler Chickens" Animals 14, no. 2: 176. https://doi.org/10.3390/ani14020176
APA StyleChe, S., Pham, P. H., Barbut, S., Bienzle, D., & Susta, L. (2024). Transcriptomic Profiles of Pectoralis major Muscles Affected by Spaghetti Meat and Woody Breast in Broiler Chickens. Animals, 14(2), 176. https://doi.org/10.3390/ani14020176





