Non-Additive and Asymmetric Allelic Expression of p38 mapk in Hybrid Tilapia (Oreochromis niloticus ♀ × O. aureus ♂)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Rearing of Experimental Fish
2.3. Sample Collection and Analysis
2.4. Extraction and Quality Control of Total RNA and DNA
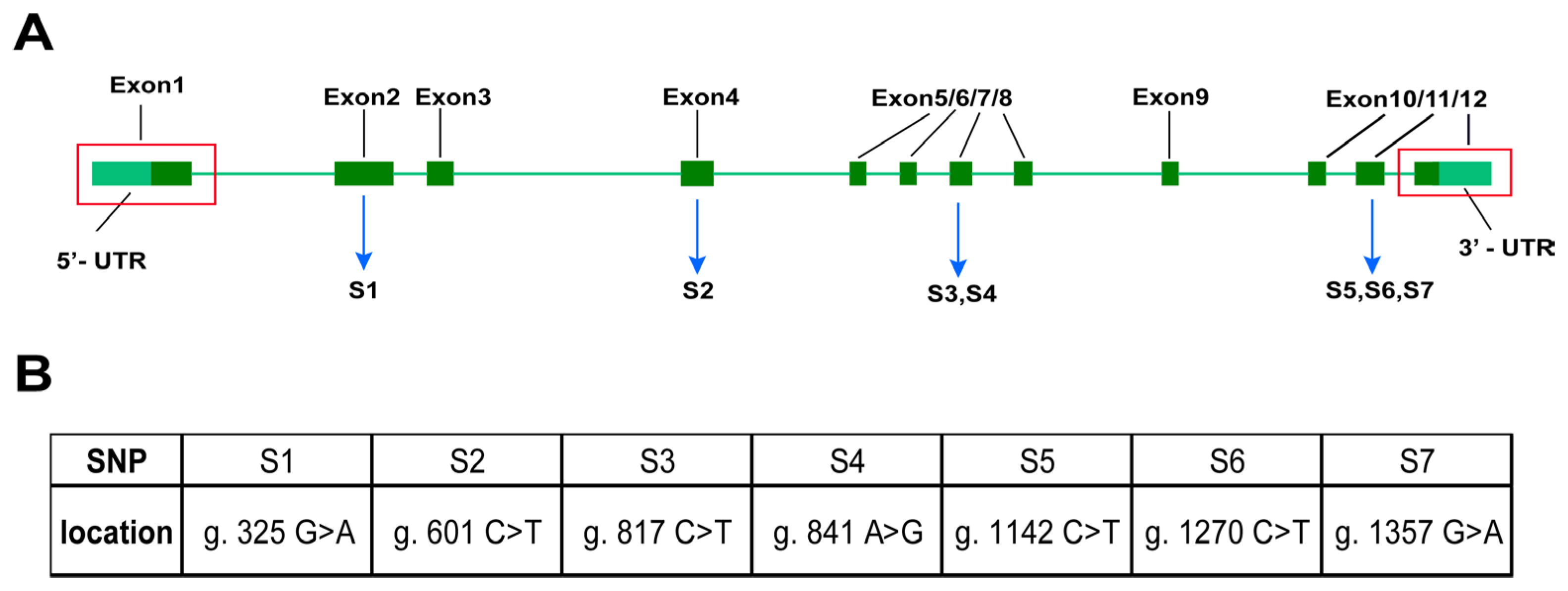
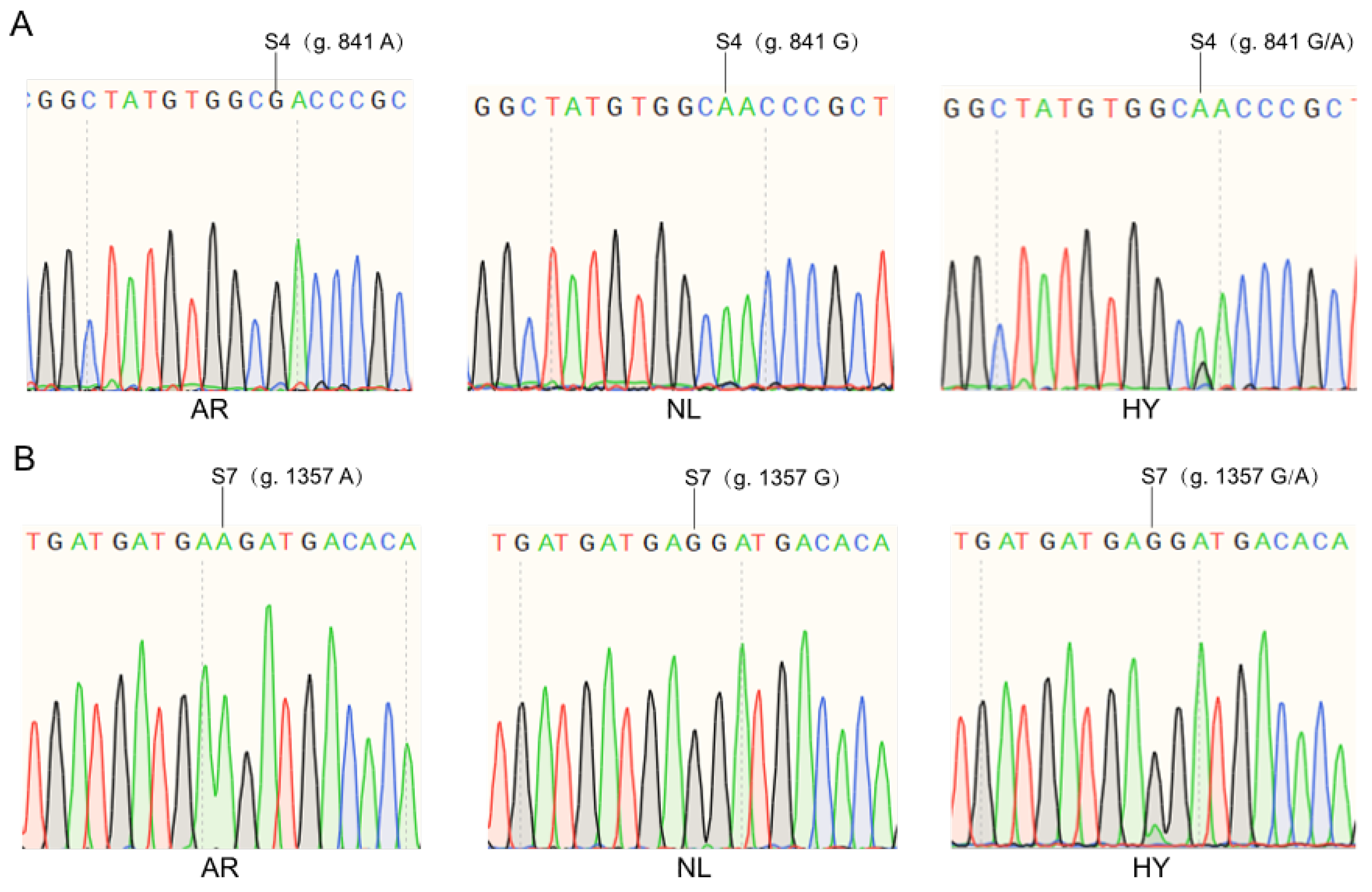
2.5. Confirmation of SNP Loci in the Coding Region of p38
2.6. Specific Expression of p38 Allele in the Hybrid Tilapia
2.7. Quantitative Analysis of p38 Expression among Different Strains
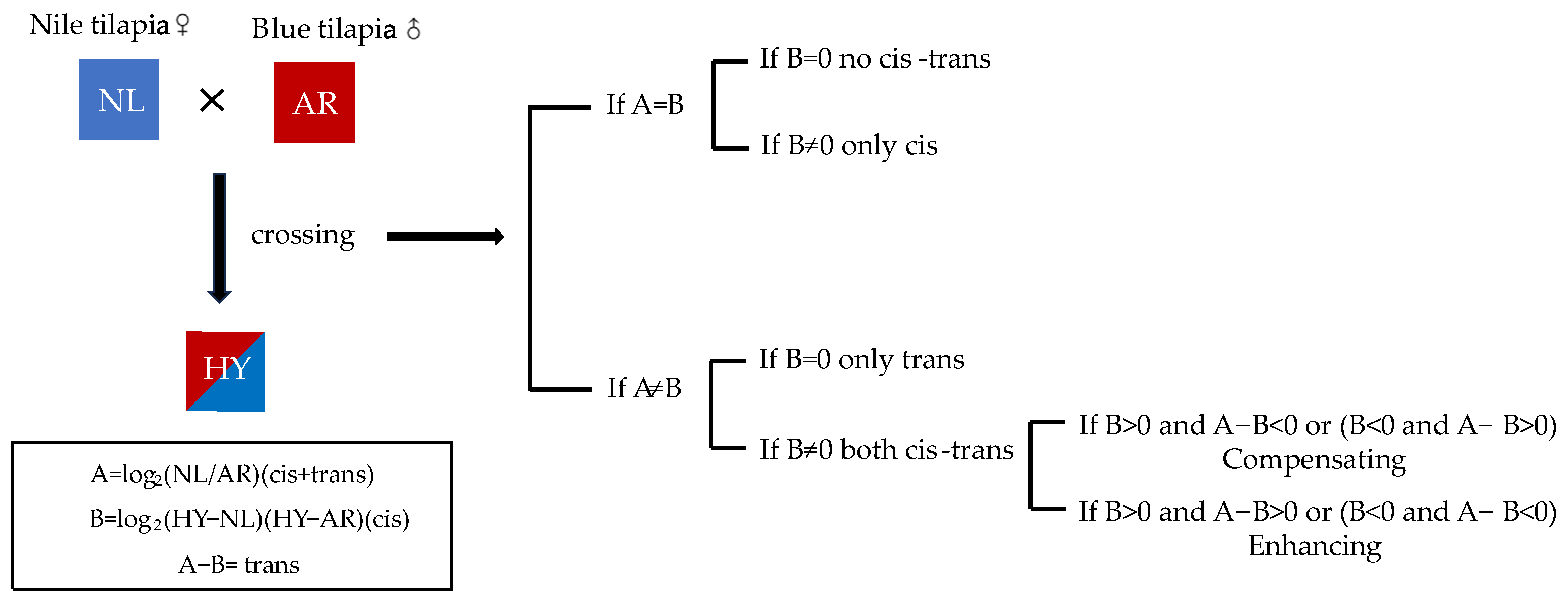
2.8. Cis-Trans Effects from the Parents in Hybrid Tilapia
2.9. Analysis of Differences in the Gene Promoter Region
2.10. Dual-Luciferase Activity Assay of SNP Site Promoters
3. Results
3.1. Gene p38 Coding Region and SNP Locus Screening
3.2. Gene p38 Allele-Specific Expression
3.3. Cis and Trans Effects between p38 Alleles in HY
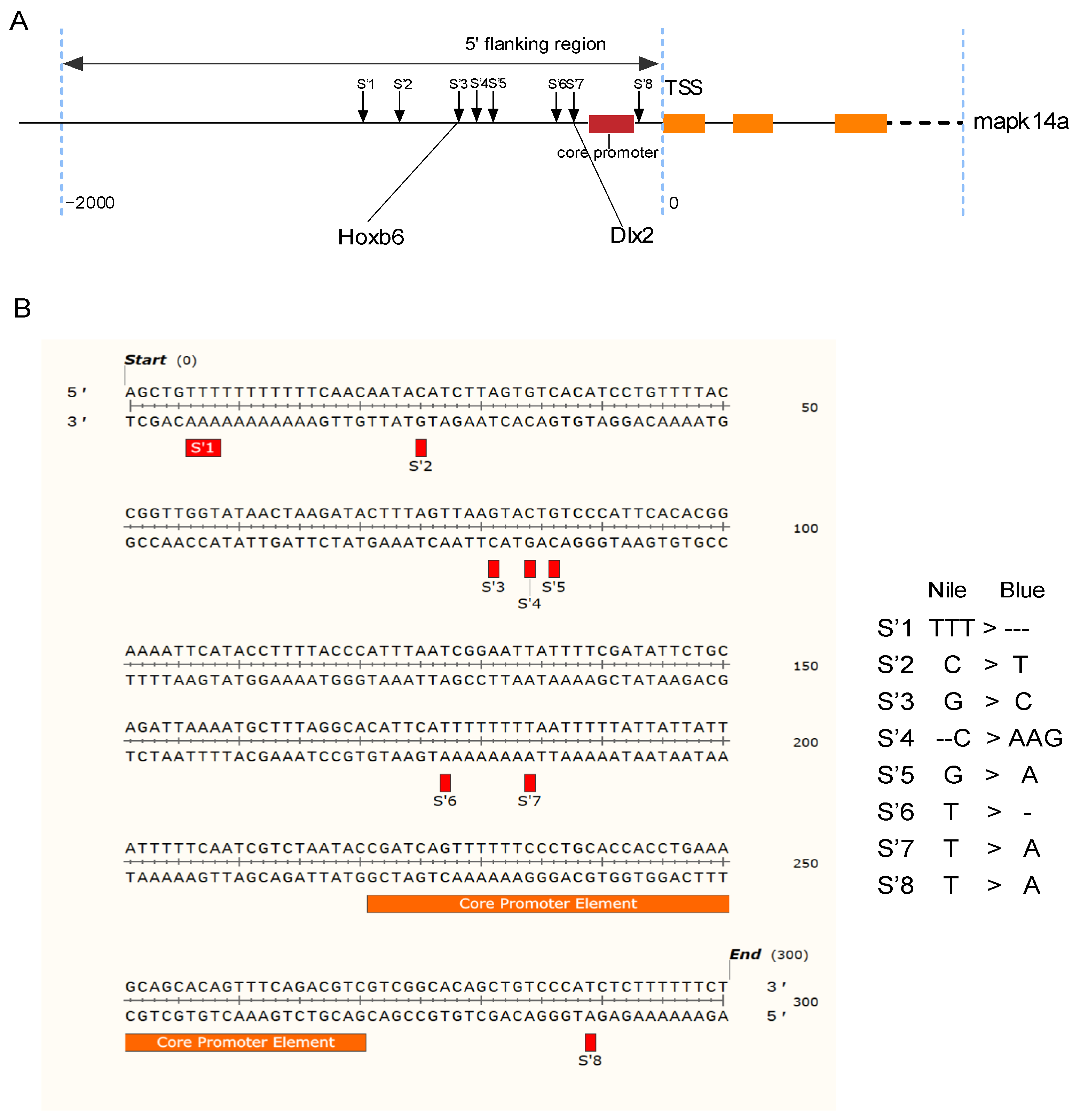
3.4. Distribution of SNP Sites in the 5′ Flanking Region of p38 Gene and Prediction of Transcription Factor Binding Sites
3.5. Promoter Relative Activity Detection
4. Discussion
4.1. The Role of Coding Region SNP Markers in Tilapia p38 Allele-Specific Expression Studies
4.2. Role of Cis-Acting Element Variants in p38 Allele-Specific Expression
4.3. Effect of Cis and Trans Compensation Effect on the Non-Additive Expression of p38 Gene
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mallet, J. Hybrid speciation. Nature 2007, 446, 279–283. [Google Scholar] [CrossRef]
- Fu, D.; Xiao, M.; Hayward, A.; Fu, Y.; Liu, G.; Jiang, G.; Zhang, H. Utilization of crop heterosis: A review. Euphytica 2014, 197, 161–173. [Google Scholar] [CrossRef]
- Hu, P.; Zou, Z.; Xiao, W.; Zhu, J.; Li, D.; Yu, J.; Yang, H.; Chen, B.; Ma, Y. Antibacterial ability of two species of tilapia is related to inducibility of the p38 mitogen-activated protein kinase pathway. Aquac. Res. 2021, 52, 456–462. [Google Scholar] [CrossRef]
- Liu, S.J. Gonadal structure of triploid crucian carp produced by crossing allotetraploid hybrids of Carassium auratus red var. (♀) × Cyprinus carpio (♂) with Japanese crucian carp (Carassius auratus Cavieri T. et S). Acta Hydrobiol. Sin. 2000, 24, 301–306. [Google Scholar] [CrossRef]
- Kiriyakit, A.; Gallardo, W.G.; Bart, A.N. Successful hybridization of groupers (Epinephelus coioides × Epinephelus lanceolatus) using cryopreserved sperm. Aquaculture 2011, 320, 106–112. [Google Scholar] [CrossRef]
- Beardmore, J.; Mair, G.; Lewis, R. Monosex male production in finfish as exemplified by tilapia: Applications, problems, and prospects. Aquaculture 2001, 197, 283–301. [Google Scholar] [CrossRef]
- Pruginin, Y.; Rothbard, S.; Wohlfarth, G.; Halevy, A.; Moav, R.; Hulata, G. All-male broods of Tilapia nilotica × T. aurea hybrids. Aquaculture 1975, 6, 11–12. [Google Scholar] [CrossRef]
- Stupar, R.M.; Gardiner, J.M.; Oldre, A.G.; Haun, W.J.; Chandler, V.L.; Springer, N.M. Gene expression analyses in maize inbreds and hybrids with varying levels of heterosis. BMC Plant Biol. 2008, 8, 33. [Google Scholar] [CrossRef]
- Xiao, Q.; Huang, Z.; Shen, Y.; Gan, Y.; Wang, Y.; Gong, S.; Lu, Y.; Luo, X.; You, W.; Ke, C. Transcriptome analysis reveals the molecular mechanisms of heterosis on thermal resistance in hybrid abalone. BMC Genom. 2021, 22, 650. [Google Scholar] [CrossRef]
- Ren, L.; Li, W.; Tao, M.; Qin, Q.; Luo, J.; Chai, J.; Tang, C.; Xiao, J.; Tang, X.; Lin, G.; et al. Homoeologue expression insights into the basis of growth heterosis at the intersection of ploidy and hybridity in Cyprinidae. Sci. Rep. 2016, 6, 27040. [Google Scholar] [CrossRef]
- Li, Q.; Li, Y.; Moose, S.P.; Hudson, M.E. Transposable elements, mRNA expression level and strand-specificity of small RNAs are associated with non-additive inheritance of gene expression in hybrid plants. BMC Plant Biol. 2015, 15, 168. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Xing, F.; Xu, C.; Zhang, Q.; Che, J.; Wang, X.; Song, J.; Li, X.; Xiao, J.; Chen, L.-L.; et al. Patterns of genome-wide allele-specific expression in hybrid rice and the implications on the genetic basis of heterosis. Proc. Natl. Acad. Sci. USA 2019, 116, 5653–5658. [Google Scholar] [CrossRef] [PubMed]
- Ren, P.; Deng, F.; Wang, Y.; Ran, J.; Li, J.; Yin, L.; Liu, X.; Chen, S.; Yang, C.; Jiang, X.; et al. Genome-wide analysis of spatiotemporal allele-specific expression in F1 hybrids of meat- and egg-type chickens. Gene 2020, 747, 144671. [Google Scholar] [CrossRef]
- Guo, M.; Yang, S.; Rupe, M.; Hu, B.; Bickel, D.R.; Arthur, L.; Smith, O. Genome-wide allele-specific expression analysis using Massively Parallel Signature Sequencing (MPSS (TM)) Reveals cis- and trans-effects on gene expression in maize hybrid meristem tissue. Plant Mol. Biol. 2008, 66, 551–563. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; Haerum, B.K.; Clark, A.G. Evolutionary changes in cis and trans gene regulation. Nature 2004, 430, 85–88. [Google Scholar] [CrossRef]
- Coolon, J.D.; Webb, W.; Wittkopp, P.J. Sex-specific effects of cis-regulatory variants in Drosophila melanogaster. Genetics 2013, 195, 1419–1422. [Google Scholar] [CrossRef]
- Zhu, F.; Schlupp, I.; Tiedemann, R. Allele-specific expression at the androgen receptor alpha gene in a hybrid unisexual fish, the Amazon molly (Poecilia formosa). PLoS ONE 2017, 12, e0186411. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Boswell, M.; Boswell, W.; Kneitz, S.; Hausmann, M.; Klotz, B.; Regneri, J.; Savage, M.; Amores, A.; Postlethwait, J.; et al. Molecular genetic analysis of the melanoma regulatory locus in Xiphophorus interspecies hybrids. Mol. Carcinog. 2017, 56, 1935–1944. [Google Scholar] [CrossRef]
- Zhong, H.; Zhang, X.; Xu, Q.; Yan, J.; Han, Z.; Zheng, H.; Xiao, J.; Tang, Z.; Wang, F.; Luo, Y.; et al. Nonadditive and asymmetric allelic expression of growth hormone in hybrid tilapia. Front. Genet. 2019, 10, 961. [Google Scholar] [CrossRef]
- Wang, S.; Ding, L.; Ji, H.; Xu, Z.; Liu, Q.; Zheng, Y. The role of p38 MAPK in the development of Diabetic Cardiomyopathy. Int. J. Mol. Sci. 2016, 17, 1037. [Google Scholar] [CrossRef]
- Ono, K.; Han, J. The p38 signal transduction pathway: Activation and function. Cell Signal 2000, 12, 1–13. [Google Scholar] [CrossRef]
- Umasuthan, N.; Bathige, S.; Noh, J.K.; Lee, J. Gene structure, molecular characterization and transcriptional expression of two p38 isoforms (MAPK11 and MAPK14) from rock bream (Oplegnathus fasciatus). Fish. Shellfish. Immunol. 2015, 47, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.E.; Jørgensen, J.B. Cloning and characterisation of p38 MAP kinase from Atlantic salmon—A kinase important for regulating salmon TNF-2 and IL-1 beta expression. Mol. Immunol. 2007, 44, 3137–3146. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.H.; Huang, Y.H.; OuYang, Z.L.; Cai, J.; Yan, Y.; Qin, Q.W. Roles of stress-activated protein kinases in the replication of Singapore grouper iridovirus and regulation of the inflammatory responses in grouper cells. J. Gen. Virol. 2011, 92, 1292–1301. [Google Scholar] [CrossRef]
- Zhu, J.L.; Zou, Z.Y.; Li, D.Y.; Xiao, W.; Yu, J.; Chen, B.L.; Xue, L.Y.; Yang, H. Transcriptome profiling revealed basis for growth heterosis in hybrid tilapia (Oreochromis niloticus ♀ × O. aureus ♂). Fishes 2022, 7, 43. [Google Scholar] [CrossRef]
- Maroni, P.; Bendinelli, P.; Zuccorononno, C.; Schiaffonati, L.; Piccoletti, R. Cellular signalling after heat shock in the liver. Cell Biol. Int. 2000, 24, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.L.; Ng, D.W.-K.; Zhang, C.Q.; Comai, L.; Ye, W.X.; Chen, Z.J. Cis- and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids. Nat. Commun. 2012, 3, 950. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Fan, J.; Liu, S.; Yang, Q.; Mu, G.; He, C. Characterization of a myostatin gene (MSTN1) from spotted halibut (Verasper variegatus) and association between its promoter polymorphism and individual growth performance. Comp. Biochem. Phys. B 2012, 161, 315–322. [Google Scholar] [CrossRef]
- Chen, B.; Xiao, W.; Zou, Z.; Zhu, J.; Li, D.; Yu, J.; Yang, H. The effects of single nucleotide polymorphisms in Neuropeptide Y and prepro-orexin on growth in Nile tilapia (Oreochromis niloticus). Aquaculture 2021, 543, 736974. [Google Scholar] [CrossRef]
- Zhou, R.T.; Yue, Z.F.; Ji, J.J. Cloning and transcriptional regulation of slitrk3 gene promoter in large yellow croaker (Larimichthys crocea). South China Fish. Sci. 2023, 19, 68–76. [Google Scholar] [CrossRef]
- Ren, L.; Cui, J.; Wang, J.; Tan, H.; Li, W.; Tang, C.; Qin, Q.; Liu, S. Analyzing homoeolog expression provides insights into the rediploidization event in gynogenetic hybrids of Carassius auratus red var. × Cyprinus carpio. Sci. Rep. 2017, 7, 13679. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Li, J.; Zhang, J.; Liang, X.; Zhang, X.; Wang, T.; Yin, S. Integrated analysis of transcriptomic, miRNA and proteomic changes of a novel hybrid yellow catfish uncovers key roles for miRNAs in heterosis. Mol. Cell Proteom. 2019, 18, 1437–1453. [Google Scholar] [CrossRef] [PubMed]
- Wei, L. Selection on synonymous mutations revealed by 1135 genomes of Arabidopsis thaliana. Evol. Bioinform. 2020, 16, 1176934320916794. [Google Scholar] [CrossRef]
- Dana, A.; Tuller, T. The effect of tRNA levels on decoding times of mRNA codons. Nucleic Acids Res. 2014, 42, 9171–9181. [Google Scholar] [CrossRef]
- Hwang, J.H.; Kwon, S.G.; Park, D.H.; Kim, T.W.; Kang, D.G.; Ha, J.; Kim, S.W.; Kim, C.W. Molecular characterization of porcine PGM1 gene associated with meat quality traits. Can. J. Anim. Sci. 2015, 95, 31–36. [Google Scholar] [CrossRef]
- Wang, Y.; Qiu, C.; Cui, Q. A large-scale analysis of the relationship of synonymous SNPs changing microRNA regulation with functionality and disease. Int. J. Mol. Sci. 2015, 16, 23545–23555. [Google Scholar] [CrossRef] [PubMed]
- Hubner, N.; Wallace, C.A.; Zimdahl, H.; Petretto, E.; Schulz, H.; Maciver, F.; Mueller, M.; Hummel, O.; Monti, J.; Zidek, V.; et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat. Genet. 2005, 37, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Pastinen, T. Genome-wide allele-specific analysis: Insights into regulatory variation. Nat. Rev. Genet. 2010, 11, 533–538. [Google Scholar] [CrossRef]
- Bell, G.D.; Kane, N.C.; Rieseberg, L.H.; Adams, K.L. RNA-seq analysis of allele-Specific expression, hybrid effects, and regulatory divergence in hybrids compared with their parents from natural populations. Genome Biol. Evol. 2013, 5, 1309–1323. [Google Scholar] [CrossRef]
- He, F.; Zhang, X.; Hu, J.; Turck, F.; Dong, X.; Goebel, U.; Borevitz, J.; de Meaux, J. Genome-wide analysis of cis-regulatory divergence between species in the Arabidopsis genus. Mol. Biol. Evol. 2012, 29, 3385–3395. [Google Scholar] [CrossRef]
- McManus, C.J.; Coolon, J.D.; Duff, M.O.; Eipper-Mains, J.; Graveley, B.R.; Wittkopp, P.J. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 2010, 20, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.H.; Zhao, J.L.; Chen, X.W. Genders and gonadal development in hybrids of Nile tilapia Oreochromis niloticus ♀ × O. aureus ♂. Fish. Sci. 2017, 36, 467–471. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; Haerum, B.K.; Clark, A.G. Regulatory changes underlying expression differences within and between Drosophila species. Nat. Genet. 2008, 40, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Meiklejohn, C.D.; Coolon, J.D.; Hartl, D.L.; Wittkopp, P.J. The roles of cis- and trans- regulation in the evolution of regulatory incompatibilities and sexually dimorphic gene expression. Genome Res. 2014, 24, 84–95. [Google Scholar] [CrossRef]
- Combes, M.-C.; Hueber, Y.; Dereeper, A.; Rialle, S.; Herrera, J.-C.; Lashermes, P. Regulatory divergence between parental alleles determines gene expression patterns in hybrids. Genome Biol. Evol. 2015, 7, 1110–1121. [Google Scholar] [CrossRef]
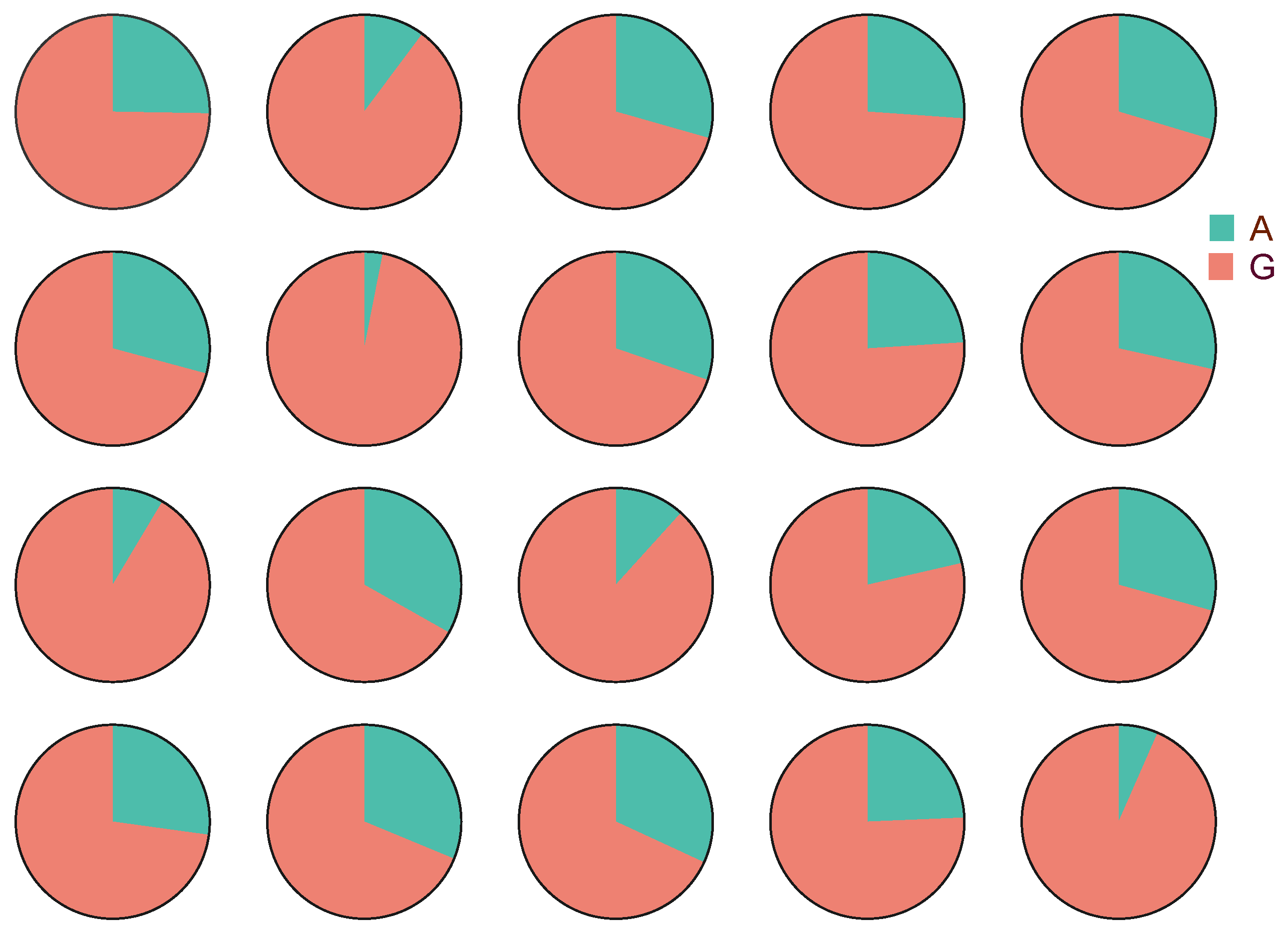


| Gene | Primers Sequence (5′-3′) | Annealing Temperature | Use | Product Length |
|---|---|---|---|---|
| p38-P1-F | GCGCCGTTTAGCGAAGCGA | 59 °C | cDNA | 731 |
| p38-P1-R | TGTATGATGCCTGCTGAGTGA | |||
| p38-P2-F | GACTCATCTAATGGGGGCAG | 57 °C | cDNA | 752 |
| p38-P2-R | CATCATCAAAGCTAGGTGGCT | |||
| p38-P3-F | CTGAAGCCGAGCCTTATGAC | 60 °C | cDNA | 888 |
| p38-P3-R | CTTAAGGGATCTTTCGATTGC | |||
| p38-F | ACATCAGCTCCTTGCCACACA | 60 °C | cDNA | 135 |
| p38-R | GTGAGCTTCAGCTGCCGTTATC | |||
| β-actin-F | CTCTGGTCGTACCACTGGTATCG | 62 °C | cDNA | 169 |
| β-actin-R | GAAGGAGTAGCCACGCTCCTC | |||
| p38-PS-F | GCTCAATACCACGATCCAGAC | 55 °C | Pyrophosphate sequencing | 193 |
| p38-PS-BR | AAAAGCTTGGGTTGAACTGGT | |||
| p38-P2-FS | AGCCACCTAGCTTTGATGATG |
| Primers | Primers’ Sequences (5′-3′) | Annealing Temperature | Product Length (bp) |
|---|---|---|---|
| p38-P4-F | GCTACTTGAGCTGACTTACCTTG | 57 °C | 387 bp |
| p38-P4-R | ACAAACAAATTCTCAGTCTTTACAT | ||
| p38-P5-F | ACAGTGAGCAAATCAGCAGTC | 59 °C | 352 bp |
| p38-P5-R | TCTCCCCCTGATGGTCTGAG | ||
| p38-P6-F | CCAGAATTAGATCCAAGCTTTC | 59 °C | 217 bp |
| p38-P6-R | ACCTATGCAGGTCTACTATGCC | ||
| p38-P7-F | AATACCTTAAGGCATAGTAGACC | 58 °C | 278 bp |
| p38-P7-R | CCAAGTATAAAAGAGGAGAAAAGATA | ||
| p38-P8-F | TATCTTTTCTCCTCTTTTATACTTGG | 56 °C | 268 bp |
| p38-P8-R | ACCAACAACCACTTTATTTTATTTC | ||
| p38-P9-F | TTCAGGTCTCCACGTTATTTGTTC | 57 °C | 382 bp |
| p38-P9-R | AATTCGTAAATAAATGTAGCTG | ||
| p38-P10-F | AATACCTTAAGGCATAGTAGACCTGC | 59 °C | 669 bp |
| p38-P10-R | TGAATTTTCCGTGTGAATGGGAC | ||
| p38-P11-F | CAGGACGTGACCTCTGATGC | 57 °C | 612 bp |
| p38-P11-R | TTCAGGTCTCCACGTTATTTGTTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Chen, B.; Zou, Z.; Li, D.; Zhu, J.; Yu, J.; Xiao, W.; Yang, H. Non-Additive and Asymmetric Allelic Expression of p38 mapk in Hybrid Tilapia (Oreochromis niloticus ♀ × O. aureus ♂). Animals 2024, 14, 266. https://doi.org/10.3390/ani14020266
Liu Z, Chen B, Zou Z, Li D, Zhu J, Yu J, Xiao W, Yang H. Non-Additive and Asymmetric Allelic Expression of p38 mapk in Hybrid Tilapia (Oreochromis niloticus ♀ × O. aureus ♂). Animals. 2024; 14(2):266. https://doi.org/10.3390/ani14020266
Chicago/Turabian StyleLiu, Zihui, Binglin Chen, Zhiying Zou, Dayu Li, Jinglin Zhu, Jie Yu, Wei Xiao, and Hong Yang. 2024. "Non-Additive and Asymmetric Allelic Expression of p38 mapk in Hybrid Tilapia (Oreochromis niloticus ♀ × O. aureus ♂)" Animals 14, no. 2: 266. https://doi.org/10.3390/ani14020266





