Prevalence and Genetic Variation Investigation of the Pseudorabies Virus in Southwest China
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Detection of PRV gE Antibody by ELISA
2.3. Detection of PRV Antigen via PCR
2.4. Partial Genome Sequencing
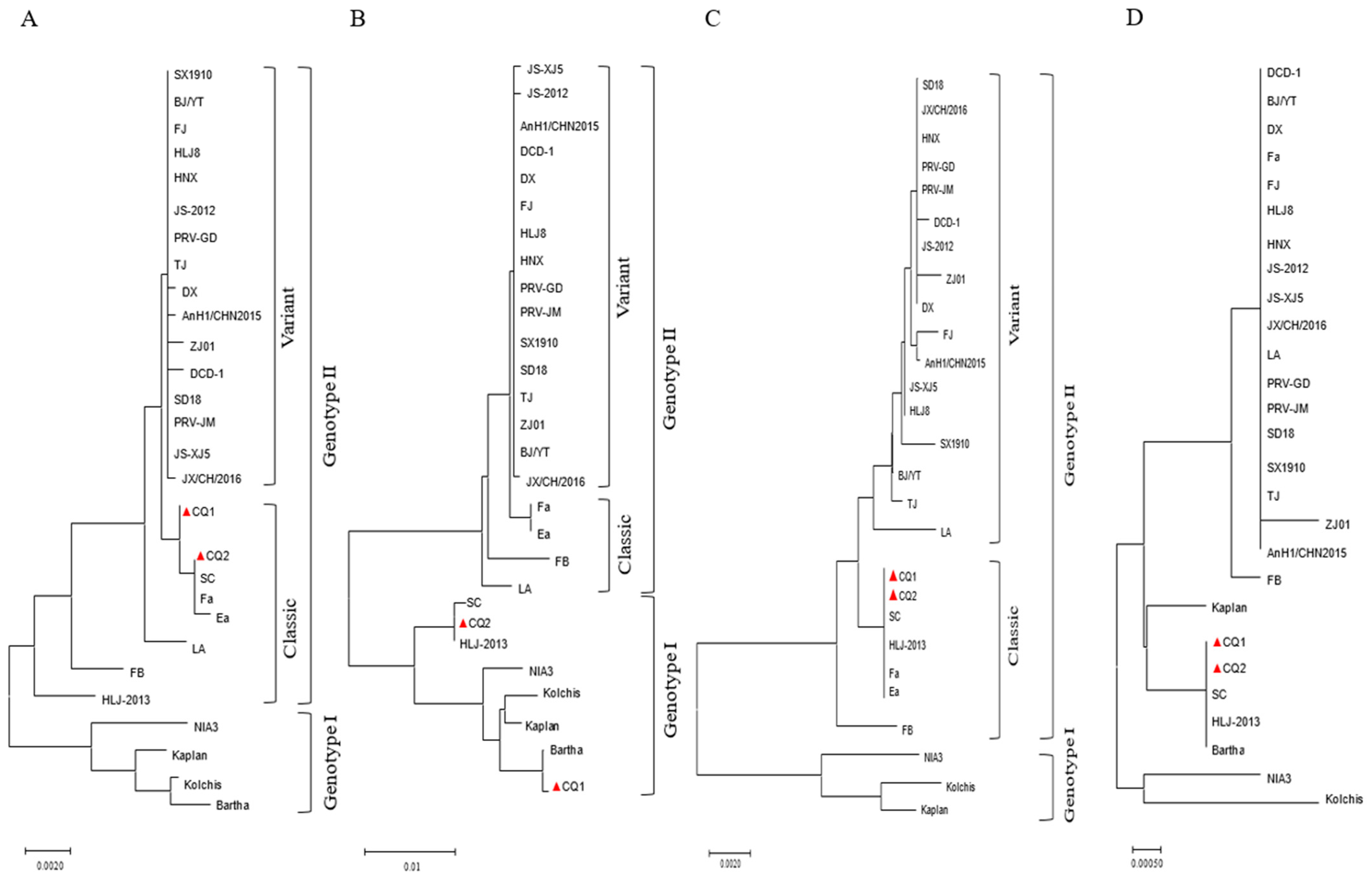
2.5. Phylogenetic and Genomic Analysis
3. Results
3.1. Prevalence Analysis of PRV in Southwest China’s Pig Populations
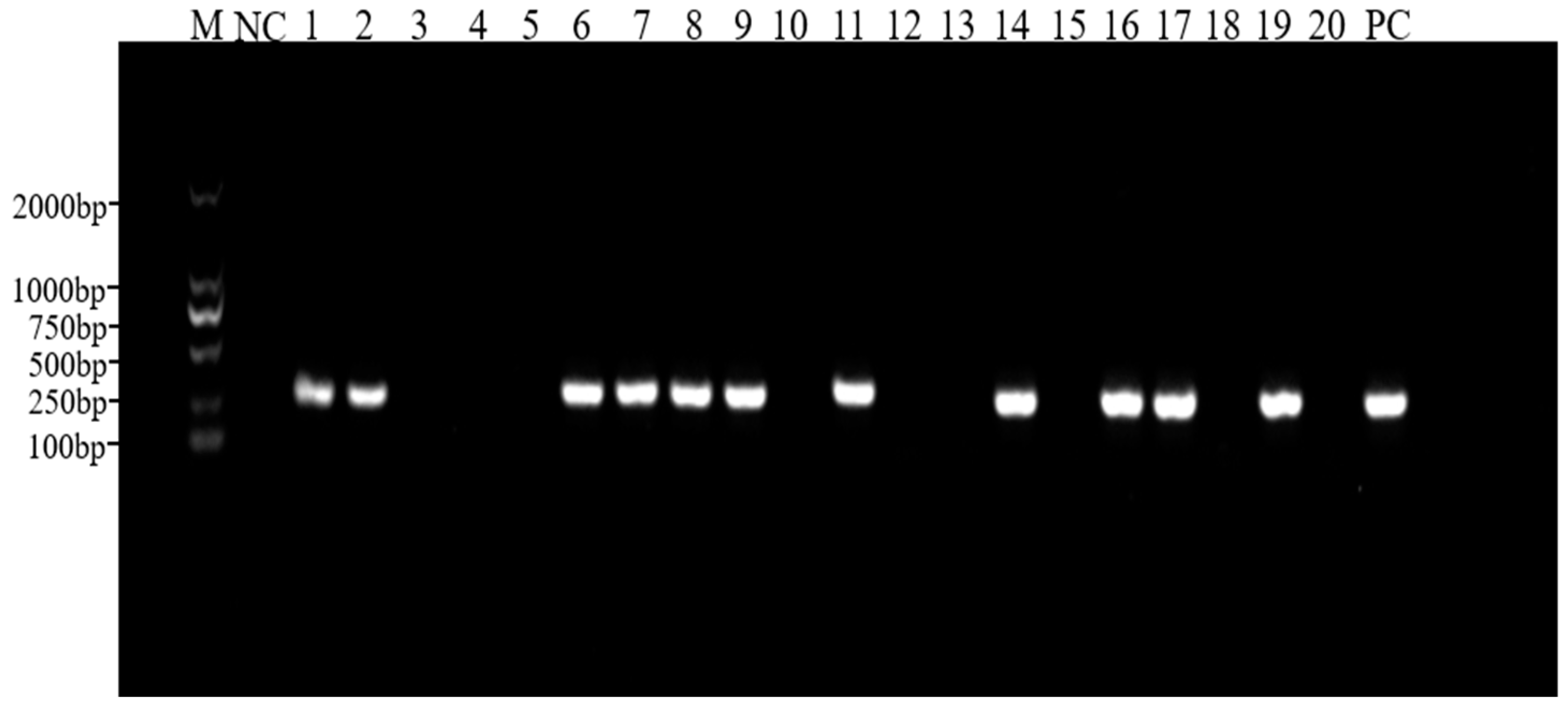
3.2. PCR Detection of PRV Antigen
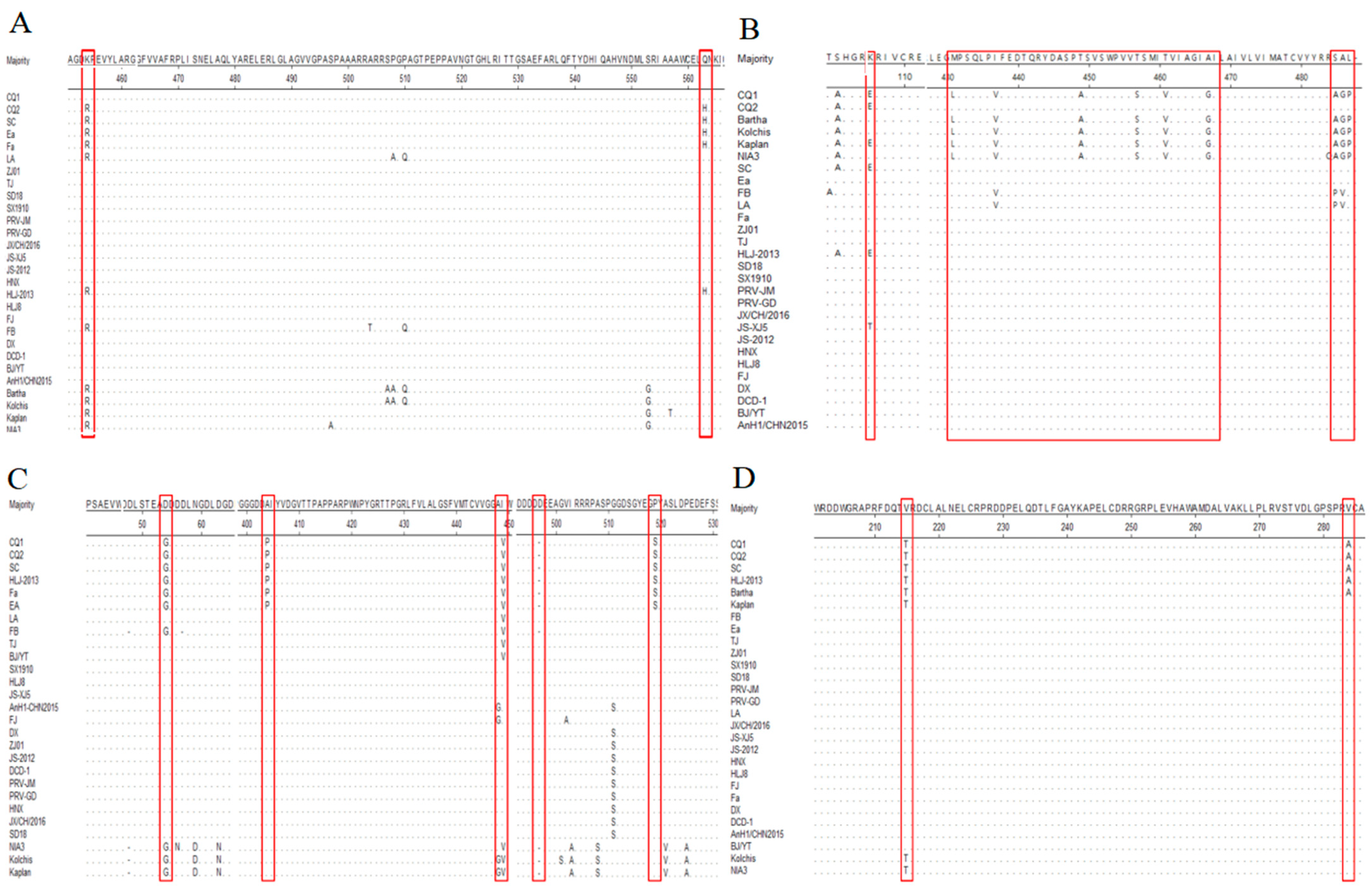
3.3. Variation Analysis of gB Gene of Epidemic PRV Strains
3.4. Variation Analysis of gC Gene of Epidemic PRV Strains
3.5. Variation Analysis of gE Gene of Epidemic PRV Strains
3.6. Variation Analysis of TK Gene of Epidemic PRV Strains
4. Discussion
4.1. Prevalence and Economic Impact of PRV Infections in Southwest China: Comparative Analysis and the Imperative for Enhanced Surveillance
4.2. Molecular Characterization of PRV Strains in Southwest China: Implications for Vaccine Efficacy and Disease Management
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, H.H.; Fu, P.F.; Chen, H.Y.; Wang, Z.Y. Pseudorabies Virus: From Pathogenesis to Prevention Strategies. Viruses 2022, 14, 1638. [Google Scholar] [CrossRef] [PubMed]
- Bo, Z.; Zhu, J.; Guo, M.; Zhang, C.; Cao, Y.; Zhang, X.; Wu, Y. Gallocatechin Gallate Inhibits the Replication of Pseudorabies Virus via Suppressing the Entry and Release Stages in Its Replication Cycle. Vet. Sci. 2023, 10, 189. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.J.; Yuan, W.Z.; Zhu, Y.P.; Hou, S.H.; Wang, X.J. Latent pseudorabies virus infection in medulla oblongata from quarantined pigs. Transbound. Emerg. Dis. 2021, 68, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Hagemoser, W.A.; Kluge, J.P.; Hill, H.T. Studies on the pathogenesis of pseudorabies in domestic cats following oral inoculation. Can. J. Comp. Med. 1980, 44, 192–202. [Google Scholar]
- Jin, H.L.; Gao, S.M.; Liu, Y.; Zhang, S.F.; Hu, R.L. Pseudorabies in farmed foxes fed pig offal in Shandong province, China. Arch. Virol. 2016, 161, 445–448. [Google Scholar] [CrossRef]
- Zhang, L.; Zhong, C.; Wang, J.; Lu, Z.; Liu, L.; Yang, W.; Lyu, Y. Pathogenesis of natural and experimental Pseudorabies virus infections in dogs. Virol. J. 2015, 12, 44. [Google Scholar] [CrossRef]
- Lian, K.; Zhang, M.; Zhou, L.; Song, Y.; Wang, G.; Wang, S. First report of a pseudorabies-virus-infected wolf (Canis lupus) in China. Arch. Virol. 2020, 165, 459–462. [Google Scholar] [CrossRef]
- McCracken, R.M.; McFerran, J.B.; Dow, C. The neural spread of pseudorabies virus in calves. J. Gen. Virol. 1973, 20, 17–28. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, X.; Xie, C.; Ding, S.; Yang, H.; Guo, S.; Li, J.; Qin, L.; Ban, F.; Wang, D.; et al. A Novel Human Acute Encephalitis Caused by Pseudorabies Virus Variant Strain. Clin. Infect. Dis. 2021, 73, e3690–e3700. [Google Scholar] [CrossRef]
- Ai, J.W.; Weng, S.S.; Cheng, Q.; Cui, P.; Li, Y.J.; Wu, H.L.; Zhu, Y.M.; Xu, B.; Zhang, W.H. Human Endophthalmitis Caused By Pseudorabies Virus Infection, China, 2017. Emerg. Infect. Dis. 2018, 24, 1087–1090. [Google Scholar] [CrossRef]
- Yang, X.; Guan, H.; Li, C.; Li, Y.; Wang, S.; Zhao, X.; Zhao, Y.; Liu, Y. Characteristics of human encephalitis caused by pseudorabies virus: A case series study. Int. J. Infect. Dis. 2019, 87, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Han, H.; Wang, H.; Cui, Y.; Liu, H.; Ding, S. A Case of Human Viral Encephalitis Caused by Pseudorabies Virus Infection in China. Front. Neurol. 2019, 10, 534. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhou, Y.; Luo, Y.; Chen, Y. Effects of gE/gI deletions on the miRNA expression of PRV-infected PK-15 cells. Virus Genes 2020, 56, 461–471. [Google Scholar] [CrossRef]
- Zheng, H.H.; Jin, Y.; Hou, C.Y.; Li, X.S.; Zhao, L.; Wang, Z.Y.; Chen, H.Y. Seroprevalence investigation and genetic analysis of pseudorabies virus within pig populations in Henan province of China during 2018–2019. Infect. Genet. Evol. 2021, 92, 104835. [Google Scholar] [CrossRef]
- Xiang, S.; Zhou, Z.; Hu, X.; Li, Y.; Zhang, C.; Wang, J.; Li, X.; Tan, F.; Tian, K. Complete Genome Sequence of a Variant Pseudorabies Virus Strain Isolated in Central China. Genome Announc. 2016, 4, e00149-16. [Google Scholar] [CrossRef]
- Yao, J.; Li, J.; Gao, L.; He, Y.; Xie, J.; Zhu, P.; Zhang, Y.; Zhang, X.; Duan, L.; Yang, S.; et al. Epidemiological Investigation and Genetic Analysis of Pseudorabies Virus in Yunnan Province of China from 2017 to 2021. Viruses 2022, 14, 895. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kuang, Y.; Li, Y.; Guo, H.; Zhou, C.; Guo, S.; Tan, C.; Wu, B.; Chen, H.; Wang, X. The Epidemiology and Variation in Pseudorabies Virus: A Continuing Challenge to Pigs and Humans. Viruses 2022, 14, 1463. [Google Scholar] [CrossRef]
- OIE. OIE World Animal Health Information System. Available online: https://wahis.oie.int/#/dashboards/ (accessed on 30 September 2024).
- Sun, Y.; Luo, Y.; Wang, C.H.; Yuan, J.; Li, N.; Song, K.; Qiu, H.J. Control of swine pseudorabies in China: Opportunities and limitations. Vet. Microbiol. 2016, 183, 119–124. [Google Scholar] [CrossRef]
- Bo, Z.; Li, X. A Review of Pseudorabies Virus Variants: Genomics, Vaccination, Transmission, and Zoonotic Potential. Viruses 2022, 14, 1003. [Google Scholar] [CrossRef]
- Yu, X.; Zhou, Z.; Hu, D.; Zhang, Q.; Han, T.; Li, X.; Gu, X.; Yuan, L.; Zhang, S.; Wang, B.; et al. Pathogenic pseudorabies virus, China, 2012. Emerg. Infect. Dis. 2014, 20, 102–104. [Google Scholar] [CrossRef]
- Tong, G.; Chen, H. Pseudorabies epidemic status and control measures in China. Chin. J. Vet. Sci. 1999, 19, 1–2. [Google Scholar]
- An, T.Q.; Peng, J.M.; Tian, Z.J.; Zhao, H.Y.; Li, N.; Liu, Y.M.; Chen, J.Z.; Leng, C.L.; Sun, Y.; Chang, D.; et al. Pseudorabies virus variant in Bartha-K61-vaccinated pigs, China, 2012. Emerg. Infect. Dis. 2013, 19, 1749–1755. [Google Scholar] [CrossRef] [PubMed]
- Mulder, W.A.; Jacobs, L.; Priem, J.; Kok, G.L.; Wagenaar, F.; Kimman, T.G.; Pol, J.M. Glycoprotein gE-negative pseudorabies virus has a reduced capability to infect second- and third-order neurons of the olfactory and trigeminal routes in the porcine central nervous system. J. Gen. Virol. 1994, 75 Pt 11, 3095–3106. [Google Scholar] [CrossRef]
- Ferrari, M.; Mettenleiter, T.C.; Romanelli, M.G.; Cabassi, E.; Corradi, A.; Dal Mas, N.; Silini, R. A comparative study of pseudorabies virus (PRV) strains with defects in thymidine kinase and glycoprotein genes. J. Comp. Pathol. 2000, 123, 152–163. [Google Scholar] [CrossRef]
- Li, J.; Fang, K.; Rong, Z.; Li, X.; Ren, X.; Ma, H.; Chen, H.; Li, X.; Qian, P. Comparison of gE/gI- and TK/gE/gI-Gene-Deleted Pseudorabies Virus Vaccines Mediated by CRISPR/Cas9 and Cre/Lox Systems. Viruses 2020, 12, 369. [Google Scholar] [CrossRef]
- Freuling, C.M.; Muller, T.F.; Mettenleiter, T.C. Vaccines against pseudorabies virus (PrV). Vet. Microbiol. 2017, 206, 3–9. [Google Scholar] [CrossRef]
- He, W.; Auclert, L.Z.; Zhai, X.; Wong, G.; Zhang, C.; Zhu, H.; Xing, G.; Wang, S.; He, W.; Li, K.; et al. Interspecies Transmission, Genetic Diversity, and Evolutionary Dynamics of Pseudorabies Virus. J. Infect. Dis. 2019, 219, 1705–1715. [Google Scholar] [CrossRef]
- Ma, Z.; Han, Z.; Liu, Z.; Meng, F.; Wang, H.; Cao, L.; Li, Y.; Jiao, Q.; Liu, S.; Liu, M. Epidemiological investigation of porcine pseudorabies virus and its coinfection rate in Shandong Province in China from 2015 to 2018. J. Vet. Sci. 2020, 21, e36. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros, C.; Pouliot, M.; Froment, R.; Maghezzi, M.S.; St-Jean, C.; Li, C.; Paquette, D.; Authier, S. Cerebrospinal Fluid Characterization in Cynomolgus Monkeys, Beagle Dogs, and Gottingen Minipigs. Int. J. Toxicol. 2020, 39, 124–130. [Google Scholar] [CrossRef]
- Song, C.; Ye, H.; Zhang, X.; Zhang, Y.; Li, Y.; Yao, J.; Gao, L.; Wang, S.; Yu, Y.; Shu, X. Isolation and Characterization of Yunnan Variants of the Pseudorabies Virus and Their Pathogenicity in Rats. Viruses 2024, 16, 233. [Google Scholar] [CrossRef]
- Ye, C.; Guo, J.C.; Gao, J.C.; Wang, T.Y.; Zhao, K.; Chang, X.B.; Wang, Q.; Peng, J.M.; Tian, Z.J.; Cai, X.H.; et al. Genomic analyses reveal that partial sequence of an earlier pseudorabies virus in China is originated from a Bartha-vaccine-like strain. Virology 2016, 491, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.; Xiao, S.; Zhou, R.; Fang, L.; He, Q.; Wu, B.; Zhou, F.; Chen, H. Protection induced by intramuscular immunization with DNA vaccines of pseudorabies in mice, rabbits and piglets. Vaccine 2002, 20, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Yuan, J.; Qin, H.Y.; Luo, Y.; Cong, X.; Li, Y.; Chen, J.; Li, S.; Sun, Y.; Qiu, H.J. A novel gE-deleted pseudorabies virus (PRV) provides rapid and complete protection from lethal challenge with the PRV variant emerging in Bartha-K61-vaccinated swine population in China. Vaccine 2014, 32, 3379–3385. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.; Liu, F.; Zheng, H.; Liang, C.; Zhou, Y.J.; Jiang, Y.F.; Shan, T.L.; Gao, F.; Li, G.X.; Tong, G.Z. Emergence of a Pseudorabies virus variant with increased virulence to piglets. Vet. Microbiol. 2015, 181, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Shi, Z.; Liu, C.; Wang, P.; Wang, M.; Wang, S.; Liu, Z.; Wei, L.; Sun, Z.; He, X.; et al. Implication of the Identification of an Earlier Pseudorabies Virus (PRV) Strain HLJ-2013 to the Evolution of Chinese PRVs. Front. Microbiol. 2020, 11, 612474. [Google Scholar] [CrossRef]
- Zhang, L.; Ren, W.; Chi, J.; Lu, C.; Li, X.; Li, C.; Jiang, S.; Tian, X.; Li, F.; Wang, L.; et al. Epidemiology of Porcine Pseudorabies from 2010 to 2018 in Tianjin, China. Viral Immunol. 2021, 34, 714–721. [Google Scholar] [CrossRef]
- Gu, Z.; Hou, C.; Sun, H.; Yang, W.; Dong, J.; Bai, J.; Jiang, P. Emergence of highly virulent pseudorabies virus in southern China. Can. J. Vet. Res. 2015, 79, 221–228. [Google Scholar]
- Gao, W.; Jiang, X.; Hu, Z.; Wang, Q.; Shi, Y.; Tian, X.; Qiao, M.; Zhang, J.; Li, Y.; Li, X. Epidemiological investigation, determination of related factors, and spatial-temporal cluster analysis of wild type pseudorabies virus seroprevalence in China during 2022. Front. Vet. Sci. 2023, 10, 1298434. [Google Scholar] [CrossRef]
- Sun, Y.; Liang, W.; Liu, Q.; Zhao, T.; Zhu, H.; Hua, L.; Peng, Z.; Tang, X.; Stratton, C.W.; Zhou, D.; et al. Epidemiological and genetic characteristics of swine pseudorabies virus in mainland China between 2012 and 2017. PeerJ 2018, 6, e5785. [Google Scholar] [CrossRef]
- Zhou, H.; Pan, Y.; Liu, M.; Han, Z. Prevalence of Porcine Pseudorabies Virus and Its Coinfection Rate in Heilongjiang Province in China from 2013 to 2018. Viral Immunol. 2020, 33, 550–554. [Google Scholar] [CrossRef]
- Lin, Y.; Tan, L.; Wang, C.; He, S.; Fang, L.; Wang, Z.; Zhong, Y.; Zhang, K.; Liu, D.; Yang, Q.; et al. Serological Investigation and Genetic Characteristics of Pseudorabies Virus in Hunan Province of China From 2016 to 2020. Front. Vet. Sci. 2021, 8, 762326. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Xue, T.; Zhao, X.; Zou, J.; Pu, H.; Hu, X.; Tian, Z. Pseudorabies Virus Associations in Wild Animals: Review of Potential Reservoirs for Cross-Host Transmission. Viruses 2022, 14, 2254. [Google Scholar] [CrossRef]
- Li, X.D.; Fu, S.H.; Chen, L.Y.; Li, F.; Deng, J.H.; Lu, X.C.; Wang, H.Y.; Tian, K.G. Detection of Pseudorabies Virus Antibodies in Human Encephalitis Cases. Biomed. Environ. Sci. 2020, 33, 444–447. [Google Scholar] [CrossRef]
- Gu, J.; Hu, D.; Peng, T.; Wang, Y.; Ma, Z.; Liu, Z.; Meng, F.; Shang, Y.; Liu, S.; Xiao, Y. Epidemiological investigation of pseudorabies in Shandong Province from 2013 to 2016. Transbound. Emerg. Dis. 2018, 65, 890–898. [Google Scholar] [CrossRef] [PubMed]
- Bo, Z.; Miao, Y.; Xi, R.; Gao, X.; Miao, D.; Chen, H.; Jung, Y.S.; Qian, Y.; Dai, J. Emergence of a novel pathogenic recombinant virus from Bartha vaccine and variant pseudorabies virus in China. Transbound. Emerg. Dis. 2021, 68, 1454–1464. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Cheng, J.; Pan, H.; Yang, F.; Zhu, X.; Wu, J.; Pan, H.; Yan, P.; Zhou, J.; Gao, Q.; et al. Analysis of the recombination and evolution of the new type mutant pseudorabies virus XJ5 in China. BMC Genom. 2024, 25, 752. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Tang, W.; Wang, X.; Zhao, J.; Peng, K.; Sun, X.; Li, S.; Kuang, S.; Zhu, L.; Zhou, Y.; et al. The Genetic Characterization of a Novel Natural Recombinant Pseudorabies Virus in China. Viruses 2022, 14, 978. [Google Scholar] [CrossRef]
- Vallbracht, M.; Brun, D.; Tassinari, M.; Vaney, M.C.; Pehau-Arnaudet, G.; Guardado-Calvo, P.; Haouz, A.; Klupp, B.G.; Mettenleiter, T.C.; Rey, F.A.; et al. Structure-Function Dissection of Pseudorabies Virus Glycoprotein B Fusion Loops. J. Virol. 2018, 92, e01203-17. [Google Scholar] [CrossRef]
- Yu, Z.Q.; Tong, W.; Zheng, H.; Li, L.W.; Li, G.X.; Gao, F.; Wang, T.; Liang, C.; Ye, C.; Wu, J.Q.; et al. Variations in glycoprotein B contribute to immunogenic difference between PRV variant JS-2012 and Bartha-K61. Vet. Microbiol. 2017, 208, 97–105. [Google Scholar] [CrossRef]
- Hu, R.M.; Zhou, Q.; Song, W.B.; Sun, E.C.; Zhang, M.M.; He, Q.G.; Chen, H.C.; Wu, B.; Liu, Z.F. Novel pseudorabies virus variant with defects in TK, gE and gI protects growing pigs against lethal challenge. Vaccine 2015, 33, 5733–5740. [Google Scholar] [CrossRef]
- Mettenleiter, T.C. Immunobiology of pseudorabies (Aujeszky’s disease). Vet. Immunol. Immunopathol. 1996, 54, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Zhang, Q.Z.; Tian, Z.J.; Zheng, H.; Zhao, K.; Liu, F.; Guo, J.C.; Tong, W.; Jiang, C.G.; Wang, S.J.; et al. Genomic characterization of emergent pseudorabies virus in China reveals marked sequence divergence: Evidence for the existence of two major genotypes. Virology 2015, 483, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Wang, H.; Zhou, L.; Ge, X.; Guo, X.; Han, J.; Yang, H. Glycoproteins C and D of PRV Strain HB1201 Contribute Individually to the Escape From Bartha-K61 Vaccine-Induced Immunity. Front. Microbiol. 2020, 11, 323. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Li, Z.; Nan, X.; Wu, Y.; Li, Y. Isolation and identification of pseudorabies virus. Chin. J. Prev. Vet. Med. 1987, 3, 10–11. [Google Scholar]
| Primer | Sequence (5′-3′) | Amplicon Size (bp) | Primer Function |
|---|---|---|---|
| gE1-F | GCGGACGCACATGCTCTCTC | 250 | Detection of gE gene |
| gE1-R | CGGTCACGCCATAGTTGGGT | ||
| gE2-F | CGTCCCCCAGCCCAAGAT | 2048 | Cloning PRV gE gene |
| gE2-R | GTCCCTTGGGGGCCAGCA | ||
| gB1-F | AGACGTGCGATCAACGGCAT | 1146 | Segmented cloning of PRV gB gene |
| gB1-R | AACAAGGACCGCACCCTGTG | ||
| gB2-F | ACCCGCCGCCCAGCTTAAAG | 1346 | |
| gB2-R | CGTCTCCAAGGCCGAGTACG | ||
| gB3-F | GCAGGCCGTAGAAGGGGGAC | 1126 | |
| gB3-R | CGGCTTCTACCGCTTCCAGA | ||
| gC-F | CGTTTCCTGATTCACGCCCAC | 1917 | Cloning PRV gC gene |
| gC-R | GCACCATCGACGCCAGCTC | ||
| TK-F | GCGCACCCCGAGGTTGACTT | 1255 | Cloning PRV TK gene |
| TK-R | GACGGGCACGGCAAACTTT |
| CQ1 Nucleotide (nt) and Amino Acid (aa) Sequence Identity% | CQ2 Nucleotide (nt) and Amino Acid (aa) Sequence Identity% | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gB | gC | gE | TK | gB | gC | gE | TK | |||||||||||||
| No | GenBank | Strain | Collection Data | Region | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa |
| 1 | MK618718.1 | AnH1/CHN2015 | 2015 | China | 99.9 | 99.5 | 95.6 | 92.7 | 99.1 | 99.0 | 99.7 | 99.1 | 99.8 | 99.3 | 95.5 | 93.2 | 99.1 | 99.0 | 99.7 | 99.1 |
| 2 | JF797217.1 | Bartha | 1961 | Hungary | 98.2 | 95.7 | 99.9 | 99.6 | / | / | 100.0 | 99.7 | 97.8 | 96.2 | 98.9 | 98.0 | / | / | 100.0 | 99.7 |
| 3 | KC981239.1 | BJ/YT | 2013 | China | 99.9 | 99.6 | 95.6 | 92.7 | 99.6 | 99.5 | 99.7 | 99.1 | 99.8 | 99.5 | 95.5 | 93.2 | 99.6 | 99.5 | 99.7 | 99.1 |
| 4 | OL639029.1 | DCD-1 | 2017 | China | 99.8 | 99.5 | 95.6 | 92.7 | 99.1 | 99.0 | 99.7 | 99.1 | 99.7 | 99.3 | 95.5 | 93.2 | 99.1 | 99.0 | 99.7 | 99.1 |
| 5 | MZ063026.1 | DX | 2012 | China | 99.9 | 99.5 | 95.6 | 92.7 | 99.5 | 99.1 | 99.7 | 99.1 | 99.8 | 99.3 | 95.5 | 93.2 | 99.5 | 99.1 | 99.7 | 99.1 |
| 6 | KU315430.1 | Ea | 1990 | China | 99.9 | 99.5 | 95.6 | 92.5 | 100.0 | 100.0 | 99.5 | 99.1 | 99.9 | 99.8 | 95.5 | 93.0 | 100.0 | 100.0 | 99.5 | 99.1 |
| 7 | ON005002.1 | FB | 1986 | China | 99.2 | 97.9 | 95.4 | 92.1 | 98.9 | 99.1 | 99.6 | 98.8 | 98.9 | 98.0 | 94.8 | 91.8 | 98.9 | 99.1 | 99.6 | 98.8 |
| 8 | KM189913.1 | Fa | 2001 | China | 99.9 | 99.7 | 95.6 | 92.5 | 100.0 | 100.0 | 99.7 | 99.1 | 100.0 | 100.0 | 95.2 | 93.0 | 100.0 | 100.0 | 99.7 | 99.1 |
| 9 | MW286330.1 | FJ | 2019 | China | 99.9 | 99.6 | 95.6 | 92.7 | 99.4 | 99.0 | 99.7 | 99.1 | 99.8 | 99.5 | 95.5 | 93.2 | 99.4 | 99.0 | 99.7 | 99.1 |
| 10 | KT824771.1 | HLJ8 | 2013 | China | 99.9 | 99.6 | 95.6 | 92.7 | 99.5 | 99.3 | 99.7 | 99.1 | 99.8 | 99.5 | 95.5 | 93.2 | 99.5 | 99.3 | 99.7 | 99.1 |
| 11 | MK080279.1 | HLJ-2013 | 2013 | China | 99.1 | 97.0 | 97.4 | 98.1 | 100.0 | 100.0 | 100.0 | 99.7 | 98.8 | 97.7 | 99.9 | 100.0 | 100.0 | 100.0 | 100.0 | 99.7 |
| 12 | KM189912.1 | HNX | 2012 | China | 99.9 | 99.6 | 95.6 | 92.7 | 99.5 | 99.1 | 99.7 | 99.1 | 99.8 | 99.5 | 95.5 | 93.0 | 99.5 | 99.1 | 99.7 | 99.1 |
| 13 | KP257591.1 | JS-2012 | 2012 | China | 99.9 | 99.6 | 95.6 | 92.5 | 99.5 | 99.1 | 99.7 | 99.1 | 99.8 | 99.5 | 95.4 | 93.2 | 99.5 | 99.1 | 99.7 | 99.1 |
| 14 | OP512542.1 | JS-XJ5 | 2015 | China | 99.9 | 99.6 | 95.6 | 92.7 | 99.5 | 99.3 | 99.7 | 99.1 | 99.8 | 99.5 | 95.4 | 93.2 | 99.5 | 99.3 | 99.7 | 99.1 |
| 15 | MK806387.1 | JX/CH/2016 | 2016 | China | 99.9 | 99.5 | 95.6 | 92.5 | 99.3 | 99.1 | 99.7 | 99.1 | 99.8 | 99.3 | 95.4 | 93.0 | 99.3 | 99.1 | 99.7 | 99.1 |
| 16 | JF797218.1 | Kaplan | 1987 | Hungary | 98.5 | 96.2 | 97.9 | 99.2 | 97.5 | 96.2 | 99.8 | 99.4 | 98.0 | 96.6 | 98.5 | 97.3 | 97.5 | 96.2 | 99.8 | 99.4 |
| 17 | KT983811.1 | Kolchis | 2010 | Greece | 98.4 | 96.0 | 97.5 | 98.1 | 97.4 | 96.2 | 99.5 | 98.8 | 98.0 | 96.4 | 98.1 | 96.3 | 97.4 | 96.2 | 99.5 | 98.8 |
| 18 | KU552118.1 | LA | 1997 | China | 99.6 | 98.8 | 95.7 | 93.1 | 99.3 | 99.5 | 99.7 | 99.1 | 99.6 | 98.9 | 93.9 | 92.0 | 99.3 | 99.5 | 99.7 | 99.1 |
| 19 | KU900059.1 | NIA3 | 1973 | Belgium | 98.4 | 96.6 | 97.2 | 97.5 | 97.5 | 95.8 | 99.6 | 99.1 | 98.3 | 96.9 | 97.7 | 95.7 | 97.5 | 95.8 | 99.6 | 99.1 |
| 20 | OK338076.1 | PRV-GD | 2021 | China | 99.9 | 99.6 | 95.6 | 92.7 | 99.5 | 99.1 | 99.7 | 99.1 | 99.8 | 99.5 | 95.5 | 93.2 | 99.5 | 99.1 | 99.7 | 99.1 |
| 21 | OK338077.1 | PRV-JM | 2021 | China | 99.9 | 99.6 | 95.6 | 92.7 | 99.5 | 99.1 | 99.7 | 99.1 | 99.8 | 99.5 | 95.5 | 93.2 | 99.5 | 99.1 | 99.7 | 99.1 |
| 22 | KT809429.1 | SC | 1986 | China | 99.9 | 99.7 | 97.5 | 98.1 | 100.0 | 100.0 | 100.0 | 99.7 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| 23 | MT949536.1 | SD18 | 2020 | China | 99.9 | 99.6 | 95.6 | 92.7 | 99.3 | 98.8 | 99.7 | 99.1 | 99.8 | 99.5 | 95.5 | 93.2 | 99.3 | 98.8 | 99.7 | 99.1 |
| 24 | OL606749.1 | SX1910 | 2022 | China | 99.9 | 99.2 | 95.6 | 92.7 | 99.5 | 99.1 | 99.7 | 99.1 | 99.7 | 99.3 | 95.5 | 93.2 | 99.5 | 99.1 | 99.7 | 99.1 |
| 25 | KJ789182.1 | TJ | 2012 | China | 99.9 | 99.6 | 95.6 | 92.7 | 99.5 | 99.3 | 99.7 | 99.1 | 99.8 | 99.5 | 95.5 | 93.2 | 99.5 | 99.3 | 99.7 | 99.1 |
| 26 | KM061380.1 | ZJ01 | 2012 | China | 99.8 | 99.5 | 95.6 | 92.7 | 98.5 | 98.4 | 99.6 | 99.1 | 99.7 | 99.3 | 95.5 | 93.2 | 98.5 | 98.4 | 99.6 | 99.1 |
| Swine Herd | PRV gE Antibodies | ||
|---|---|---|---|
| Number of Detections | Positive Number | Positive Rate (%) | |
| breeding pig | 9 | 1 | 1.11% |
| suckling pig | 66 | 35 | 53.03% |
| nursery pig | 46 | 25 | 54.35% |
| nursing sow | 80 | 60 | 75.00% |
| pregnant sow | 268 | 204 | 76.12% |
| total | 469 | 325 | 69.30% |
| Year | PRV gE Antibodies | PRV gE Antigens | ||||
|---|---|---|---|---|---|---|
| Number of Detections | Positive Number | Positive Rate (%) | Number of Detections | Positive Number | Positive Rate (%) | |
| 2022 | 3757 | 209 | 5.56% | 283 | 3 | 1.06% |
| 2023 | 1330 | 115 | 8.65% | 297 | 7 | 2.36% |
| 2024 | 465 | 11 | 2.36% | / | / | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Zhang, J.; Zhou, J.; Luo, Y.; Wang, X.; Yang, R.; Zhu, J.; Jia, M.; Zhang, L.; Fu, L.; et al. Prevalence and Genetic Variation Investigation of the Pseudorabies Virus in Southwest China. Animals 2024, 14, 3103. https://doi.org/10.3390/ani14213103
Wu J, Zhang J, Zhou J, Luo Y, Wang X, Yang R, Zhu J, Jia M, Zhang L, Fu L, et al. Prevalence and Genetic Variation Investigation of the Pseudorabies Virus in Southwest China. Animals. 2024; 14(21):3103. https://doi.org/10.3390/ani14213103
Chicago/Turabian StyleWu, Jiaqi, Juan Zhang, Jun Zhou, Yi Luo, Xinrong Wang, Rui Yang, Junhai Zhu, Meiyu Jia, Longxiang Zhang, Lizhi Fu, and et al. 2024. "Prevalence and Genetic Variation Investigation of the Pseudorabies Virus in Southwest China" Animals 14, no. 21: 3103. https://doi.org/10.3390/ani14213103
APA StyleWu, J., Zhang, J., Zhou, J., Luo, Y., Wang, X., Yang, R., Zhu, J., Jia, M., Zhang, L., Fu, L., Yan, N., & Wang, Y. (2024). Prevalence and Genetic Variation Investigation of the Pseudorabies Virus in Southwest China. Animals, 14(21), 3103. https://doi.org/10.3390/ani14213103






