Investigating the Impact of Varied Dietary Protein Levels on Litopenaeus vannamei: An Exploration of the Intestinal Microbiota and Transcriptome Responses
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Diets
2.2. Shrimp and Feeding Management
2.3. Growth Performance Analysis
2.4. Sample Collection
2.5. Challenge Tests
2.6. Non-Specific Immune Indices
2.7. Digestive Enzyme Activity
2.8. Intestinal Microbial Analysis
2.9. Transcriptome Analysis
2.10. Statistical Analysis
3. Results
3.1. Growth Performance and Feed Utilization
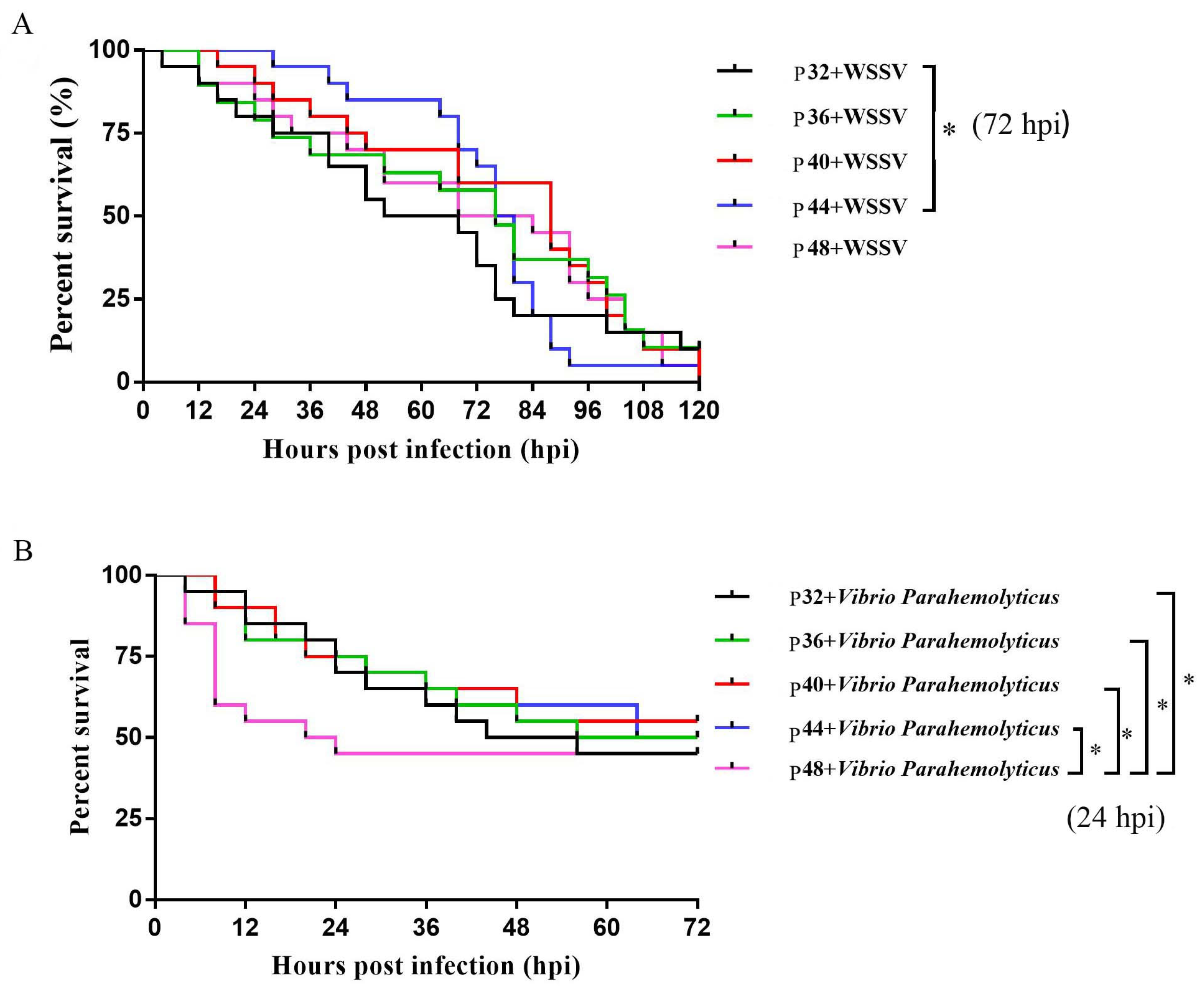
3.2. Survival Rates of L. vannamei after WSSV and V. parahaemolyticus Infections
3.3. Non-Specific Immune Indices
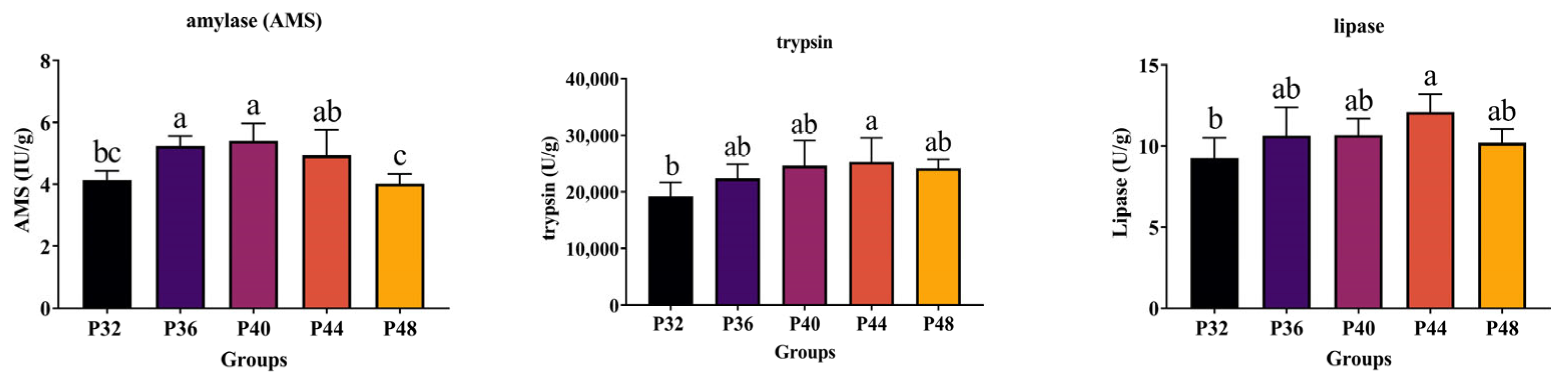
3.4. Digestive Enzyme Activity
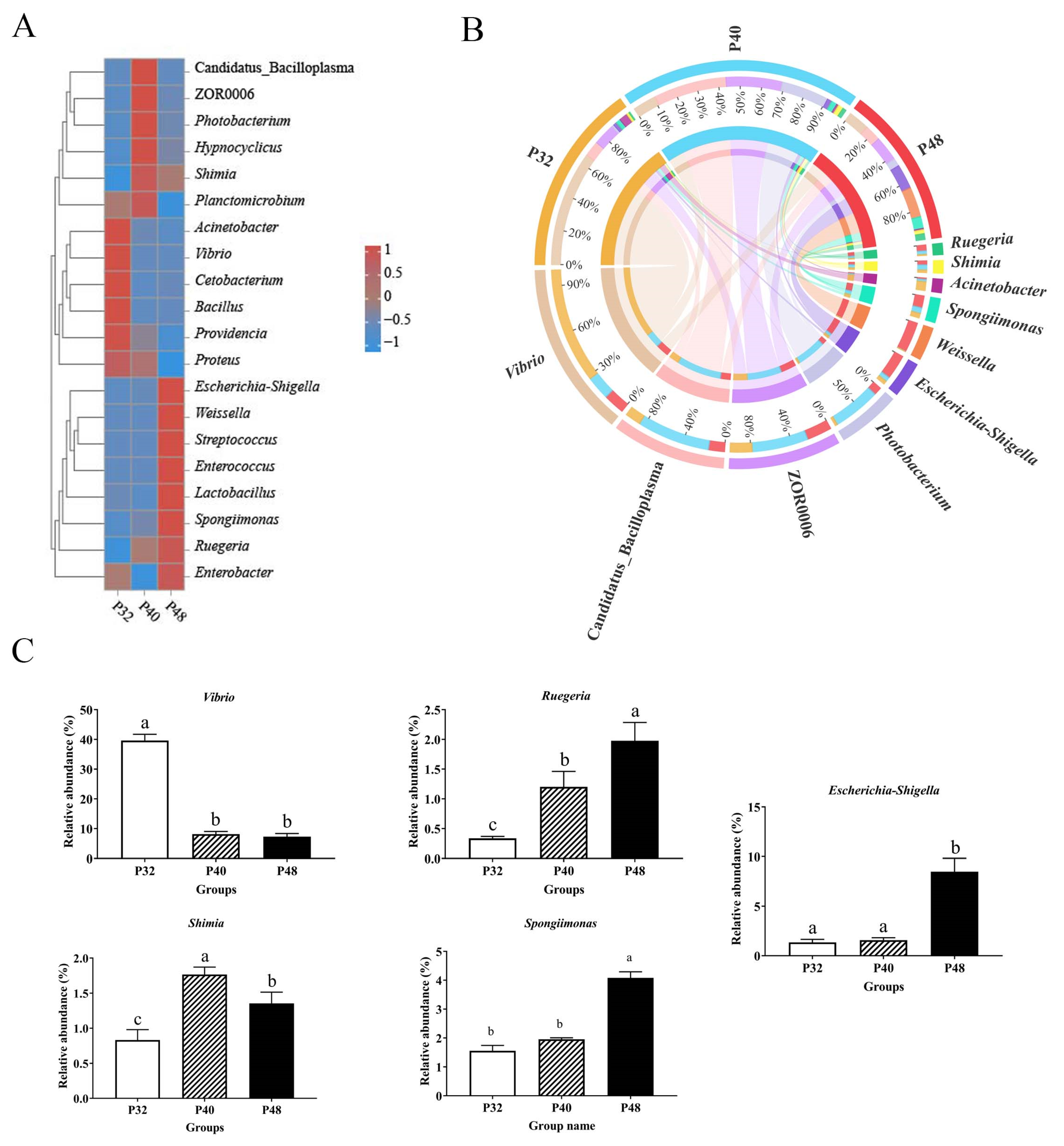
3.5. Intestinal Microbiota Analysis
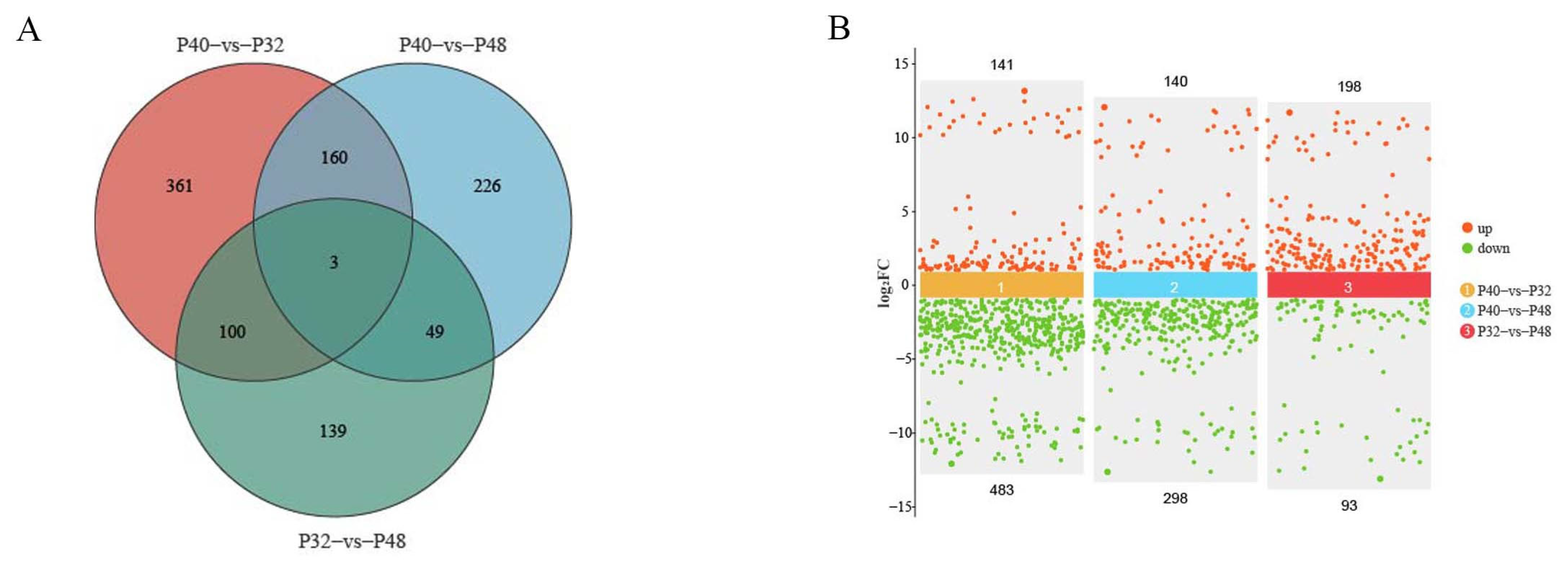
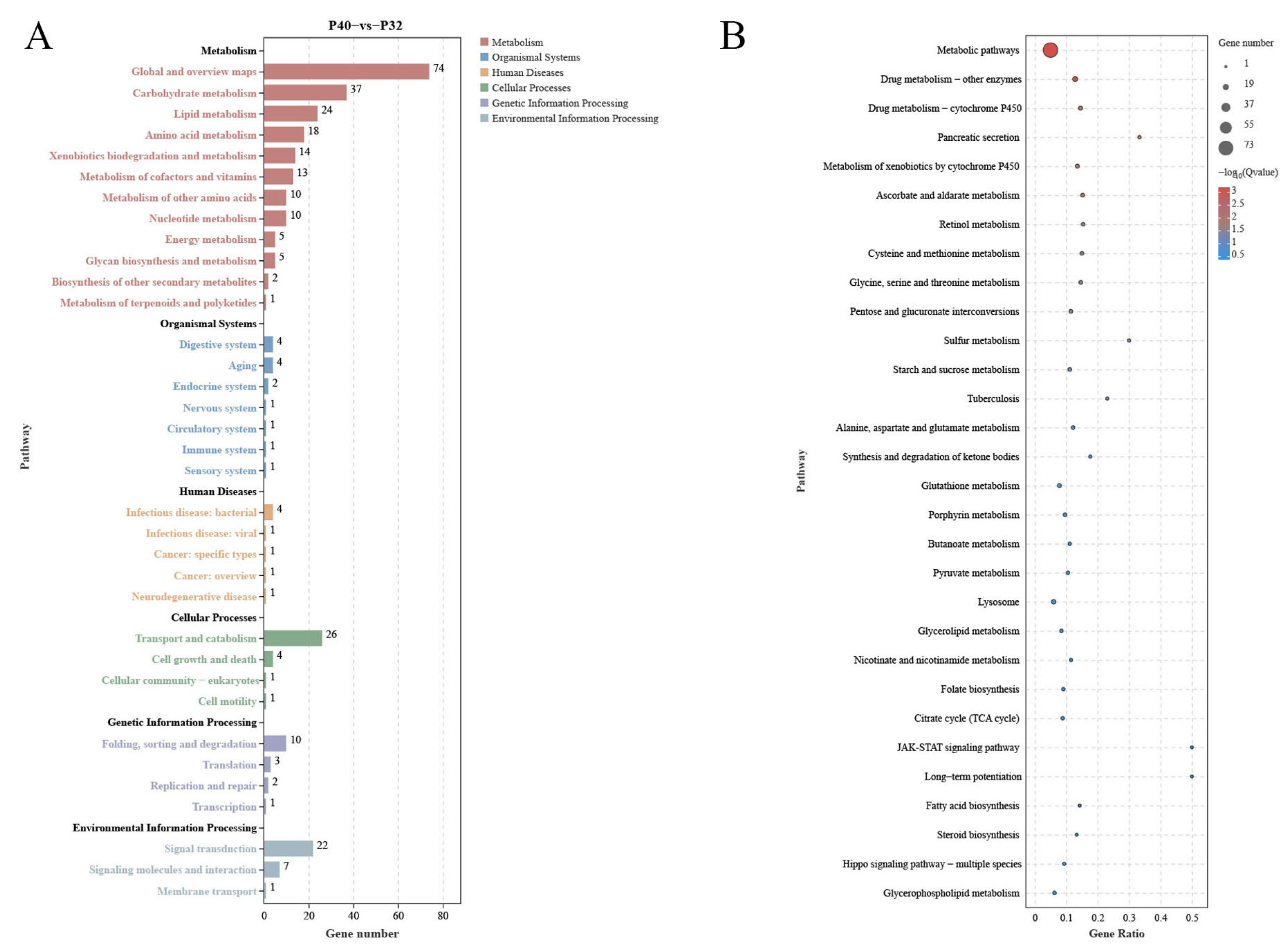
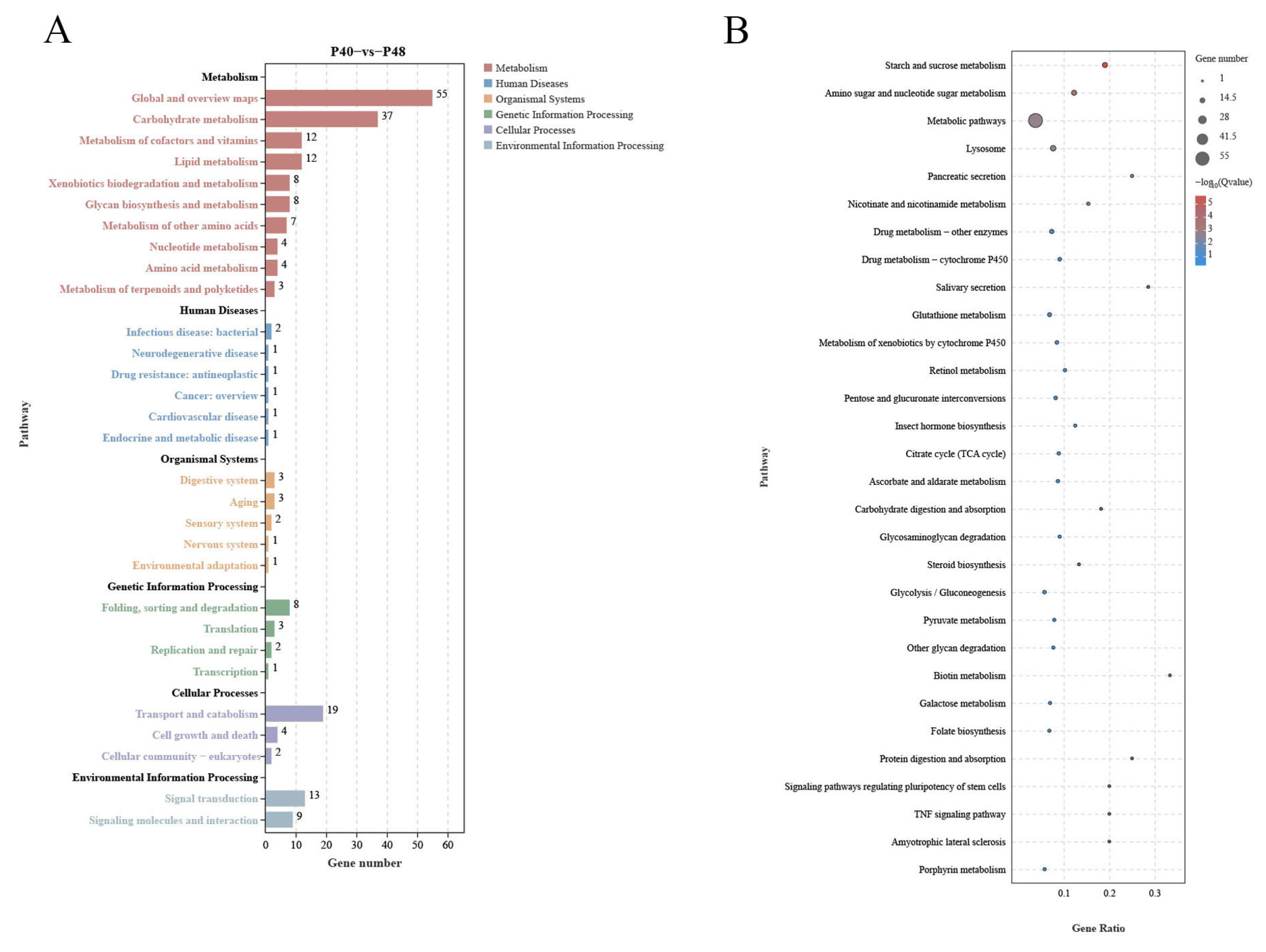
3.6. Transcriptome Level Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gatlin, D.M., III.; Barrows, F.T.; Brown, P.; Dabrowski, K.; Gaylord, T.G.; Hardy, R.W.; Herman, E.; Hu, G.; Krogdahl, Å.; Nelson, R.; et al. Expanding the Utilization of Sustainable Plant Products in Aquafeeds: A Review. Aquac. Res. 2007, 38, 551–579. [Google Scholar] [CrossRef]
- Alloul, A.; Wille, M.; Lucenti, P.; Bossier, P.; Van Stappen, G.; Vlaeminck, S.E. Purple Bacteria as Added-Value Protein Ingredient in Shrimp Feed: Penaeus vannamei Growth Performance, and Tolerance against vibrio and Ammonia Stress. Aquaculture 2021, 530, 735788. [Google Scholar] [CrossRef]
- Haitao, Z.; Xuesong, P.; Qihui, Y.; Beiping, T.; Xiaohui, D.; Shuyan, C.; Hongyu, L.; Shuang, Z. Effects of replacing fishmeal with high-protein cotton meal on growth performance, non-specific im-mune indexes, and disease resistance of Penaeus vannamei. J. Guangdong Ocean. Univ. 2018, 4, 20–26. [Google Scholar] [CrossRef]
- Kandathil Radhakrishnan, D.; AkbarAli, I.; Schmidt, B.V.; John, E.M.; Sivanpillai, S.; Thazhakot Vasunambesan, S. Improvement of Nutritional Quality of Live Feed for Aquaculture: An Overview. Aquac. Res. 2020, 51, 1–17. [Google Scholar] [CrossRef]
- Xie, S.; Wei, D.; Fang, W.; Yin, P.; Liu, Y.; Niu, J.; Tian, L. Survival and Protein Synthesis of Post-Larval White shrimp, Litopenaeus vannamei Were Affected by Dietary Protein Level. Anim. Feed Sci. Technol. 2020, 263, 114462. [Google Scholar] [CrossRef]
- Schveitzer, R.; Arantes, R.; Costódio, P.F.S.; do Espírito Santo, C.M.; Arana, L.V.; Seiffert, W.Q.; Andreatta, E.R. Effect of Different Biofloc Levels on Microbial Activity, Water Quality and Performance of Litopenaeus vannamei in a Tank System Operated with No Water Exchange. Aquac. Eng. 2013, 56, 59–70. [Google Scholar] [CrossRef]
- Roques, S.; Deborde, C.; Richard, N.; Skiba-Cassy, S.; Moing, A.; Fauconneau, B. Metabolomics and Fish Nutrition: A Review in the Context of Sustainable Feed Development. Rev. Aquac. 2020, 12, 261–282. [Google Scholar] [CrossRef]
- Martinez-Cordova, L.R.; Campaña Torres, A.; Porchas-Cornejo, M.A. Dietary Protein Level and Natural Food Management in the Culture of Blue (Litopenaeus stylirostris) and White Shrimp (Litopenaeus vannamei) in Microcosms. Aquac. Nutr. 2003, 9, 155–160. [Google Scholar] [CrossRef]
- Yearbook, F. Fishery and Aquaculture Statistics 2019/FAO Annuaire. Statistiques Des Pêches et de L’aquaculture 2019/FAO Anuario; FAO: Roma, Italy, 2019. [Google Scholar]
- Wu, Y.C.; Li, R.M.; Shen, G.R.; Huang, F.; Yang, Q.H.; Tan, B.P.; Chi, S.Y. Effects of small peptides on growth, antioxidant capacity, non-specific immunity, and intestinal flora structure of Penaeus vannamei. J. Guangdong Ocean. Univ. 2021, 5, 9. [Google Scholar] [CrossRef]
- Xu, W.-J.; Pan, L.-Q. Evaluation of Dietary Protein Level on Selected Parameters of Immune and Antioxidant Systems, and Growth Performance of Juvenile Litopenaeus vannamei Reared in Zero-Water Exchange Biofloc-Based Culture Tanks. Aquaculture 2014, 426–427, 181–188. [Google Scholar] [CrossRef]
- Sainz Hernández, J.C.; Cordova Murueta, J.H. Activity of Trypsin from Litopenaeus vannamei. Aquaculture 2009, 290, 190–195. [Google Scholar] [CrossRef]
- Jang, I.-K.; Shahkar, E.; Kyoung Kim, S.; Yun, H.; Katya, K.; Park, G.; Bai, S.C. Evaluation of Optimum Dietary Protein Level for Juvenile Whiteleg Shrimp (Litopenaeus vannamei). J. Crustac. Biol. 2014, 34, 552–558. [Google Scholar] [CrossRef]
- Jatobá, A.; da Silva, B.C.; da Silva, J.S.; Vieira, F.d.N.; Mouriño, J.L.P.; Seiffert, W.Q.; Toledo, T.M. Protein Levels for Litopenaeus vannamei in Semi-Intensive and Biofloc Systems. Aquaculture 2014, 432, 365–371. [Google Scholar] [CrossRef]
- Yao, W.; Li, X.; Zhang, C.; Wang, J.; Cai, Y.; Leng, X. Effects of Dietary Synbiotics Supplementation Methods on Growth, Intestinal Health, Non-Specific Immunity and Disease Resistance of Pacific White Shrimp, Litopenaeus Vannamei. Fish. Shellfish. Immunol. 2021, 112, 46–55. [Google Scholar] [CrossRef]
- Yuanfa, H.; Shuyan, C.; Beiping, T.; Hanle, Z.; Xiaohui, D.; Qihui, Y.; Hongyu, L.; Shuang, Z. In-fluence of yeast cultures on the structure of intestinal flora of Penaeus vannamei. J. Guangdong Ocean. Univ. 2017, 4, 21–27. [Google Scholar] [CrossRef]
- Qiao, Y.; Zhou, L.; Qu, Y.; Lu, K.; Han, F.; Li, E. Effects of Different Dietary β-Glucan Levels on Antioxidant Capacity and Immunity, Gut Microbiota and Transcriptome Responses of White Shrimp (Litopenaeus vannamei) under Low Salinity. Antioxidants 2022, 11, 2282. [Google Scholar] [CrossRef]
- Zheng, Y.; Hou, C.; Chen, J.; Wang, H.; Yuan, H.; Hu, N.; Shi, L.; Zhang, S. Integrating Microbiome and Transcriptome Analyses to Understand the Effect of Replacing Fishmeal with Tenebrio molitor Meal in Pacific White Shrimp (Litopenaeus vannamei) Diets. Aquaculture 2023, 575, 739818. [Google Scholar] [CrossRef]
- Nakov, R.; Velikova, T. Chemical Metabolism of Xenobiotics by Gut Microbiota. Curr. Drug. Metab. 2020, 21, 260–269. [Google Scholar] [CrossRef]
- Huang, X.M.; Wen, C.Q.; Liang, H.F.; Hong, J.K.; Xue, M. Comparison of bacterial community structure in nursery pond water during the mysid stage of healthy and morbid Penaeus Vannamei. J. Guangdong Ocean. Univ. 2018, 4, 27–34. [Google Scholar] [CrossRef]
- Levy, M.; Blacher, E.; Elinav, E. Microbiome, Metabolites and Host Immunity. Curr. Opin. Microbiol. 2017, 35, 8–15. [Google Scholar] [CrossRef]
- Yang, T.L.; Huang, Y.; Ru, X.Y.; Li, J.; Zhu, K.F.; Yang, J.H.; Chen, P.P.; Zhu, C.H. Ef-fects of chilled fish replacement by combination feeds on the growth and liver transcriptome of Seriola dumerili juveniles. J. Guangdong Ocean. Univ. 2023, 5, 92–99. [Google Scholar] [CrossRef]
- Fu, Z.; Han, F.; Huang, K.; Zhang, J.; Qin, J.G.; Chen, L.; Li, E. Impact of Imidacloprid Exposure on the Biochemical Responses, Transcriptome, Gut Microbiota and Growth Performance of the Pacific White Shrimp Litopenaeus Vannamei. J. Hazard. Mater. 2022, 424, 127513. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Han, F.; Huang, K.; Zhang, J.; Qin, J.G.; Chen, L.; Li, E. Combined Toxic Effects of Thiamethoxam on Intestinal Flora, Transcriptome and Physiology of Pacific White Shrimp Litopenaeus vannamei. Sci. Total Environ. 2022, 830, 154799. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yao, X.; Lin, Y.; Chi, S.; Zhang, S.; Cao, J.; Tan, B.; Xie, S. Schizochytrium limacinum Altered Antioxidant Capacity and Transcriptome Profiles in Pacific White Shrimp Fed a Low-Fishmeal Diet. Aquac. Rep. 2022, 27, 101399. [Google Scholar] [CrossRef]
- Yao, Y.; Shi, L.; Xiao, W.; Guo, S.; Liu, S.; Li, H.; Zhang, S. Phenylalanine Hydroxylase (PAH) Plays a Positive Role during WSSV and Vibrio parahaemolyticus Infection in Litopenaeus vannamei. Fish Shellfish Immunol. 2022, 120, 515–525. [Google Scholar] [CrossRef]
- Chen, J.; Wang, H.; Yuan, H.; Hu, N.; Zheng, Y.; Tan, B.; Shi, L.; Zhang, S. Tapping Chlorella Vulgaris Potential for Enhanced Growth, Immunity, Digestion, Microbiota, and Immunometabolism in Litopenaeus vannamei Feeding across Varied Salinities. Aquaculture 2024, 581, 740469. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly Accurate OTU Sequences from Microbial Amplicon Reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Env. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A Comprehensive Online Resource for Quality Checked and Aligned Ribosomal RNA Sequence Data Compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Aßhauer, K.P.; Wemheuer, B.; Daniel, R.; Meinicke, P. Tax4Fun: Predicting Functional Profiles from Metagenomic 16S rRNA Data. Bioinformatics 2015, 31, 2882–2884. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Kureshy, N.; Davis, D.A. Protein Requirement for Maintenance and Maximum Weight Gain for the Pacific White Shrimp, Litopenaeus vannamei. Aquaculture 2002, 204, 125–143. [Google Scholar] [CrossRef]
- Gómez-Jiménez, S.; González-Félix, M.L.; Perez-Velazquez, M.; Trujillo-Villalba, D.A.; Esquerra-Brauer, I.R.; Barraza-Guardado, R. Effect of Dietary Protein Level on Growth, Survival and Ammonia Efflux Rate of Litopenaeus vannamei (Boone) Raised in a Zero Water Exchange Culture System. Aquac. Res. 2005, 36, 834–840. [Google Scholar] [CrossRef]
- Talukdar, A.; Dharmendra Deo, A.; Prasad Sahu, N.; Sardar, P.; Aklakur, M.; Harikrishna, V.; Prakash, S.; Shamna, N.; Jana, P. Effects of Different Levels of Dietary Protein on the Growth Performance, Nutrient Utilization, Digestive Enzymes and Physiological Status of White Shrimp, Litopenaeus vannamei Juveniles Reared in Inland Saline Water. Aquac. Nutr. 2021, 27, 77–90. [Google Scholar] [CrossRef]
- Xia, S.; Li, Y.; Wang, W.; Rajkumar, M.; Kumaraguru Vasagam, K.P.; Wang, H. Influence of Dietary Protein Levels on Growth, Digestibility, Digestive Enzyme Activity and Stress Tolerance in White-Leg Shrimp, Litopenaeus vannamei (Boone, 1931), Reared in High-Density Tank Trials. Aquac. Res. 2010, 41, 1845–1854. [Google Scholar] [CrossRef]
- Ravindran, V. Feed Enzymes: The Science, Practice, and Metabolic Realities1 1Presented as a Part of the Informal Nutrition Symposium “Metabolic Responses to Nutrition and Modifiers” at the Poultry Science Association’s Annual Meeting in Athens, Georgia, July 9, 2012. J. Appl. Poult. Res. 2013, 22, 628–636. [Google Scholar] [CrossRef]
- Xue, Y.; Wei, F.; Jiang, Y.; Li, L.; Dong, S.; Tian, X. Transcriptome Signatures of the Pacific White Shrimp Litopenaeus vannamei Hepatopancreas in Response to Stress in Biofloc Culture Systems. Fish Shellfish Immunol. 2019, 91, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Marinho-Soriano, E.; Panucci, R.A.; Carneiro, M.A.A.; Pereira, D.C. Evaluation of Gracilaria Caudata J. Agardh for Bioremediation of Nutrients from Shrimp Farming Wastewater. Bioresour. Technol. 2009, 100, 6192–6198. [Google Scholar] [CrossRef]
- Liu, X.; Ye, J.; Wang, K.; Kong, J.; Yang, W.; Zhou, L. Partial Replacement of Fish Meal with Peanut Meal in Practical Diets for the Pacific White Shrimp, Litopenaeus vannamei. Aquac. Res. 2012, 43, 745–755. [Google Scholar] [CrossRef]
- Söderhäll, K.; Cerenius, L. Role of the Prophenoloxidase-Activating System in Invertebrate Immunity. Curr. Opin. Immunol. 1998, 10, 23–28. [Google Scholar] [CrossRef]
- Ngo, H.-V.-T.; Huang, H.-T.; Lee, P.-T.; Liao, Z.-H.; Chen, H.-Y.; Nan, F.-H. Effects of Phyllanthus amarus Extract on Nonspecific Immune Responses, Growth, and Resistance to Vibrio alginolyticus in White Shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 2020, 107, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Krupesha Sharma, S.R.; Rathore, G.; Verma, D.K.; Sadhu, N.; Philipose, K.K. Vibrio alginolyticus Infection in Asian Seabass (Lates calcarifer, Bloch) Reared in Open Sea Floating Cages in India. Aquac. Res. 2012, 44, 86–92. [Google Scholar] [CrossRef]
- Li, Y.; Zhan, F.; Li, F.; Lu, Z.; Shi, F.; Xu, Z.; Yang, Y.; Zhao, L.; Qin, Z.; Lin, L. Immune Function of Cytosolic Manganese Superoxide Dismutase from Macrobrachium rosenbergii in Response to Bacterial Infection. Aquaculture 2021, 541, 736771. [Google Scholar] [CrossRef]
- Li, F.; Liang, Z.; Zheng, X.; Zhao, W.; Wu, M.; Wang, Z. Toxicity of Nano-TiO2 on Algae and the Site of Reactive Oxygen Species Production. Aquat. Toxicol. 2015, 158, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A Core Gut Microbiome in Obese and Lean Twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef]
- Liu, H.; Dong, X.; Tan, B.; Du, T.; Zhang, S.; Yang, Y.; Chi, S.; Yang, Q.; Liu, H. Effects of Two Dietary Protein Levels on Growth, Body Composition, Intestinal Microflora and Expression of TOR, IGF-I, LPL and HSP70 of Juvenile Silver Sillago Sillago Sihama. Aquac. Nutr. 2021, 27, 2218–2230. [Google Scholar] [CrossRef]
- Fan, Y.; Luo, K.; Guo, Y.; Gao, W.; Xu, Q.; Zhang, W.; Mai, K. Replacement of Fish Meal by Enzyme-Treated Soybean on the Growth Performance, Intestine Microflora, Immune Responses and Disease Resistance of Pacific White Shrimp Litopenaeus vannamei. Aquac. Res. 2021, 52, 4619–4628. [Google Scholar] [CrossRef]
- Huang, Z.; Li, X.; Wang, L.; Shao, Z. Changes in the Intestinal Bacterial Community during the Growth of White Shrimp, Litopenaeus vannamei. Aquac. Res. 2016, 47, 1737–1746. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, J.; Li, J.; Zou, J.; Fan, L. The Immune Defense Response of Pacific White Shrimp (Litopenaeus vannamei) to Temperature Fluctuation. Fish Shellfish Immunol. 2020, 103, 103–110. [Google Scholar] [CrossRef]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial Signature of Dysbiosis in Gut Microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Sung, H.-H.; Hsu, S.-F.; Chen, C.-K.; Ting, Y.-Y.; Chao, W.-L. Relationships between Disease Outbreak in Cultured Tiger Shrimp (Penaeus monodon) and the Composition of Vibrio Communities in Pond Water and Shrimp Hepatopancreas during Cultivation. Aquaculture 2001, 192, 101–110. [Google Scholar] [CrossRef]
- Aguirre-Guzmán, G.; Vázquez-Juárez, R.; Ascencio, F. Differences in the Susceptibility of American White Shrimp Larval Substages (Litopenaeus vannamei) to Four Vibrio Species. J. Invertebr. Pathol. 2001, 78, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Gu, Y.; Zhou, H.; Xu, L.; Cao, H.; Gai, C. Acinetobacter Venetianus, a Potential Pathogen of Red Leg Disease in Freshwater-Cultured Whiteleg Shrimp Penaeus vannamei. Aquac. Rep. 2020, 18, 100543. [Google Scholar] [CrossRef]
- Barreto-Curiel, F.; Ramirez-Puebla, S.T.; Ringø, E.; Escobar-Zepeda, A.; Godoy-Lozano, E.; Vazquez-Duhalt, R.; Sanchez-Flores, A.; Viana, M.T. Effects of Extruded Aquafeed on Growth Performance and Gut Microbiome of Juvenile Totoaba macdonaldi. Anim. Feed Sci. Technol. 2018, 245, 91–103. [Google Scholar] [CrossRef]
- Duan, Y.; Wang, Y.; Liu, Q.; Dong, H.; Li, H.; Xiong, D.; Zhang, J. Changes in the Intestine Microbial, Digestion and Immunity of Litopenaeus vannamei in Response to Dietary Resistant Starch. Sci. Rep. 2019, 9, 6464. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S. Reactive Oxygen Species and Cellular Defense System. In Free Radicals in Human Health and Disease; Springer: New Delhi, India, 2015; pp. 17–29. ISBN 978-81-322-2035-0. [Google Scholar]
- Lobo, G.; Pereira, L.F.; Gonçalves, J.F.M.; Peixoto, M.J.; Ozório, R.O.A. Effect of Dietary Seaweed Supplementation on Growth Performance, Antioxidant and Immune Responses in European Seabass (Dicentrarchus labrax) Subjected to Rearing Temperature and Salinity Oscillations. Int. Aquat. Res. 2018, 10, 321–331. [Google Scholar] [CrossRef]
- Kumar, N.; Gupta, S.K.; Bhushan, S.; Singh, N.P. Impacts of Acute Toxicity of Arsenic (III) Alone and with High Temperature on Stress Biomarkers, Immunological Status and Cellular Metabolism in Fish. Aquat. Toxicol. 2019, 214, 105233. [Google Scholar] [CrossRef]
- Xiao, J.; Li, Q.-Y.; Tu, J.-P.; Chen, X.-L.; Chen, X.-H.; Liu, Q.-Y.; Liu, H.; Zhou, X.-Y.; Zhao, Y.-Z.; Wang, H.-L. Stress Response and Tolerance Mechanisms of Ammonia Exposure Based on Transcriptomics and Metabolomics in Litopenaeus vannamei. Ecotoxicol. Environ. Saf. 2019, 180, 491–500. [Google Scholar] [CrossRef]
- McDonnell, A.M.; Dang, C.H. Basic Review of the Cytochrome P450 System. J. Adv. Pract. Oncol. 2013, 4, 263–268. [Google Scholar]
- Uno, T.; Ishizuka, M.; Itakura, T. Cytochrome P450 (CYP) in Fish. Environ. Toxicol. Pharmacol. 2012, 34, 1–13. [Google Scholar] [CrossRef]
- Liu, C.; Li, J.; Qi, X.; Wang, L.; Sun, D.; Zhang, J.; Zhang, K.; Li, J.; Li, Y.; Wen, H. Cytochrome P450 Superfamily in Spotted Sea Bass: Genome-Wide Identification and Expression Profiles under Trichlorfon and Environmental Stresses. Comp. Biochem. Physiol. Part D Genom. Proteom. 2023, 46, 101078. [Google Scholar] [CrossRef] [PubMed]
- Snyder, M.J.; Mulder, E.P. Environmental Endocrine Disruption in Decapod Crustacean Larvae: Hormone Titers, Cytochrome P450, and Stress Protein Responses to Heptachlor Exposure. Aquat. Toxicol. 2001, 55, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Appelqvist, H.; Wäster, P.; Kågedal, K.; Öllinger, K. The Lysosome: From Waste Bag to Potential Therapeutic Target. J. Mol. Cell Biol. 2013, 5, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Lüllmann-Rauch, R. History and Morphology of the Lysosome. In Lysosomes; Saftig, P., Ed.; Medical Intelligence Unit; Springer US: Boston, MA, USA, 2005; pp. 1–16. ISBN 978-0-387-28957-1. [Google Scholar]
- Vogt, G. Functional Cytology of the Hepatopancreas of Decapod Crustaceans. J. Morphol. 2019, 280, 1405–1444. [Google Scholar] [CrossRef]
- Jiang, X.; Yao, W.; Yang, H.; Tan, S.; Leng, X.; Li, X. Dietary Effects of Clostridium Autoethanogenum Protein Substituting Fish Meal on Growth, Intestinal Histology and Immunity of Pacific White Shrimp (Litopenaeus vannamei) Based on Transcriptome Analysis. Fish Shellfish Immunol. 2021, 119, 635–644. [Google Scholar] [CrossRef]
- Fu, Z.D.; Selwyn, F.P.; Cui, J.Y.; Klaassen, C.D. RNA-Seq Unveiled Section-Specific Host Response to Lack of Gut Microbiota in Mouse Intestine. Toxicol. Appl. Pharmacol. 2021, 433, 115775. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, X.; Yu, Y.; Li, F.; Xiang, J. RNA-Seq Reveals the Dynamic and Diverse Features of Digestive Enzymes during Early Development of Pacific White Shrimp Litopenaeus vannamei. Comp. Biochem. Physiol. Part D Genom. Proteom. 2014, 11, 37–44. [Google Scholar] [CrossRef]
- Petrera, A.; Lai, Z.W.; Schilling, O. Carboxyterminal Protein Processing in Health and Disease: Key Actors and Emerging Technologies. J. Proteome Res. 2014, 13, 4497–4504. [Google Scholar] [CrossRef]
- Castro, P.F.; Freitas, A.C.V., Jr.; Santana, W.M.; Costa, H.M.S.; Carvalho, L.B., Jr.; Bezerra, R.S. Comparative Study of Amylases from the Midgut Gland of Three Species of Penaeid Shrimp. J. Crustac. Biol. 2012, 32, 607–613. [Google Scholar] [CrossRef][Green Version]
- Haworth, A.S.; Brackenbury, W.J. Emerging Roles for Multifunctional Ion Channel Auxiliary Subunits in Cancer. Cell Calcium 2019, 80, 125–140. [Google Scholar] [CrossRef]
- Itoh, R.; Kawamoto, S.; Miyamoto, Y.; Kinoshita, S.; Okubo, K. Isolation and Characterization of a Ca2+-Activated Chloride Channel from Human Corneal Epithelium. Curr. Eye Res. 2000, 21, 918–925. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Ramena, G.; Yin, Y.; Premkumar, L.; Elble, R.C. CLCA2 Is a Positive Regulator of Store-Operated Calcium Entry and TMEM16A. PLoS ONE 2018, 13, e0196512. [Google Scholar] [CrossRef] [PubMed]



| Ingredient (g/kg) | Groups | ||||
|---|---|---|---|---|---|
| P32 | P36 | P40 | P44 | P48 | |
| Fishmeal | 471 | 530 | 589 | 647.5 | 706 |
| Corn starch | 150 | 150 | 150 | 150 | 150 |
| Fish oil | 9.5 | 6.55 | 3.6 | 0.68 | 0 |
| Corn oil | 9.5 | 6.55 | 3.6 | 0.68 | 0 |
| Soyabean lecithin | 10 | 10 | 10 | 10 | 5.5 |
| Vitamin and mineral premix a | 12 | 12 | 12 | 12 | 12 |
| Choline chloride | 5 | 5 | 5 | 5 | 5 |
| Antioxidant b | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Attractant c | 1 | 1 | 1 | 1 | 1 |
| CaH2PO4·H2O | 12 | 12 | 12 | 12 | 12 |
| Vitamin C | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Cellulose | 319 | 265.9 | 212.8 | 160.14 | 107.5 |
| Total | 1000.0 | 1000.0 | 1000.0 | 1000.0 | 1000.0 |
| Nutrient index d | Proximate composition (%) | ||||
| Crude protein | 33.02 | 37.27 | 40.73 | 45.94 | 47.76 |
| Crude lipid | 6.15 | 6.41 | 6.69 | 6.87 | 7.56 |
| Crude ash | 8.4 | 9.2 | 10.4 | 11.6 | 12.7 |
| Moisture | 9 | 9 | 9 | 9 | 9 |
| Item | Groups | ||||
|---|---|---|---|---|---|
| P32 | P36 | P40 | P44 | P48 | |
| IBW (g) | 0.63 ± 0.02 | 0.63 ± 0.02 | 0.63 ± 0.02 | 0.63 ± 0.02 | 0.63 ± 0.02 |
| FBW (g) | 6.58 ± 0.30 | 7.03 ± 0.36 | 7.6 ± 0.75 | 8.44 ± 0.16 | 7.89 ± 0.07 |
| WGR (%) | 934.81 ± 47.00 c | 1011.25 ± 56.86 bc | 1095.83 ± 116.90 abc | 1231.5 ± 30.81 a | 1142.76 ± 8.05 ab |
| SR (%) | 91.67 ± 1.44 | 96.67 ± 1.44 | 93.33 ± 6.29 | 94.17 ± 5.20 | 94.17 ± 5.77 |
| SGR (%) | 4.17 ± 0.08 c | 4.3 ± 0.09 bc | 4.43 ± 0.18 abc | 4.62 ± 0.04 a | 4.5 ± 0.01 ab |
| FCR | 4.42 ± 0.32 a | 3.94 ± 0.20 ab | 3.42 ± 0.31 bc | 3.06 ± 0.15 c | 2.99 ± 0.17 c |
| PER (%) | 7.27 ± 0.52 c | 9.15 ± 0.48 c | 11.76 ± 1.07 b | 14.4 ± 0.68 a | 16.09 ± 0.92 a |
| Index | Groups | ||
|---|---|---|---|
| P32 | P40 | P48 | |
| Sobs | 423.33 ± 20.55 b | 383 ± 23.64 b | 507.67 ± 29.02 a |
| Shannon | 3.08 ± 0.09 c | 3.64 ± 0.09 b | 3.92 ± 0.06 a |
| Simpson | 0.8 ± 0.03 | 0.78 ± 0.05 | 0.87 ± 0.04 |
| Chao | 657.87 ± 35.72 b | 659.51 ± 33.75 b | 747.61 ± 23.67 a |
| Ace | 741.15 ± 21.33 b | 730.47 ± 16.39 b | 792.87 ± 19.63 a |
| Sample | All Reads-Raw Data (bp) | Q20 (%) | Q30 (%) | GC (%) | Filter-Clean Data (%) |
|---|---|---|---|---|---|
| P32-1 | 6,111,628,800 | 97.88% | 93.75% | 43.42% | 40,568,650 (99.57%) |
| P32-2 | 5,780,577,000 | 97.86% | 93.70% | 44.59% | 38,366,864 (99.56%) |
| P32-3 | 5,490,795,900 | 97.75% | 93.53% | 44.25% | 36,386,546 (99.40%) |
| P40-1 | 5,771,514,600 | 97.98% | 94.07% | 43.26% | 38,295,220 (99.53%) |
| P40-2 | 6,010,638,000 | 98.02% | 94.13% | 44.31% | 39,829,030 (99.40%) |
| P40-3 | 5,179,257,000 | 97.57% | 93.13% | 45.92% | 34,328,232 (99.42%) |
| P48-1 | 5,367,786,300 | 98.00% | 94.11% | 43.48% | 35,587,522 (99.45%) |
| P48-2 | 5,600,367,300 | 97.92% | 93.97% | 45.02% | 37,140,840 (99.48%) |
| P48-3 | 6,363,420,300 | 98.05% | 94.16% | 42.75% | 42,212,616 (99.50%) |
| DEGs | Description | P32 Mean (fpkm) | P40 Mean (fpkm) | P48 Mean (fpkm) |
|---|---|---|---|---|
| ROT61949.1 | amy | 0.17 | 1.00 | 0.03 |
| ROT75446.1 | enpp3 | 3.62 | 8.00 | 1.51 |
| MSTRG.31007 | amy | 0.33 | 3.89 | 0.33 |
| MSTRG.31768 | celd | 0.50 | 4.92 | 0.90 |
| ROT62696.1 | rrm1 | 2.80 | 24.23 | 5.62 |
| ROT66027.1 | pck2 | 9.77 | 27.33 | 6.42 |
| ROT67232.1 | pnlip | 1.33 | 6.61 | 1.45 |
| ROT67236.1 | inpp4a | 11.10 | 5.32 | 12.72 |
| ROT68435.1 | tpi1a | 4.21 | 0.00 | 2.80 |
| ROT68492.1 | rgn | 1.84 | 8.90 | 3.73 |
| ROT71552.1 | acss3 | 0.22 | 2.25 | 0.18 |
| ROT72812.1 | smpd1 | 0.08 | 1.47 | 0.24 |
| ROT73315.1 | ugt2b16 | 2.15 | 6.91 | 2.50 |
| ROT76522.1 | chia | 1.18 | 6.07 | 1.53 |
| ROT77738.1 | sam-s | 5.31 | 78.44 | 26.02 |
| ROT77960.1 | amy1 | 0.96 | 6.31 | 0.97 |
| ROT79971.1 | ugt8 | 1.93 | 5.92 | 1.56 |
| ROT80223.1 | akr1b1 | 5.85 | 21.78 | 6.80 |
| ROT81932.1 | -- | 0.86 | 4.75 | 1.27 |
| ROT82637.1 | pnliprp2 | 0.11 | 2.55 | 0.33 |
| MSTRG.26644 | hexb | 5.71 | 13.32 | 6.04 |
| MSTRG.30076 | rrm1 | 1.26 | 10.50 | 2.78 |
| MSTRG.4293 | scsalpha1 | 1.68 | 0.05 | 3.20 |
| ROT79533.1 | gstd1 | 2.83 | 14.90 | 4.57 |
| ROT61670.1 | gpx | 0.66 | 7.74 | 0.94 |
| MSTRG.22898 | se | 6.28 | 14.56 | 6.88 |
| ROT70506.1 | lip3 | 0.75 | 4.92 | 0.74 |
| ROT80984.1 | lipf | 20.37 | 57.51 | 14.35 |
| Gene ID | log2(fc) | Symbol | Description |
|---|---|---|---|
| Glutathione Metabolism | |||
| P40 vs. P32 | |||
| ROT61670.1 | −3.54 | gpx | Glutathione peroxidase 3 (Penaeus monodon) |
| ROT62696.1 | −3.11 | rrm1 | PREDICTED: ribonucleoside-diphosphate reductase large subunit-like (Hyalella azteca) |
| ROT79532.1 | −1.25 | gstd1 | Delta-class glutathione S-transferase (Fenneropenaeus chinensis) |
| ROT79533.1 | −2.40 | gstd1 | Delta-class glutathione S-transferase (Fenneropenaeus chinensis) |
| ROT81832.1 | 0.87 | gclc | PREDICTED: glutamate--cysteine ligase catalytic subunit-like (Hyalella azteca) |
| MSTRG.22898 | −1.21 | se | Pyrimidodiazepine synthase-like (Penaeus vannamei) |
| MSTRG.30076 | −3.06 | rrm1 | Ribonucleoside diphosphate reductase large subunit-like (Penaeus vannamei) |
| P40 vs. P48 | |||
| ROT61670.1 | −3.04 | gpx | Glutathione peroxidase 3 (Penaeus monodon) |
| ROT62696.1 | −2.11 | rrm1 | PREDICTED: ribonucleoside diphosphate reductase large subunit-like (Hyalella azteca) |
| ROT71141.1 | −1.95 | gstd1 | Delta-class glutathione S-transferase (Fenneropenaeus chinensis) |
| ROT72062.1 | −0.48 | anpep | PREDICTED: aminopeptidase N-like (Hyalella azteca) |
| ROT78750.1 | −1.07 | gstm1 | Glutathione S-transferase (Litopenaeus vannamei) |
| ROT79533.1 | −1.71 | gstd1 | Delta-class glutathione S-transferase (Fenneropenaeus chinensis) |
| MSTRG.22898 | −1.08 | se | Pyrimidodiazepine synthase-like (Penaeus vannamei) |
| MSTRG.30076 | −1.92 | rrm1 | Ribonucleoside diphosphate reductase large subunit-like (Penaeus vannamei) |
| Drug Metabolism—Cytochrome P450 | |||
| P40 vs. P32 | |||
| ROT71141.1 | −3.74 | gstd1 | Delta-class glutathione S-transferase (Fenneropenaeus chinensis) |
| ROT72759.1 | −4.29 | ugt2b13 | PREDICTED: UDP-glucuronosyltransferase 2B14-like (Hyalella azteca) |
| ROT73315.1 | −1.68 | ugt2b16 | PREDICTED: UDP-glucuronosyltransferase-like isoform X2 (Hyalella azteca) |
| ROT74229.1 | −4.52 | ugt8 | PREDICTED: 2-hydroxyacylsphingosine 1-beta-galactosyltransferase-like (Hyalella azteca) |
| ROT78750.1 | −2.52 | gstm1 | Glutathione S-transferase (Litopenaeus vannamei) |
| ROT79533.1 | −2.40 | gstd1 | Delta-class glutathione S-transferase (Fenneropenaeus chinensis) |
| ROT79971.1 | −1.62 | ugt8 | PREDICTED: UDP-glucuronosyltransferase 2B19-like isoform X1 (Hyalella azteca) |
| ROT80959.1 | −3.98 | ugt1a8 | PREDICTED: UDP-glucuronosyltransferase 2B19-like isoform X1 (Hyalella azteca) |
| P40 vs. P48 | |||
| ROT73315.1 | −1.47 | ugt2b16 | PREDICTED: UDP-glucuronosyltransferase-like isoform X2 (Hyalella azteca) |
| ROT74228.1 | −3.40 | ugt8 | PREDICTED: 2-hydroxyacylsphingosine 1-beta-galactosyltransferase-like (Hyalella azteca) |
| ROT79532.1 | −2.99 | gstd1 | Delta-class glutathione S-transferase (Fenneropenaeus chinensis) |
| ROT79533.1 | −1.71 | gstd1 | Delta-class glutathione S-transferase (Fenneropenaeus chinensis) |
| ROT79971.1 | −1.92 | ugt8 | PREDICTED: UDP-glucuronosyltransferase 2B19-like isoform X1 (Hyalella azteca) |
| Lysosome | |||
| P40 vs. P32 | |||
| ROT62942.1 | −2.46 | ctsc | Cathepsin C (Fenneropenaeus chinensis) |
| ROT64929.1 | −9.56 | asm-2 | PREDICTED: sphingomyelin phosphodiesterase-like (Hyalella azteca) |
| ROT70506.1 | −2.72 | lip3 | Triacylglycerol lipase (Litopenaeus vannamei) |
| ROT72812.1 | −4.26 | smpd1 | PREDICTED: sphingomyelin phosphodiesterase-like (Hyalella azteca) |
| ROT73988.1 | −9.94 | lcp2 | Cathepsin L (Marsupenaeus japonicus) |
| ROT79070.1 | −2.90 | ctsc | Cathepsin C (Fenneropenaeus chinensis) |
| ROT80984.1 | −1.50 | lipf | Triacylglycerol lipase (Portunus trituberculatus) |
| ROT83980.1 | −2.28 | arsa | PREDICTED: arylsulfatase A-like (Hyalella azteca) |
| ROT85091.1 | −1.99 | man2b1 | PREDICTED: lysosomal alpha-mannosidase-like (Hyalella azteca) |
| ROT85637.1 | −4.11 | npc1 | PREDICTED: Niemann–Pick C1 protein-like (Hyalella azteca) |
| MSTRG.26644 | −1.22 | hexb | Beta-hexosaminidase subunit alpha-like (Penaeus vannamei) |
| P40 vs. P48 | |||
| ROT61198.1 | −2.96 | aael006169 | Cathepsin D-like protein (Homarus americanus) |
| ROT62149.1 | −1.97 | brafldraft_56888 | PREDICTED: alpha-L-fucosidase-like (Hyalella azteca) |
| ROT62942.1 | −2.61 | ctsc | Cathepsin C (Fenneropenaeus chinensis) |
| ROT66187.1 | −4.72 | arsb | PREDICTED: arylsulfatase B-like (Branchiostoma belcheri) |
| ROT67034.1 | −2.55 | arsb | PREDICTED: arylsulfatase B-like (Branchiostoma belcheri) |
| ROT70506.1 | −2.73 | lip3 | Triacylglycerol lipase (Litopenaeus vannamei) |
| ROT70922.1 | −2.51 | smpd1 | PREDICTED: sphingomyelin phosphodiesterase-like (Diachasma alloeum) |
| ROT72812.1 | −2.62 | smpd1 | PREDICTED: sphingomyelin phosphodiesterase-like (Hyalella azteca) |
| ROT73985.1 | −3.71 | lcp2 | Cathepsin l, partial (Litopenaeus vannamei) |
| ROT73986.1 | −2.78 | lcp2 | Cathepsin l (Litopenaeus vannamei) |
| ROT80984.1 | −2.00 | lipf | Triacylglycerol lipase (Portunus trituberculatus) |
| ROT84188.1 | −9.15 | slc17a2 | PREDICTED: sialin-like (Hyalella azteca) |
| ROT85091.1 | −2.40 | man2b1 | PREDICTED: lysosomal alpha-mannosidase-like (Hyalella azteca) |
| MSTRG.26644 | −1.14 | hexb | Beta-hexosaminidase subunit alpha-like (Penaeus vannamei) |
| Pancreatic Secretion | |||
| P40 vs. P32 | |||
| MSTRG.26333 | −5.19 | -- | Group 3 secretory phospholipase A2-like (Penaeus vannamei) |
| MSTRG.31007 | −3.56 | amy | Amylase (Penaeus vannamei) |
| MSTRG.31195 | −3.54 | cpa1 | Carboxypeptidase B-like (Penaeus vannamei) |
| MSTRG.31546 | −1.77 | clca2 | Calcium-activated chloride channel regulator 4A-like isoform X2 (Penaeus vannamei) |
| P40 vs. P48 | |||
| MSTRG.31001 | −3.15 | amy | LOW-QUALITY PROTEIN: alpha-amylase-like (Penaeus vannamei) |
| MSTRG.31007 | −3.57 | amy | Amylase (Penaeus vannamei) |
| MSTRG.31195 | −2.00 | cpa1 | Carboxypeptidase B-like (Penaeus vannamei) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.; Yuan, H.; Fu, Z.; Luo, X.; Xue, Z.; Zhang, S. Investigating the Impact of Varied Dietary Protein Levels on Litopenaeus vannamei: An Exploration of the Intestinal Microbiota and Transcriptome Responses. Animals 2024, 14, 372. https://doi.org/10.3390/ani14030372
Li G, Yuan H, Fu Z, Luo X, Xue Z, Zhang S. Investigating the Impact of Varied Dietary Protein Levels on Litopenaeus vannamei: An Exploration of the Intestinal Microbiota and Transcriptome Responses. Animals. 2024; 14(3):372. https://doi.org/10.3390/ani14030372
Chicago/Turabian StyleLi, Gongyu, Hang Yuan, Zhibin Fu, Xinghui Luo, Zhihao Xue, and Shuang Zhang. 2024. "Investigating the Impact of Varied Dietary Protein Levels on Litopenaeus vannamei: An Exploration of the Intestinal Microbiota and Transcriptome Responses" Animals 14, no. 3: 372. https://doi.org/10.3390/ani14030372
APA StyleLi, G., Yuan, H., Fu, Z., Luo, X., Xue, Z., & Zhang, S. (2024). Investigating the Impact of Varied Dietary Protein Levels on Litopenaeus vannamei: An Exploration of the Intestinal Microbiota and Transcriptome Responses. Animals, 14(3), 372. https://doi.org/10.3390/ani14030372





