MassArray Genotyping as a Selection Tool for Extending the Shelf-Life of Fresh Gilthead Sea Bream and European Seabass
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animal Selection for Whole Genome Sequencing
2.3. Animal Selection for Genotyping
2.4. Enzymatic Phenotyping
2.5. DNA Extraction
2.6. Data Filtering and Association Analysis
2.7. SIFT Algorithm for Amino Acid Substitution Prediction
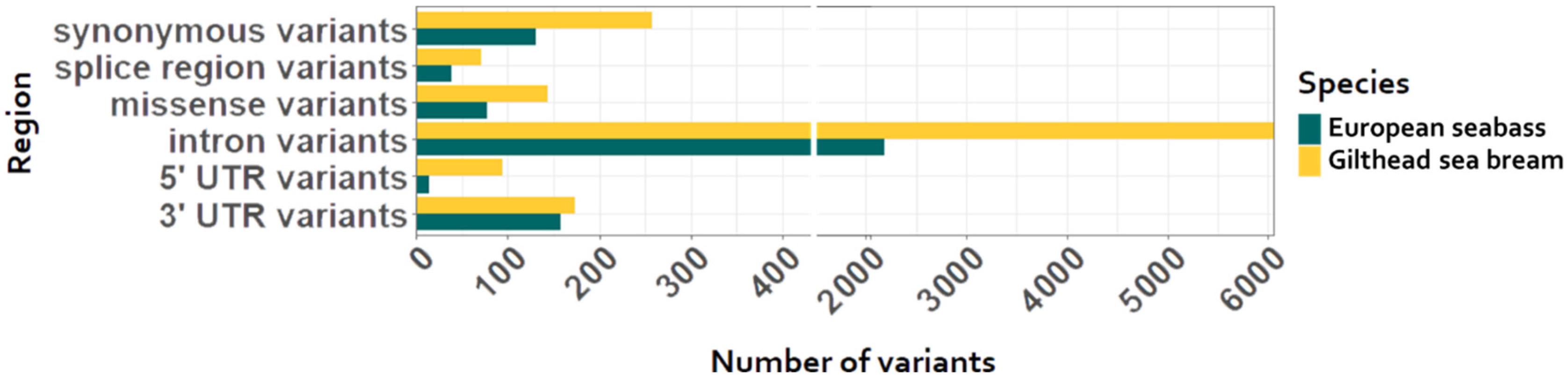
3. Results
Whole Genome Sequencing and Genotyping
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ravishankar, N.C. Advances in Processing and Packaging of Fish and Fishery Products. Adv. Agric. Res. Technol. J. 2019, 3, 168–181. [Google Scholar]
- Yu, D.; Wu, L.; Regenstein, J.M.; Jiang, Q.; Yang, F.; Xu, Y.; Xia, W. Recent Advances in Quality Retention of Non-Frozen Fish and Fishery Products: A Review. Crit. Rev. Food Sci. Nutr. 2019, 60, 1747–1759. [Google Scholar] [CrossRef]
- Hao, R.; Roy, K.; Pan, J.; Shah, B.R.; Mraz, J. Critical Review on the Use of Essential Oils against Spoilage in Chilled Stored Fish: A Quantitative Meta-Analyses. Trends Food Sci. Technol. 2021, 111, 175–190. [Google Scholar] [CrossRef]
- Ntzimani, A.; Angelakopoulos, R.; Semenoglou, I.; Dermesonlouoglou, E.; Tsironi, T.; Moutou, K.; Taoukis, P. Slurry Ice as an Alternative Cooling Medium for Fish Harvesting and Transportation: Study of the Effect on Seabass Flesh Quality and Shelf Life. Aquac. Fish. 2021, 8, 385–392. [Google Scholar] [CrossRef]
- Singh, A.; Benjakul, S. Proteolysis and Its Control Using Protease Inhibitors in Fish and Fish Products: A Review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 496–509. [Google Scholar] [CrossRef]
- Delbarre-Ladrat, C.; Chéret, R.; Taylor, R.; Verrez-Bagnis, V.; Taylor, P.; Taylor, R.; Verrez-Bagnis, V. Trends in Postmortem Aging in Fish: Understanding of Proteolysis and Disorganization of the Myofibrillar Structure. Crit. Rev. Food Sci. Nutr. 2006, 46, 409–421. [Google Scholar] [CrossRef]
- Sriket, C. Proteases in Fish and Shellfish: Role on Muscle Softening and Prevention. Int. Food Res. J. 2014, 21, 433–445. [Google Scholar]
- Fanjul-Fernández, M.; Folgueras, A.R.; Cabrera, S.; López-Otín, C. Matrix Metalloproteinases: Evolution, Gene Regulation and Functional Analysis in Mouse Models. Biochim. Biophys. Acta—Mol. Cell Res. 2010, 1803, 3–19. [Google Scholar] [CrossRef]
- Pedersen, M.E.; Vuong, T.T.; Rønning, S.B.; Kolset, S.O. Matrix Metalloproteinases in Fish Biology and Matrix Turnover. Matrix Biol. 2015, 44–46, 86–93. [Google Scholar] [CrossRef]
- Turk, V.; Stoka, V.; Vasiljeva, O.; Renko, M.; Sun, T.; Turk, B.; Turk, D. Cysteine Cathepsins: From Structure, Function and Regulation to New Frontiers. Biochim. Biophys. Acta—Proteins Proteom. 2012, 1824, 68–88. [Google Scholar] [CrossRef]
- Sorimachi, H.; Hata, S.; Ono, Y. Calpain Chronicle—An Enzyme Family under Multidisciplinary Characterization. Proc. Jpn. Acad. Ser. B 2011, 87, 287–327. [Google Scholar] [CrossRef] [PubMed]
- Ono, Y.; Sorimachi, H. Calpains—An Elaborate Proteolytic System. Biochim. Biophys. Acta—Proteins Proteom. 2012, 1824, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Georges, M.; Charlier, C.; Hayes, B. Harnessing Genomic Information for Livestock Improvement. Nat. Rev. Genet. 2018, 20, 135–156. [Google Scholar] [CrossRef]
- Soliva-Fortuny, R.C.; Martín-Belloso, O. New Advances in Extending the Shelf-Life of Fresh-Cut Fruits: A Review. Trends Food Sci. Technol. 2003, 14, 341–353. [Google Scholar] [CrossRef]
- Deegan, L.H.; Cotter, P.D.; Hill, C.; Ross, P. Bacteriocins: Biological Tools for Bio-Preservation and Shelf-Life Extension. Int. Dairy J. 2006, 16, 1058–1071. [Google Scholar] [CrossRef]
- Banerjee, A.; Chatterjee, S.; Variyar, P.S.; Sharma, A. Shelf Life Extension of Minimally Processed Ready-to-Cook (RTC) Cabbage by Gamma Irradiation. J. Food Sci. Technol. 2016, 53, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Odueke, O.B.; Farag, K.W.; Baines, R.N.; Chadd, S.A. Irradiation Applications in Dairy Products: A Review. Food Bioprocess Technol. 2016, 9, 751–767. [Google Scholar] [CrossRef]
- Cavaliere, A.; Ventura, V. Mismatch between Food Sustainability and Consumer Acceptance toward Innovation Technologies among Millennial Students: The Case of Shelf Life Extension. J. Clean. Prod. 2018, 175, 641–650. [Google Scholar] [CrossRef]
- No, H.K.; Meyers, S.P.; Prinyawiwatkul, W.; Xu, Z. Applications of Chitosan for Improvement of Quality and Shelf Life of Foods: A Review. J. Food Sci. 2007, 72, R87–R100. [Google Scholar] [CrossRef]
- Peñaloza, C.; Manousaki, T.; Franch, R.; Tsakogiannis, A.; Sonesson, A.K.; Aslam, M.L.; Allal, F.; Bargelloni, L.; Houston, R.D.; Tsigenopoulos, C.S. Development and Testing of a Combined Species SNP Array for the European Seabass (Dicentrarchus Labrax) and Gilthead Seabream (Sparus Aurata). Genomics 2021, 113, 2096–2107. [Google Scholar] [CrossRef]
- Houston, R.D.; Bean, T.P.; Macqueen, D.J.; Gundappa, M.K.; Jin, Y.H.; Jenkins, T.L.; Selly, S.L.C.; Martin, S.A.M.; Stevens, J.R.; Santos, E.M.; et al. Harnessing Genomics to Fast-Track Genetic Improvement in Aquaculture. Nat. Rev. Genet. 2020, 21, 389–409. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.; Toro, M.; Sonesson, A.K.; Villanueva, B. Optimizing the Creation of Base Populations for Aquaculture Breeding Programs Using Phenotypic and Genomic Data and Its Consequences on Genetic Progress. Front. Genet. 2014, 5, 414. [Google Scholar] [CrossRef] [PubMed]
- Tine, M.; Kuhl, H.; Gagnaire, P.A.; Louro, B.; Desmarais, E.; Martins, R.S.T.; Hecht, J.; Knaust, F.; Belkhir, K.; Klages, S.; et al. European Sea Bass Genome and Its Variation Provide Insights into Adaptation to Euryhalinity and Speciation. Nat. Commun. 2014, 5, 5770. [Google Scholar] [CrossRef] [PubMed]
- Pauletto, M.; Manousaki, T.; Ferraresso, S.; Babbucci, M.; Tsakogiannis, A.; Louro, B.; Vitulo, N.; Quoc, V.H.; Carraro, R.; Bertotto, D.; et al. Genomic Analysis of Sparus Aurata Reveals the Evolutionary Dynamics of Sex-Biased Genes in a Sequential Hermaphrodite Fish. Commun. Biol. 2018, 1, 119. [Google Scholar] [CrossRef] [PubMed]
- Oeth, P.; del Mistro, G.; Marnellos, G.; Shi, T.; van den Boom, D. Qualitative and Quantitative Genotyping Using Single Base Primer Extension Coupled with Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MassARRAY). Methods Mol. Biol. 2009, 578, 307–343. [Google Scholar] [CrossRef] [PubMed]
- Zenger, K.R.; Khatkar, M.S.; Jones, D.B.; Khalilisamani, N.; Jerry, D.R.; Raadsma, H.W. Genomic Selection in Aquaculture: Application, Limitations and Opportunities with Special Reference to Marine Shrimp and Pearl Oysters. Front. Genet. 2019, 10, 693. [Google Scholar] [CrossRef] [PubMed]
- Kruglyak, L. The Road to Genome-Wide Association Studies. Nat. Rev. Genet. 2008, 9, 314–318. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/ (accessed on 28 June 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-Based Variant Detection from Short-Read Sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar] [CrossRef]
- Angelakopoulos, R.; Dimitroglou, A.; Papaharisis, L.; Moutou, K.A. Electrical Stunning Has the Potential to Delay Fillet Degradation Post-Harvest in Red Seabream (Pagrus Major). Aquac. J. 2022, 2, 302–315. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A Web Tool for the Analysis of Association Studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef] [PubMed]
- Lake, S.L.; Lyon, H.; Tantisira, K.; Silverman, E.K.; Weiss, S.T.; Laird, N.M.; Schaid, D.J. Estimation and Tests of Haplotype-Environment Interaction When Linkage Phase Is Ambiguous. Hum. Hered. 2003, 55, 56–65. [Google Scholar] [CrossRef]
- Sim, N.L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT Web Server: Predicting Effects of Amino Acid Substitutions on Proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Robert, F.; Pelletier, J. Exploring the Impact of Single-Nucleotide Polymorphisms on Translation. Front. Genet. 2018, 9, 507. [Google Scholar] [CrossRef]
- Cordell, H.J.; Clayton, D.G. Genetic Association Studies; Elsevier B.V.: Amsterdam, The Netherlands, 2005; Volume 366, pp. 1121–1131. [Google Scholar]
- Ishigaki, K.; Sakaue, S.; Terao, C.; Luo, Y.; Sonehara, K.; Yamaguchi, K.; Amariuta, T.; Too, C.L.; Laufer, V.A.; Scott, I.C.; et al. Multi-Ancestry Genome-Wide Association Analyses Identify Novel Genetic Mechanisms in Rheumatoid Arthritis. Nat. Genet. 2022, 54, 1640–1651. [Google Scholar] [CrossRef]
- Sharma, S.; Upadhyaya, H.D.; Varshney, R.K.; Gowda, C.L.L. Pre-Breeding for Diversification of Primary Gene Pool and Genetic Enhancement of Grain Legumes. Front. Plant Sci. 2013, 4, 309. [Google Scholar] [CrossRef]
- Ahmed, Z.; Donkor, O.; Street, W.A.; Vasiljevic, T. Calpains- and Cathepsins-Induced Myofibrillar Changes in Post-Mortem Fish: Impact on Structural Softening and Release of Bioactive Peptides. Trends Food Sci. Technol. 2015, 45, 130–146. [Google Scholar] [CrossRef]
- Yu, H.; He, Y.; Wang, X.; Zhang, Q.; Bao, Z.; Guo, X. Polymorphism in a Serine Protease Inhibitor Gene and Its Association with Disease Resistance in the Eastern Oyster (Crassostrea Virginica Gmelin). Fish Shellfish Immunol. 2011, 30, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Bernatchez, L. On the Maintenance of Genetic Variation and Adaptation to Environmental Change: Considerations from Population Genomics in Fishes. J. Fish Biol. 2016, 89, 2519–2556. [Google Scholar] [CrossRef] [PubMed]
- Geddes, J.; Velez, G.; Schaefer, K.; Yu, C.; Bassuk, A.; Bondada, V.; Mashburn, C.; Cox, A.; Borcherding, N.; Tsang, S.; et al. Calpain-5 Expression in the Retina Localizes to Photoreceptor Synapses. Investig. Ophthalmol. Vis. Sci. 2016, 57, 2509–2521. [Google Scholar] [CrossRef]
- Croall, D.E.; Vanhooser, L.M.; Cashon, R.E. Detecting the Active Conformation of Calpain with Calpastatin-Based Reagents. Biochim. Biophys. Acta—Proteins Proteom. 2008, 1784, 1676–1686. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Macqueen, D.J.; Delbridge, M.L.; Manthri, S.; Johnston, I.A. A Newly Classified Vertebrate Calpain Protease, Directly Ancestral to CAPN1 and 2, Episodically Evolved a Restricted Physiological Function in Placental Mammals. Mol. Biol. Evol. 2010, 27, 1886–1902. [Google Scholar] [CrossRef]
- Maki, M.; Maemoto, Y.; Osako, Y.; Shibata, H. Evolutionary and Physical Linkage between Calpains and Penta-EF-Hand Ca2+-Binding Proteins. FEBS J. 2012, 279, 1414–1421. [Google Scholar] [CrossRef]
- Hosseini, M.; Najmabadi, H.; Kahrizi, K. Calpains: Diverse Functions but Enigmatic. Arch. Iran. Med. 2018, 21, 170–179. [Google Scholar]
- Yeh, H.-Y.; Klesius, P.H. Channel Catfish, Ictalurus Punctatus, Cysteine Proteinases: Cloning, Characterisation and Expression of Cathepsin H and L. Fish Shellfish Immunol. 2009, 26, 332–338. [Google Scholar] [CrossRef]
- Yu, C.; Cha, Y.; Wu, F.; Xu, X.; Qin, L.; Du, M. Molecular Cloning and Functional Characterization of Cathepsin D from Sea Cucumber Apostichopus Japonicus. Fish Shellfish Immunol. 2017, 70, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Herrero, M.M.; Duflos, G.; Malle, P.; Bouquelet, S. Collagenase Activity and Protein Hydrolysis as Related to Spoilage of Iced Cod (Gadus Morhua). Food Res. Int. 2003, 36, 141–147. [Google Scholar] [CrossRef]
- Murugan, A.K.; Dong, J.; Xie, J.; Xing, M. Uncommon Gnaq, MMP8, AKT3, EGFR, and PIK3R1 Mutations in Thyroid Cancers. Endocr. Pathol. 2011, 22, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Dolmatov, I.Y.; Nizhnichenko, V.A.; Dolmatova, L.S. Matrix Metalloproteinases and Tissue Inhibitors of Metalloproteinases in Echinoderms: Structure and Possible Functions. Cells 2021, 10, 2331. [Google Scholar] [CrossRef] [PubMed]
- Parks, W.C.; Wilson, C.L.; López-Boado, Y.S. Matrix Metalloproteinases as Modulators of Inflammation and Innate Immunity. Nat. Rev. Immunol. 2004, 4, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Hadler-Olsen, E.; Fadnes, B.; Sylte, I.; Uhlin-Hansen, L.; Winberg, J.O. Regulation of Matrix Metalloproteinase Activity in Health and Disease. FEBS J. 2011, 278, 28–45. [Google Scholar] [CrossRef]
- Moorehead, C.; Prudnikova, K.; Marcolongo, M. The Regulatory Effects of Proteoglycans on Collagen Fibrillogenesis and Morphology Investigated Using Biomimetic Proteoglycans. J. Struct. Biol. 2019, 206, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, P.W. What Is the Evidence for Heterozygote Advantage Selection? Trends Ecol. Evol. 2012, 27, 698–704. [Google Scholar] [CrossRef]
- Sellis, D.; Callahan, B.J.; Petrov, D.A.; Messer, P.W. Heterozygote Advantage as a Natural Consequence of Adaptation in Diploids. Proc. Natl. Acad. Sci. USA 2011, 108, 20666–20671. [Google Scholar] [CrossRef]
- Casci, T. Small but Dominant RNA. Nat. Rev. Genet. 2010, 11, 671. [Google Scholar] [CrossRef]
- Chen, Y.; Lun, A.T.L.; Smyth, G.K. From Reads to Genes to Pathways: Differential Expression Analysis of RNA-Seq Experiments Using Rsubread and the EdgeR Quasi-Likelihood Pipeline. F1000Research 2016, 5, 1438. [Google Scholar] [CrossRef] [PubMed]
- Savage, A.E.; Zamudio, K.R. Adaptive Tolerance to a Pathogenic Fungus Drives Major Histocompatibility Complex Evolution in Natural Amphibian Populations. Proc. R. Soc. B Biol. Sci. 2016, 283, 20153115. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, P.W. Conservation Genetics: Where Are We Now? Trends Ecol. Evol. 2001, 16, 629–636. [Google Scholar] [CrossRef]
| Gene ID | Gene Name | Expression | Gene ID | Gene Name | Expression |
|---|---|---|---|---|---|
| ENSSAUG00010000077 | capn11a | Low | ENSSAUG00010005776 | CTSAa | Low |
| ENSSAUG00010008141 | capn11b | No | ENSSAUG00010008071 | CTSBa | High |
| ENSSAUG00010025995 | capn11c | High | ENSSAUG00010003083 | CTSBb | Low |
| ENSSAUG00010003429 | capn14a | High | ENSSAUG00010007964 | CTSC | High |
| ENSSAUG00010016749 | capn14b | Low | ENSSAUG00010015701 | CTSDa | High |
| ENSSAUG00010016757 | capn14c | Low | ENSSAUG00010016838 | CTSDb | High |
| ENSSAUG00010012619 | capn15a | Low | ENSSAUG00010016344 | CTSDc | High |
| ENSSAUG00010016176 | capn15b | Low | ENSSAUG00010024233 | CTSF | High |
| ENSSAUG00010000032 | capn2a | High | ENSSAUG00010015817 | CTSHa | No |
| ENSSAUG00010026026 | capn2b | Low | ENSSAUG00010021061 | CTSHb | High |
| ENSSAUG00010000030 | capn2c | Low | ENSSAUG00010011634 | CTSK | High |
| ENSSAUG00010006640 | capn2d | High | ENSSAUG00010016582 | CTSLa | High |
| ENSSAUG00010000034 | capn2e | High | ENSSAUG00010010127 | CTSLb | No |
| ENSSAUG00010017861 | capn3a | Low | ENSSAUG00010011634 | CTSLc | No |
| ENSSAUG00010012311 | capn3b | High | ENSSAUG00010002932 | CTSO | Low |
| ENSSAUG00010002636 | capn5a | No | ENSSAUG00010011098 | CTSSa | High |
| ENSSAUG00010005676 | capn5b | High | ENSSAUG00010011632 | CTSSb | No |
| ENSSAUG00010025836 | capn6a | High | ENSSAUG00010011115 | CTSSc | No |
| ENSSAUG00010014146 | capn6b | High | ENSSAUG00010011147 | CTSSd | No |
| ENSSAUG00010000033 | capn8a | Low | ENSSAUG00010017292 | CTSSe | Low |
| ENSSAUG00010006205 | capn8b | No | ENSSAUG00010011128 | CTSSf | No |
| ENSSAUG00010026019 | capn8c | Low | ENSSAUG00010011634 | CTSSg | No |
| ENSSAUG00010007897 | capn1 | High | ENSSAUG00010025140 | CTSZa | High |
| ENSSAUG00010013017 | capn7 | Low | ENSSAUG00010014606 | CTSZb | Low |
| ENSSAUG00010001056 | capn9 | High | ENSSAUG00010010858 | CTSZc | High |
| ENSSAUG00010013339 | capn12 | Low | ENSSAUG00010014101 | MMP13a | High |
| ENSSAUG00010017388 | capns1a | High | ENSSAUG00010010684 | MMP13b | High |
| ENSSAUG00010002445 | capns1b | High |
| Gene ID | Gene Name | Expression | Gene ID | Gene Name | Expression |
|---|---|---|---|---|---|
| ENSDLAG00005017924 | capn1 | High | ENSDLAG00005013147 | CTSAa | High |
| ENSDLAG00005000250 | capn10 | Low | ENSDLAG00005010980 | CTSAb | Low |
| ENSDLAG00005001439 | capn11a | High | ENSDLAG00005004816 | CTSBa | High |
| ENSDLAG00005016201 | capn11b | No | ENSDLAG00005013196 | CTSBb | No |
| ENSDLAG00005000961 | capn12 | No | ENSDLAG00005017730 | CTSC | High |
| ENSDLAG00005024962 | capn14a | No | ENSDLAG00005022128 | CTSDa | High |
| ENSDLAG00005005672 | capn14b | Low | ENSDLAG00005004808 | CTSDb | High |
| ENSDLAG00005009199 | capn15b | Low | ENSDLAG00005006074 | CTSF | High |
| ENSDLAG00005022265 | capn15a | No | ENSDLAG00005023385 | CTSH | High |
| ENSDLAG00005002296 | capn2b | High | ENSDLAG00005014479 | CTSK | High |
| ENSDLAG00005000590 | capn2a | No | ENSDLAG00005022121 | CTSLa | High |
| ENSDLAG00005015494 | capn3a | Low | ENSDLAG00005007883 | CTSLb | No |
| ENSDLAG00005011625 | capn3b | High | ENSDLAG00005022875 | CTSO | Low |
| ENSDLAG00005005420 | capn5a | High | ENSDLAG00005005416 | CTSSb | High |
| ENSDLAG00005004342 | capn5b | High | ENSDLAG00005014499 | CTSSa | Low |
| ENSDLAG00005001788 | capn6a | Low | ENSDLAG00005004507 | CTSZa | High |
| ENSDLAG00005014943 | capn6b | High | ENSDLAG00005011006 | CTSZb | Low |
| ENSDLAG00005006030 | capn7 | High | ENSDLAG00005026027 | CTSZb.2 | High |
| ENSDLAG00005000702 | capn8 | High | ENSDLAG00005008130 | MMP13a | High |
| ENSDLAG00005018075 | capn9 | High | ENSDLAG00005008348 | MMP13b | High |
| ENSDLAG00005012396 | capns1a | High | |||
| ENSDLAG00005006529 | capns1b | High |
| Species | SNP ID | Gene | Reference Allele | Alternative Allele | Reference Allele Frequency % | Alternative Allele Frequency % |
|---|---|---|---|---|---|---|
| S. aurata | CTSDb_9 | CTSDb | A | G | 61 | 39 |
| S. aurata | capn10_11 | capn10 | T | A | 92 | 8 |
| S. aurata | capn10_14 | capn10 | T | A | 62 | 38 |
| S. aurata | capn2b_3 | capn2b | A | T | 79 | 21 |
| S. aurata | capn5a_1 | capn5a | G | A | 92 | 8 |
| S. aurata | capn5a_2 | capn5a | A | G | 86 | 14 |
| D. labrax | capn2b_1 | capn2b | A | C | 60 | 40 |
| D. labrax | capn14b_1 | capn14b | T | A | 92 | 8 |
| D. labrax | capn5b_3 | capn5b | G | T | 68 | 32 |
| D. labrax | capn5b_5 | capn5b | A | G | 67 | 33 |
| D. labrax | capn15b_1 | capn15b | A | G | 56 | 44 |
| D. labrax | capn14b_4 | capn14b | G | A | 89 | 11 |
| D. labrax | MMP13b_1 | MMP13b | G | A | 66 | 34 |
| D. labrax | MMP13b_2 | MMP13b | A | G | 79 | 21 |
| D. labrax | MMP13a_1.1 | MMP13a | T | C | 81 | 19 |
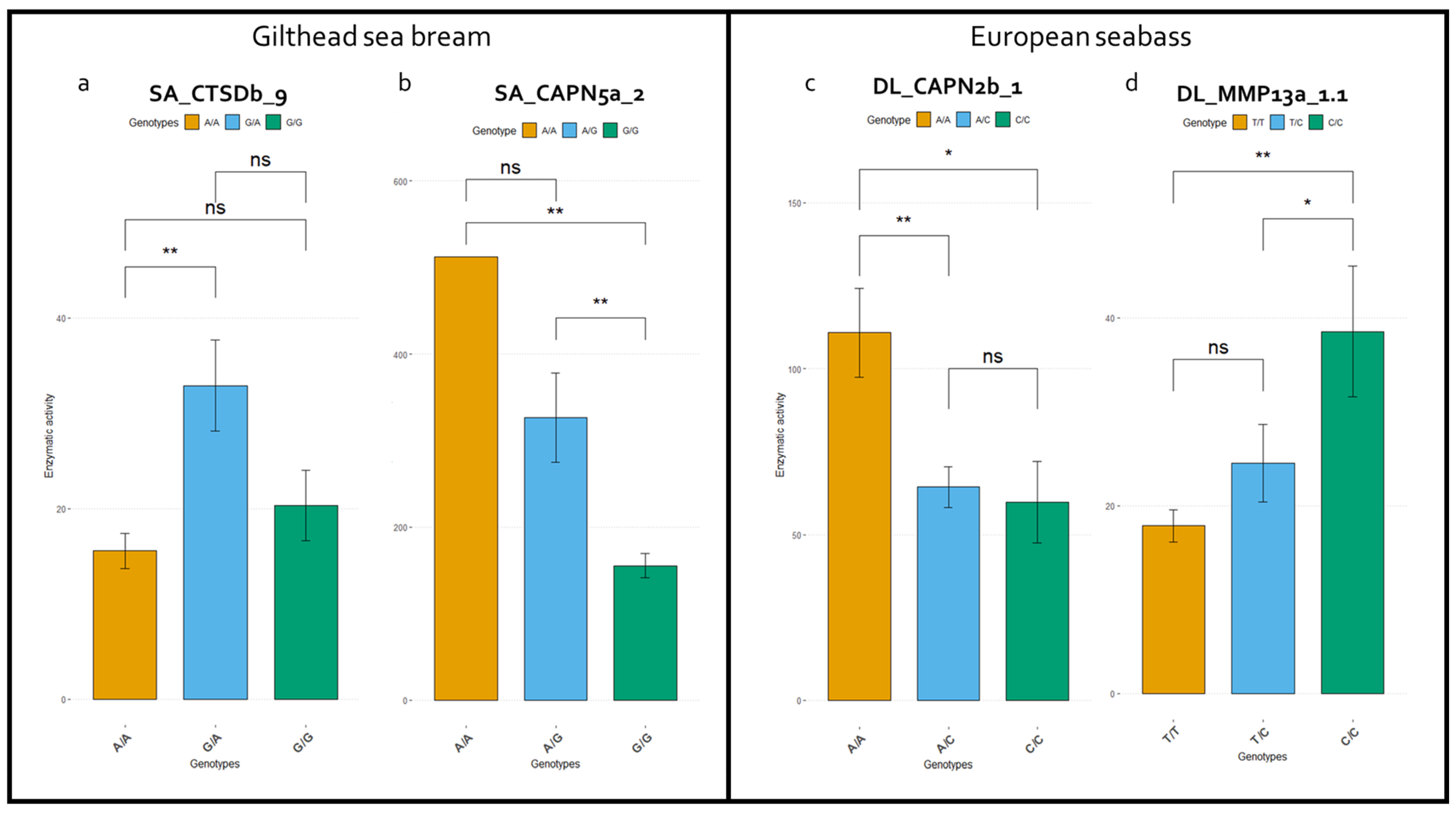
| Species | SNP ID | Gene | Alleles | Protein Domain | Aminoacid Change | Model of Inheritance | Genotype | Enzymatic Activity Mean (s.e.) | Enzymatic Activity Difference (95% CI) | p-Value | AIC | BIC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. aurata | CTSDb_9 | CTSDb | A/G | PEPTIDASE_A1 | p.Ile314Val | Overdominant | A/A-G/G | 17.21 (1.77) | 15.70 (7.65–23.75) | 0.0002 | 1247.4 | 1256.2 |
| G/A | 32.91 (4.76) | |||||||||||
| S. aurata | capn10_11 | capn10 | T/A | CysPC domain | p.Asp59Val | Dominant | T/T | 272.78 (21.31) | −129.97 (−240.56–−19.39) | 0.023 | 2081.3 | 2090.4 |
| A/T-A/A | 142.8 (36.07) | |||||||||||
| S. aurata | capn10_14 | capn10 | T/A | CysPC domain | p.Asn3Ile | Recessive | A/A-A/T | 275.33 (22.59) | −88.29 (−172.64–−3.93) | 0.042 | 2048.6 | 2057.6 |
| T/T | 187.04 (34.09) | |||||||||||
| S. aurata | capn2b_3 | capn2b | A/T | EF-hand | p.Gln574Leu | Log-additive | --- | --- | −76.80 (−138.53–−15.07) | 0.016 | 2079.1 | 2088.1 |
| S. aurata | capn5a_1 | capn5a | G/A | C2 domain | p.Ala414Thr | --- | G/G | 227.52 (20.28) | 109.19 (10.02–208.37) | 0.032 | 2131.9 | 2141.1 |
| A/G | 336.72 (46.86) | |||||||||||
| S. aurata | capn5a_2 | capn5a | A/G | C2 domain | p.Met431Val | Log-additive | --- | --- | 98.30 (1.61–194.99) | 0.049 | 1480.1 | 1488.1 |
| D. labrax | capn2b_1 | capn2b | A/C | Out of domain | p.Gln12Leu | Dominant | A/A | 96.05 (17.34) | −32.23 (−63.79–−0.66) | 0.047 | 1766.7 | 1775.7 |
| C/A-C/C | 63.82 (7.5) | |||||||||||
| D. labrax | capn14b_1 | capn14b | T/A | CysPC domain | p.Ser118Pro | Recessive | T/T-A/T | 71.4 (7.07) | 153.77 (53.06–254.48) | 0.0032 | 1808.7 | 1817.7 |
| A/A | 225.17 (96.73) | |||||||||||
| D. labrax | capn5b_3 | capn5b | G/T | CysPC domain | p.Gly227Cys | Dominant | G/G | 44.03 (7.96) | 52.20 (23.69–80.71) | 0.0005 | 1520.8 | 1529.4 |
| G/T-T/T | 96.23 (11.61) | |||||||||||
| D. labrax | capn5b_5 | capn5b | A/G | C2 domain | p.Met388Val | Overdominant | A/A-G/G | 94.07 (10.91) | −48.39 (−78.47–−18.31) | 0.002 | 1643.8 | 1652.6 |
| G/A | 45.68 (8.68) | |||||||||||
| D. labrax | capn15b_1 | capn15b | A/G | Zinc finger | p.Ser21Gly | Dominant | A/A | 96.34 (17.35) | −38.72 (−75.58–−1.86) | 0.042 | 1047.6 | 1055.1 |
| A/G-G/G | 57.62 (10.2) | |||||||||||
| D. labrax | capn14b_4 | capn14b | G/A | Out of domain | p.Ala357Thr | Recessive | G/G-A/G | 71.29 (7.17) | 107.14 (18.49–195.79) | 0.019 | 1788.9 | 1798 |
| A/A | 178.43 (82.84) | |||||||||||
| D. labrax | MMP13b_1 | MMP13b | G/A | Catalytic domain | p.Gly103Arg | Overdominant | G/G-A/A | 27.69 (3.01) | −13.59 (−20.16–−7.02) | 0.0001 | 1304.1 | 1313.1 |
| A/G | 14.1 (1.81) | |||||||||||
| D. labrax | MMP13b_2 | MMP13b | A/G | Peptidoglycan binding-like | p.Asn34Ser | Overdominant | A/A-G/G | 26.82 (2.87) | −15.55 (−24.92–−6.17) | 0.0016 | 895.7 | 903.5 |
| G/A | 11.28 (2.99) | |||||||||||
| D. labrax | MMP13a_1.1 | MMP13a | T/C | Peptidoglycan binding-like | p.Ser26Gly | Recessive | T/T-T/C | 18.13 (1.71) | 24.41 (7.98–40.84) | 0.0042 | 1259.1 | 1268 |
| C/C | 42.54 (9.61) |
| Species | SNP ID | Mutation | SIFT Score |
|---|---|---|---|
| S. aurata | capn5a_1 | p.Ala414Thr | 0.01 |
| S. aurata | capn5a_2 | p.Met431Val | 0.86 |
| S. aurata | capn2b_3 | p.Gln574Leu | 1 |
| S. aurata | capn10 | p.Met254Lys | 0.63 |
| S. aurata | capn10_11 | p.Asp59Val | 0.03 |
| S. aurata | CTSDb_9 | p.Ile314Val | 0.27 |
| D. labrax | capn2b_1 | p.Gln12Leu | 0.01 * |
| D. labrax | capn5b_3 | p.Gly227Cys | 0.68 |
| D. labrax | capn5b_5 | p.Met388Val | 0.66 |
| D. labrax | capn14b_4 | p.Ala357Thr | 0.71 |
| D. labrax | capn14b_1 | p.Ser118Pro | 0.26 |
| D. labrax | capn15b_1 | p.Ser21Gly | 0.02 * |
| D. labrax | MMP13a | p.Ser26Gly | 0.42 |
| D. labrax | MMP13b_1 | p.Gly103Arg | 0.02 |
| D. labrax | MMP13b_2 | p.Asn34Ser | 0.48 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Angelakopoulos, R.; Tsipourlianos, A.; Giannoulis, T.; Mamuris, Z.; Moutou, K.A. MassArray Genotyping as a Selection Tool for Extending the Shelf-Life of Fresh Gilthead Sea Bream and European Seabass. Animals 2024, 14, 205. https://doi.org/10.3390/ani14020205
Angelakopoulos R, Tsipourlianos A, Giannoulis T, Mamuris Z, Moutou KA. MassArray Genotyping as a Selection Tool for Extending the Shelf-Life of Fresh Gilthead Sea Bream and European Seabass. Animals. 2024; 14(2):205. https://doi.org/10.3390/ani14020205
Chicago/Turabian StyleAngelakopoulos, Rafael, Andreas Tsipourlianos, Themistoklis Giannoulis, Zissis Mamuris, and Katerina A. Moutou. 2024. "MassArray Genotyping as a Selection Tool for Extending the Shelf-Life of Fresh Gilthead Sea Bream and European Seabass" Animals 14, no. 2: 205. https://doi.org/10.3390/ani14020205
APA StyleAngelakopoulos, R., Tsipourlianos, A., Giannoulis, T., Mamuris, Z., & Moutou, K. A. (2024). MassArray Genotyping as a Selection Tool for Extending the Shelf-Life of Fresh Gilthead Sea Bream and European Seabass. Animals, 14(2), 205. https://doi.org/10.3390/ani14020205







