Genome-Wide Association Study of Insertions and Deletions Identified Novel Loci Associated with Milk Production Traits in Dairy Cattle
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. GWAS Population
2.2. GWAS Analysis
2.3. Conditional GWAS
2.4. Gene Annotation
3. Results
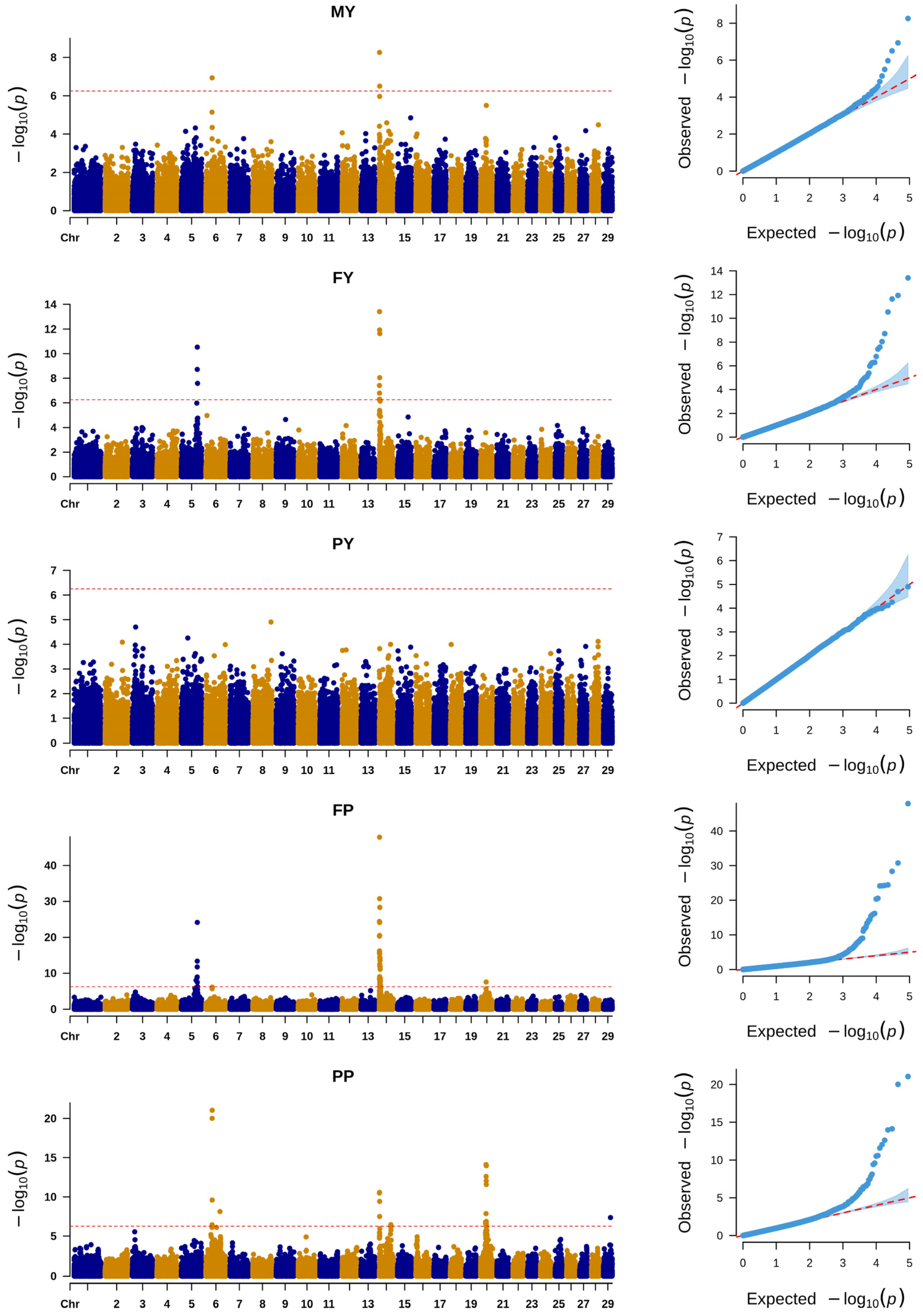
3.1. SNP-Based GWAS
3.2. INDEL-Based GWAS
3.3. Conditional Association Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mills, R.E.; Luttig, C.T.; Larkins, C.E.; Beauchamp, A.; Tsui, C.; Pittard, W.S.; Devine, S.E. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006, 16, 1182–1190. [Google Scholar] [CrossRef] [PubMed]
- Hayes, B.J.; Daetwyler, H.D. 1000 Bull Genomes Project to Map Simple and Complex Genetic Traits in Cattle: Appli-cations and Outcomes. Annu. Rev. Anim. Biosci. 2019, 7, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Mullaney, J.M.; Mills, R.E.; Pittard, W.S.; Devine, S.E. Small insertions and deletions (INDELs) in human genomes. Hum. Mol. Genet. 2010, 19, R131–R136. [Google Scholar] [CrossRef] [PubMed]
- Mills, R.E.; Pittard, W.S.; Mullaney, J.M.; Farooq, U.; Creasy, T.H.; Mahurkar, A.A.; Kemeza, D.M.; Strassler, D.S.; Ponting, C.P.; Webber, C.; et al. Natural genetic variation caused by small insertions and deletions in the human ge-nome. Genome Res. 2011, 21, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Shaw, K.; Phillips, A.; Cooper, D.N. The Human Gene Mutation Database: Building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014, 133, 1–9. [Google Scholar] [CrossRef]
- Hoffmann, T.J.; Van Den Eeden, S.K.; Sakoda, L.C.; Jorgenson, E.; Habel, L.A.; Graff, R.E.; Passarelli, M.N.; Cario, C.L.; Emami, N.C.; Chao, C.R.; et al. A large multiethnic genome-wide association study of prostate cancer identifies novel risk variants and substantial ethnic differences. Cancer Discov. 2015, 5, 878–891. [Google Scholar] [CrossRef]
- Tao, R.; Hu, S.; Wang, S.; Zhou, X.; Zhang, Q.; Wang, C.; Zhao, X.; Zhou, W.; Zhang, S.; Li, C.; et al. Association be-tween indel polymorphism in the promoter region of lncRNA GAS5 and the risk of hepatocellular carcinoma. Carcinogenesis 2015, 36, 1136–1143. [Google Scholar] [CrossRef]
- Sun, T.; Gao, Y.; Tan, W.; Ma, S.; Shi, Y.; Yao, J.; Guo, Y.; Yang, M.; Zhang, X.; Zhang, Q.; et al. A six-nucleotide inser-tion-deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat. Genet. 2007, 39, 605–613. [Google Scholar] [CrossRef]
- Dai, J.; Huang, M.; Amos, C.I.; Hung, R.J.; Tardon, A.; Andrew, A.; Chen, C.; Christiani, D.C.; Albanes, D.; Rennert, G.; et al. Genome-wide association study of INDELs identified four novel susceptibility loci associated with lung cancer risk. Int. J. Cancer 2020, 146, 2855–2864. [Google Scholar] [CrossRef]
- Ju, X.; Huang, X.; Zhang, M.; Lan, X.; Wang, D.; Wei, C.; Jiang, H. Effects of eight InDel variants in FHIT on milk traits in Xinjiang brown cattle. Anim. Biotechnol. 2021, 32, 486–494. [Google Scholar] [CrossRef]
- Jiang, J.; Liu, L.; Gao, Y.; Shi, L.; Li, Y.; Liang, W.; Sun, D. Determination of genetic associations between indels in 11 candidate genes and milk composition traits in Chinese Holstein population. BMC Genet. 2019, 20, 48. [Google Scholar] [CrossRef] [PubMed]
- Mesbah-Uddin, M.; Guldbrandtsen, B.; Capitan, A.; Lund, M.S.; Boichard, D.; Sahana, G. Genome-wide association study with imputed whole-genome sequence variants including large deletions for female fertility in 3 Nordic dairy cattle breeds. J. Dairy Sci. 2022, 105, 1298–1313. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Sun, Z.; Wang, J.; Huang, H.; Kocher, J.P.; Wang, L. CrossMap: A versatile tool for coordinate conversion be-tween genome assemblies. Bioinformatics 2014, 30, 1006–1007. [Google Scholar] [CrossRef] [PubMed]
- Teng, J.; Wang, D.; Zhao, C.; Zhang, X.; Chen, Z.; Liu, J.; Sun, D.; Tang, H.; Wang, W.; Li, J.; et al. Longitudinal ge-nome-wide association studies of milk production traits in Holstein cattle using whole-genome sequence data imputed from medium-density chip data. J. Dairy Sci. 2023, 106, 2535–2550. [Google Scholar] [CrossRef] [PubMed]
- Garrick, D.J.; Taylor, J.F.; Fernando, R.L. Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet. Sel. Evol. 2009, 41, 55. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Vanraden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Ning, C.; Wang, D.; Zhou, L.; Wei, J.; Liu, Y.; Kang, H.; Zhang, S.; Zhou, X.; Xu, S.; Liu, J.F. Efficient multivariate analysis algorithms for longitudinal genome-wide association studies. Bioinformatics 2019, 35, 4879–4885. [Google Scholar] [CrossRef]
- Jiang, J.; Gao, Y.; Hou, Y.; Li, W.; Zhang, S.; Zhang, Q.; Sun, D. Whole-Genome Resequencing of Holstein Bulls for In-del Discovery and Identification of Genes Associated with Milk Composition Traits in Dairy Cattle. PLoS ONE 2016, 11, e168946. [Google Scholar] [CrossRef]
- Tribout, T.; Croiseau, P.; Lefebvre, R.; Barbat, A.; Boussaha, M.; Fritz, S.; Boichard, D.; Hoze, C.; Sanchez, M.P. Con-firmed effects of candidate variants for milk production, udder health, and udder morphology in dairy cattle. Genet. Sel. Evol. 2020, 52, 55. [Google Scholar] [CrossRef]
- Orellana, E.A.; Siegal, E.; Gregory, R.I. tRNA dysregulation and disease. Nat. Rev. Genet. 2022, 23, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Du, Z.W.; Zhang, J.W. Identification and characterization of a novel zinc finger protein (HZF1) gene and its function in erythroid and megakaryocytic differentiation of K562 cells. Leukemia 2006, 20, 1109–1116. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, X.B.; Su, R.; Song, L.; Wang, F.; Zhang, J.W. ZNF16 (HZF1) promotes erythropoiesis and megakaryocyto-poiesis via regulation of the c-KIT gene. Biochem. J. 2014, 458, 171–183. [Google Scholar] [CrossRef] [PubMed]
- George, C.L.; Diaz-Martinez, L.A. A Novel Role for the ZNF16 Protein in rRNA Transcription. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Buitenhuis, B.; Janss, L.L.; Poulsen, N.A.; Larsen, L.B.; Larsen, M.K.; Sorensen, P. Genome-wide association and biolog-ical pathway analysis for milk-fat composition in Danish Holstein and Danish Jersey cattle. BMC Genom. 2014, 15, 1112. [Google Scholar] [CrossRef]
- Bohlouli, M.; Halli, K.; Yin, T.; Gengler, N.; Konig, S. Genome-wide associations for heat stress response suggest poten-tial candidate genes underlying milk fatty acid composition in dairy cattle. J. Dairy Sci. 2022, 105, 3323–3340. [Google Scholar] [CrossRef]
- Hosseinzadeh, S.; Rafat, S.A.; Javanmard, A.; Fang, L. Identification of candidate genes associated with milk production and mastitis based on transcriptome-wide association study. Anim. Genet. 2024, 55, 430–439. [Google Scholar] [CrossRef]
- Wang, C.; Zhao, F.; Liu, J.; Liu, H. The ubiquitin ligase Nedd4-2 mediates the regulation of PepT2 by mTORC1 in bovine mammary epithelial cells. Anim. Nutr. 2022, 10, 12–18. [Google Scholar] [CrossRef]
- Czarnowski, D. Syndecans in cancer: A review of function, expression, prognostic value, and therapeutic significance. Cancer Treat. Res. Commun. 2021, 27, 100312. [Google Scholar] [CrossRef]
- Ning, C.; Wang, D.; Zheng, X.; Zhang, Q.; Zhang, S.; Mrode, R.; Liu, J.F. Eigen decomposition expedites longitudinal genome-wide association studies for milk production traits in Chinese Holstein. Genet. Sel. Evol. 2018, 50, 12. [Google Scholar] [CrossRef]
- Sousa, S.B.; Jenkins, D.; Chanudet, E.; Tasseva, G.; Ishida, M.; Anderson, G.; Docker, J.; Ryten, M.; Sa, J.; Saraiva, J.M.; et al. Gain-of-function mutations in the phosphatidylserine synthase 1 (PTDSS1) gene cause Lenz-Majewski syndrome. Nat. Genet. 2014, 46, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Lee, H.Y.; Weinzimer, S.A.; Powell, D.R.; Clifford, J.L.; Kurie, J.M.; Cohen, P. Direct functional interactions between insulin-like growth factor-binding protein-3 and retinoid X receptor-alpha regulate transcriptional signaling and apoptosis. J. Biol. Chem. 2000, 275, 33607–33613. [Google Scholar] [CrossRef] [PubMed]
- Marini, C.; Porro, A.; Rastetter, A.; Dalle, C.; Rivolta, I.; Bauer, D.; Oegema, R.; Nava, C.; Parrini, E.; Mei, D.; et al. HCN1 mutation spectrum: From neonatal epileptic encephalopathy to benign generalized epilepsy and beyond. Brain 2018, 141, 3160–3178. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ma, L.; Prakapenka, D.; Vanraden, P.M.; Cole, J.B.; Da, Y. A Large-Scale Genome-Wide Association Study in U.S. Holstein Cattle. Front. Genet. 2019, 10, 412. [Google Scholar] [CrossRef]

| INDEL | Conditional SNP | Distance | Traits | p-Value |
|---|---|---|---|---|
| INDEL (Chr6:36865910) | SNP (Chr6:36956973) | 91,063 | PP | 3.73 × 10−4 |
| INDEL (Chr6:36443645) | SNP (Chr6:36601824) | 158,179 | PP | 1.67 × 10−3 |
| INDEL (Chr14:266639) | SNP (Chr14:269574) | 2935 | FP | 7.56 × 10−4 |
| INDEL (Chr14:920017) | SNP (Chr14:512818) | 407,199 | MY | 3.43 × 10−3 |
| INDEL (Chr14:920017) | SNP (Chr14:1045680) | 125,663 | FY | 1.57 × 10−3 |
| INDEL (Chr14:920017) | SNP (Chr14:1045680) | 125,663 | FP | 3.22 × 10−8 |
| INDEL (Chr20:31976989) | SNP (Chr20:31273619) | 703,370 | FP | 2.54 × 10−3 |
| Traits | INDELs | Nearest sig. SNPs | Distance | ||
|---|---|---|---|---|---|
| ID | p-Value | ID | p-Value | ||
| FP | INDEL (Chr5:86639620) | 9.36 × 10−9 | SNP (Chr5:87857269) | 8.11 × 10−9 | 1,217,649 |
| PP | INDEL (Chr14:67765289) | 3.58 × 10−7 | SNP (Chr14:65872284) | 1.11 × 10−8 | 1,893,005 |
| PP | INDEL (Chr20:28876195) | 1.61 × 10−7 | SNP (Chr20:30149443) | 1.31 × 10−14 | 1,273,248 |
| PP | INDEL (Chr20:29002625) | 2.56 × 10−7 | SNP (Chr20:30149443) | 1.31 × 10−14 | 1,146,818 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; Teng, J.; Ning, C.; Zhang, Q. Genome-Wide Association Study of Insertions and Deletions Identified Novel Loci Associated with Milk Production Traits in Dairy Cattle. Animals 2024, 14, 3556. https://doi.org/10.3390/ani14243556
Zhao L, Teng J, Ning C, Zhang Q. Genome-Wide Association Study of Insertions and Deletions Identified Novel Loci Associated with Milk Production Traits in Dairy Cattle. Animals. 2024; 14(24):3556. https://doi.org/10.3390/ani14243556
Chicago/Turabian StyleZhao, Lu, Jun Teng, Chao Ning, and Qin Zhang. 2024. "Genome-Wide Association Study of Insertions and Deletions Identified Novel Loci Associated with Milk Production Traits in Dairy Cattle" Animals 14, no. 24: 3556. https://doi.org/10.3390/ani14243556
APA StyleZhao, L., Teng, J., Ning, C., & Zhang, Q. (2024). Genome-Wide Association Study of Insertions and Deletions Identified Novel Loci Associated with Milk Production Traits in Dairy Cattle. Animals, 14(24), 3556. https://doi.org/10.3390/ani14243556




