1. Introduction
Nucleated red blood cells (nRBCs) are usually absent in the peripheral blood of cats and dogs, as erythropoiesis mostly occurs in the bone marrow. During erythropoiesis, hematopoietic stem cells proliferate under the influence of growth and transcription factors, resulting in a continuous mitotic division from rubriblasts to prorubricytes, basophilic rubricytes, and polychromatophilic rubricytes. Polychromatophilic rubricytes are post-mitotic and undergo maturation to metarubricytes before extruding their nucleus and developing into reticulocytes [
1]. In dogs and cats, reticulocytes mature partially still within the bone marrow and also after their release into the peripheral bloodstream, preferentially in the spleen, where hemoglobinization, membrane remodeling, and cytoskeletal stabilization are completed [
2]. In the case of the release of erythroid precursors from bone marrow, splenic macrophages also contribute to the quick removal of nuclei, nuclear remnants, and organelles. The exact mechanism that prevents nRBCs from being released into peripheral blood in health is not yet fully understood. It is suspected that the adhesion properties of specialized reticular cells of the blood–bone marrow barrier and the rigidity of nRBCs may prevent nRBCs from being released [
3].
The presence of nRBCs in the peripheral blood, independent of their maturation stage, is commonly termed normoblastosis (term used for this publication) or normoblastemia. Physiological normoblastosis may be seen in response to severe regenerative anemia (appropriate normoblastosis), e.g., secondary to blood loss or immune-mediated hemolytic anemia. Consequently, the nRBCs are accompanied by reticulocytosis, which can be appreciated as polychromasia and anisocytosis upon blood smear evaluation. Inappropriate normoblastosis in dogs and cats is commonly associated with different pathological processes such as lead toxicity, heatstroke, splenic disorders, or bone marrow injury secondary to, e.g., neoplasia, hypoxia, septicemia, or drug administration [
4,
5,
6,
7,
8]. Additionally, inappropriate normoblastosis is described in congenital dyserythropoiesis in English Springer Spaniel and familial Poodle macrocytosis [
9]. Mild normoblastosis is also reported in dogs and cats suffering from cardiovascular disease, respiratory disease, trauma, inflammatory conditions, or hyperadrenocorticism, and extramedullary hematopoiesis may also contribute to normoblastosis [
10].
In recent publications, normoblastosis has been examined as a potential prognostic biomarker involving canine and feline patients. In veterinary critical care, novel, readily available, and cost-efficient prognostic biomarkers are continuously investigated to further improve the assessment of morbidity and mortality, as well as to monitor treatment response or disease progression. One study showed that normoblastosis in cats can be associated with more severe acute clinical signs, longer hospitalization, higher treatment costs, and lower survival rates and can therefore alert the clinician to the need to implement intensive care and add additional information regarding the prognosis of the patient [
11]. In dogs, normoblastosis is most often associated with regenerative anemia and has been shown to be a negative prognostic indicator in these patients [
12,
13]. An association between normoblastosis and increased mortality has also been found in dogs with systemic inflammatory response syndrome (SIRS) after the exclusion of patients suffering from additional diseases commonly causing normoblastosis (e.g., hemolysis or hemorrhage) [
14]. In dogs suffering from heatstroke, normoblastosis has been found to be a sensitive and specific predictor of death and secondary complications. Therefore, the enumeration of peripheral nRBCs has been suggested to be a useful additional analyte in the prediction of the outcome in dogs affected by heatstroke [
15].
In the past, the automated enumeration of nRBCs was unavailable in veterinary medicine, and microscopical enumeration by blood smear evaluation is still most often used for the enumeration of nRBCs. Although simple, the manual method is time-consuming, known to be imprecise, and may miss low numbers of nRBCs [
16,
17]. Most available veterinary automated hematology analyzers do not provide an enumeration of nRBCs, thus counting nRBCs within the white blood cell (WBC) count. This necessitates an assessment of possible normoblastosis by blood smear evaluation with subsequent correction of automated WBC counts by the manually determined nRBC counts/100 WBCs. The equation used for the correction of WBC counts is as follows: corrected WBC (10
3/µL) = (measured WBC cell count × 100)/(nRBC + 100) [
18]. This previously unavoidable practice results in increased hands-on time and risk of error. Additionally, the failure to recognize significant normoblastosis can subsequently lead to erroneously high WBC counts, which may affect clinical decisions and the outcome of these patients [
4,
19].
A fast, accurate, and precise determination of nRBCs would represent an important advancement in automated veterinary hematology, add prognostic value to hematology profiles, save time, and reduce the risk of unnoticed normoblastosis. The Sysmex XN-V automated hematology analyzer is the first veterinary hematology analyzer providing automated nRBC counts alongside WBC counts for different species. The authors aimed to evaluate the automated nRBC enumeration performance of the Sysmex XN-V hematology analyzer for canine and feline patients, including method comparison, repeatability, reproducibility, linearity, stability, and carry-over.
2. Materials and Methods
2.1. Selection of Specimens
For method comparison, the study included canine and feline K3-ethylenediamine tetra-acetic acid (EDTA) whole blood specimens sent for routine diagnostic analysis to the Clinical Laboratory of the Vetsuisse Faculty, University of Zurich, Switzerland, between August 2019 and June 2022. All samples with an automated and manual nRBC count available were included in the study. Data analysis was performed retrospectively. The blood collection procedure was not standardized, and patients were included regardless of age, sex, breed, or disease. Most (>97%) specimens were collected in-house at the Small Animal Clinic of the Vetsuisse Faculty (K3-EDTA whole blood in MiniCollect, Greiner Bio-One tubes, Kremsmünster, Austria) and were readily submitted at room temperature. Few specimens (<3%) were collected at privately owned clinics in Switzerland and readily submitted by mail at ambient temperature, together with freshly prepared blood smears. After arrival at the Clinical Laboratory, all specimens were kept at room temperature, placed on a rotator (Labinco LD-76; Labinco, Breeda, The Netherlands) for at least five minutes, and gently homogenized manually before processing.
2.2. Manual Count by Blood Smear Evaluation
Blood smear preparation of in-house submitted samples was performed within one hour after sample collection by trained laboratory technicians. The smears were air-dried for 20 min before staining. Staining was performed on a HEMA-TEK 2000 slide stainer (Siemens Healthcare GmbH, Erlangen, Germany) with modified Wright’s stain. Experienced laboratory technicians counted the nRBCs by microscopic blood smear evaluation using a Leica DM LB2 microscope (Leica Microsystems AG, Heerbrugg, Switzerland) with 50× and 100× oil immersion objectives. The nRBCs were counted alongside the WBC differentiation (100 WBC differential). In cases of a left shift (presence of band neutrophils), a second 100 WBC count was performed by a second laboratory technician, and the means were calculated. This procedure was performed according to the laboratory’s non-statistical quality assurance and standard operating procedures to increase the precision of the manual differential. Manual nRBC counts were reported as nRBCs per 100 WBCs (nRBCm).
2.3. Automated Count by Sysmex XN-V
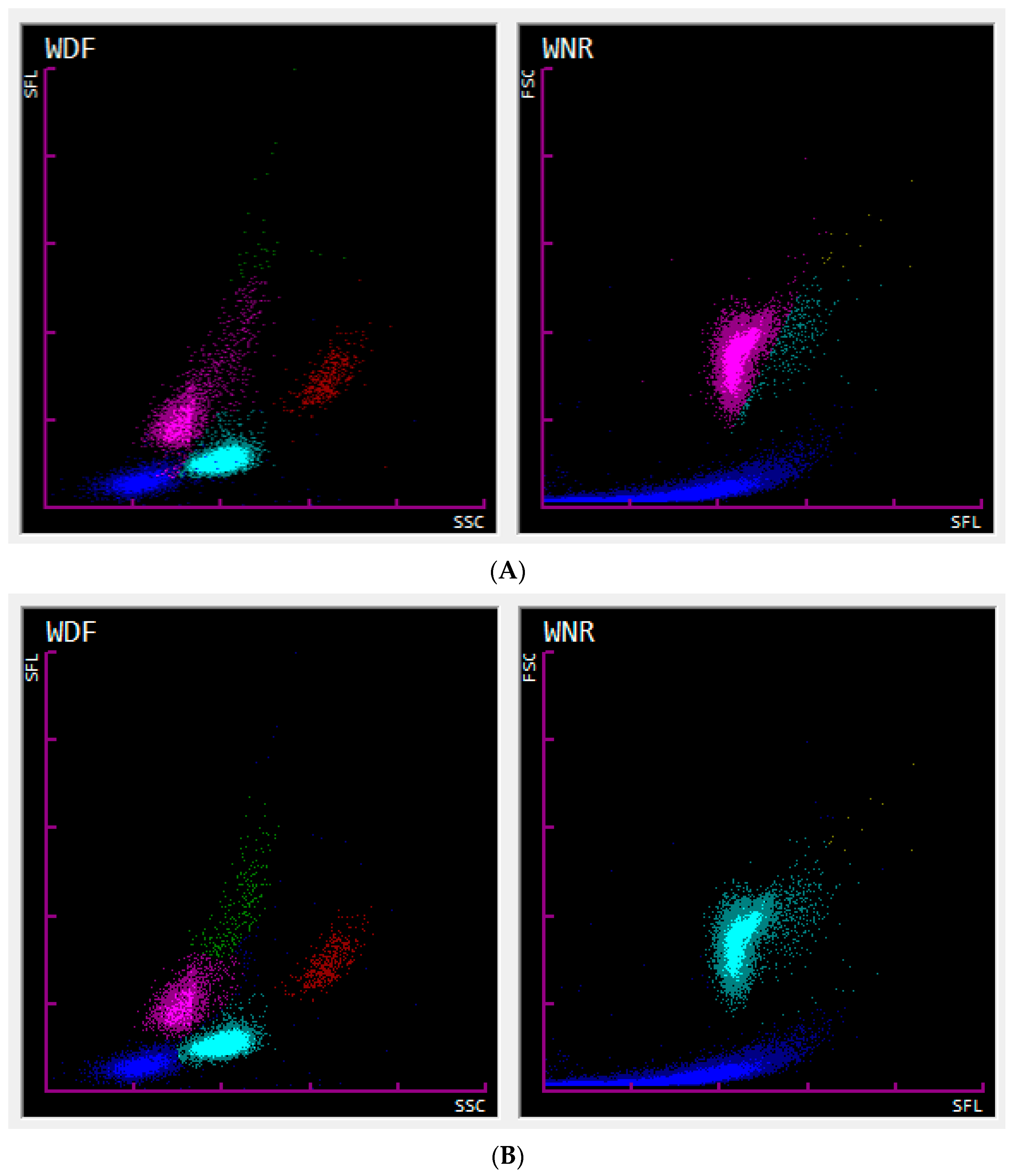
Automated enumeration of the nRBCs was performed on the Sysmex XN-1000V analyzer (Sysmex Corporation, Kobe, Japan, Version 3.03 (00-08)) according to the manufacturer’s instructions using the species-specific preinstalled profiles for cats and dogs. In-house collected samples were processed within one hour after collection. The nRBCs were counted alongside the WBCs in the WNR (WBC/basophil/nRBC) channel of the analyzer. During the process, red blood cell (RBC) membranes are initially lysed by the hemolytic Lysercell WNR reagent. This reagent also shrinks WBCs while permeating their membrane, except for the basophil’s membrane, which, in certain species (humans and rabbits), remains relatively unaffected. As RBCs and nRBCs are the most affected by Lysercell WNR treatment, further RNA and nuclear degradation is suspected in nRBCs compared to WBCs. After lysis, nucleic acids are stained using Fluorocell WNR reagent. Subsequently, the forward scattered light (FSC) and side fluorescence light (SFL) properties of the cells are determined by flow cytometric analysis. The FSC and SFL properties display the size and nucleic acid content of the cells, respectively. This allows the analyzer to discriminate nRBCs from WBCs since nRBCs show less nucleic acid content (weak SFL) and are similar to those of a slightly larger size (medium FSC) compared to WBCs. The causes for nRBCs showing lower nucleic acid content (weaker SFL) are believed to be due to the loss of intracytoplasmic organelles/nucleic acids, smaller, more condensed nuclei, and RNA degradation by the Lysercell WNR reagent affecting the erythroid lineage to a higher extent than leukocytes [
20]. An illustration of all of the acquired events is generated and shown as the associated WNR scattergram (FSC—
Y-axis; SFL—
X-axis) for each patient (
Figure 1). At the Clinical Laboratory of the Vetsuisse Faculty Zurich, all of the WBC and nRBC counts were further investigated by performing an inspection for flags and evaluation of the associated scattergrams by experienced laboratory technicians. In the case of uncertain results, a blood smear evaluation was performed, and a clinical pathologist’s review was additionally requested. The nRBCs were measured as absolute counts (nRBCs/µL), and relative counts (nRBCs/100 WBCs; nRBCa) were calculated.
2.4. Method Comparison
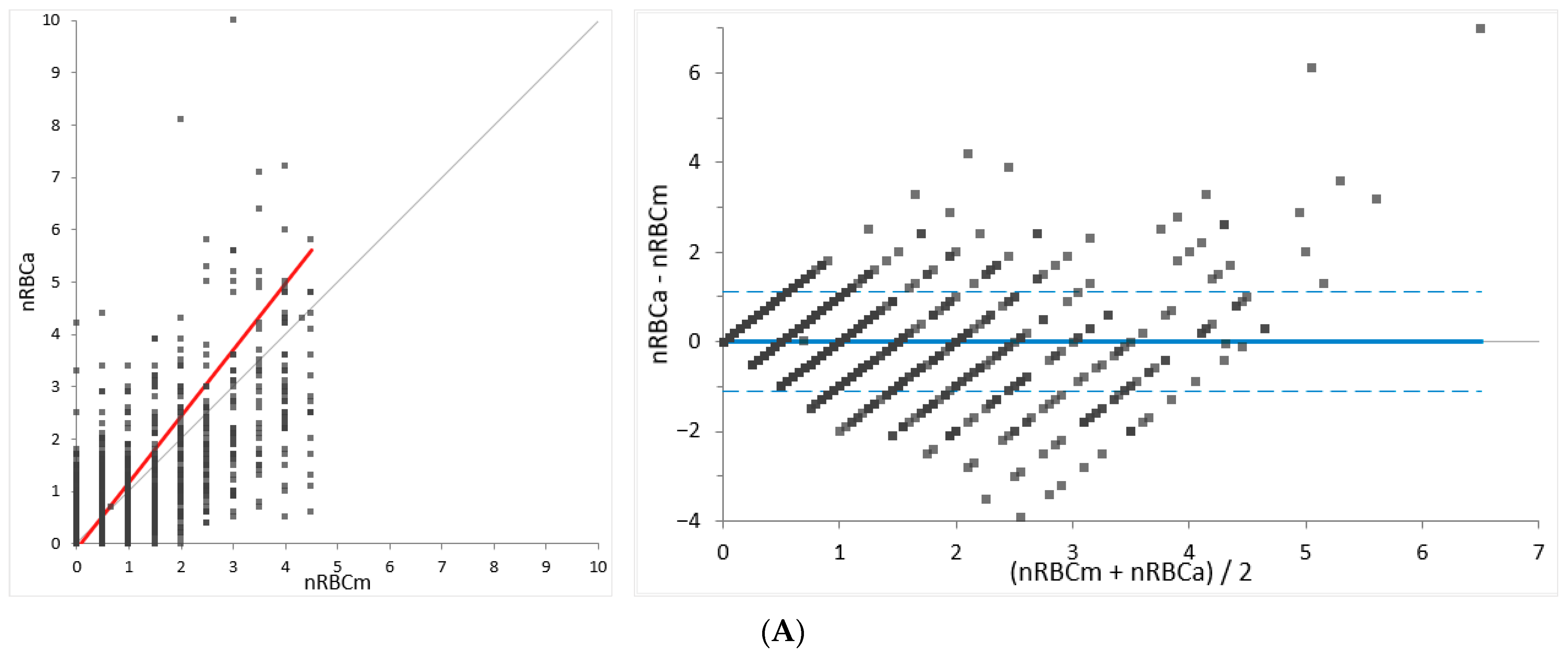
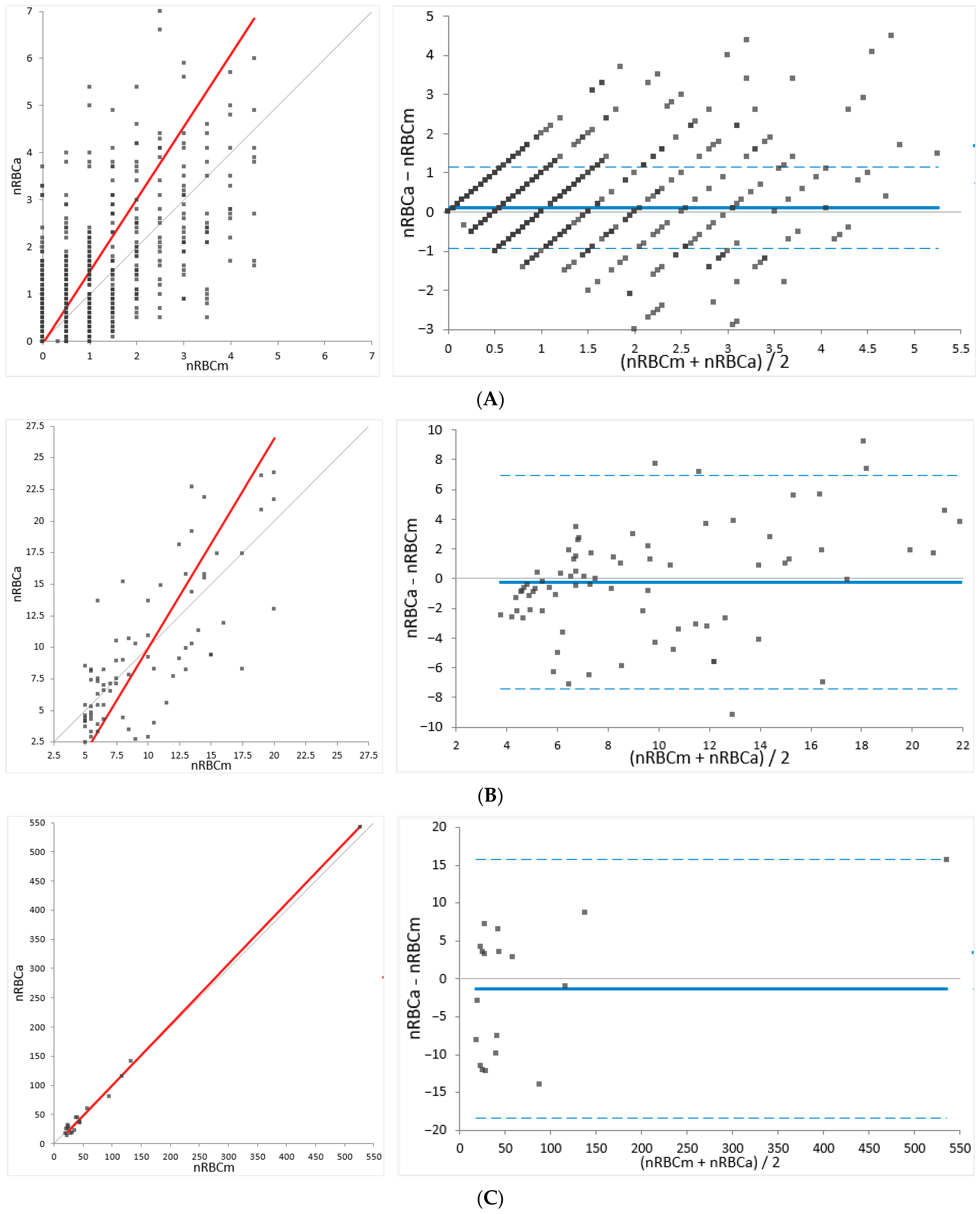
For method comparison (determination of agreement), the relative nRBC counts established by Sysmex XN-V (nRBCa) were compared to the manually determined nRBC counts (nRBCm) using a total of 2844 feline and 3810 canine samples. In cases of significant differences between the automated and manual analysis (outliers), blood smears and automated data, including scattergrams, were revised and re-gating or manual re-counting was performed if necessary. For a few of the outliers, where a review of slides and/or automated data and scattergrams was not possible (e.g., due to missing slides), the data were dismissed (dogs, n = 3; cats, n = 1). The results were subdivided into three groups depending on the number of nRBCs/severity of normoblastosis determined by the manual method (low: <5; moderate: 5–20; high: >20 nRBCs/100 WBCs) to provide proper insight into the performance of different levels of normoblastosis.
2.5. Repeatability
The precision (repeatability) of the Sysmex XN-V automated count was evaluated from five consecutive measurements of three canine and two feline specimens. Additionally, manual precision was determined by five consecutive counts (nRBCs/100 WBCs) from four canine and four feline samples using blood smear evaluation. The counts were performed by the same observer (first author) on the same slide using an Olympus BX43 microscope (Evident Europe GmbH, Hamburg, Germany) with 50× and 100× oil immersion objectives.
2.6. Reproducibility
The precision from day to day (reproducibility) of Sysmex XN-V was assessed by using the data from the manufacturer’s control material of three different levels (XN-Check QC1 Lot. QC-23121101, QC2 Lot. QC-23121102, and QC3 Lot. QC-23121103; Sysmex Europe GmbH, Norderstedt, Germany), which were measured on 20 consecutive days. Quality control material was analyzed daily before analyzing the patient samples.
2.7. Linearity
The linearity was evaluated on Sysmex XN-V by using a dilution series (100%, 75%, 50%, 25%, and 0%) of fresh EDTA-anticoagulated blood diluted by isotonic saline (NaCl, sodium chloride 0.9%, Bichsel AG, Interlaken, Switzerland). Each level was measured in duplicate.
2.8. Stability
The stability (cell aging) was determined using two canine and two feline samples stored at either room temperature or refrigerated (4 °C). For both temperature levels, the samples were analyzed immediately after collection and 4, 8, 24, and 48 h later using the Sysmex XN-V.
2.9. Carry-Over
The carry-over was evaluated using two consecutive measurements of the patients’ EDTA-anticoagulated blood followed by three measurements of the blank (NaCl, sodium chloride 0.9%, Bichsel AG, Interlaken, Switzerland). Thereby, two canine EDTA samples with moderate (10.6 nRBCs/100 WBCs; 530 nRBCs/µL and) and high (42 nRBCs/100 WBCs; 9270 nRBCs/µL) nRBC counts were analyzed using the Sysmex XN-V.
2.10. Statistical Analysis
Data were collected in a Microsoft
® Excel
® spreadsheet (Microsoft 365 MSO Version 2306 Build 16.0.16529.20100, Microsoft Corp., Redmond, WA, USA) and statistical analysis was performed using the Analyse-it
® 6.01.1 add-in (Analyse-it Software Ltd., Leeds, UK). For precision analysis, the standard deviation (SD) and coefficient of variation (CV) were calculated for each level of nRBCs and species. The percentage of carry-over was determined by using the following formula: % carry-over = {[(result empty cycle 1) − (result empty cycle 3)]/[(sample 2) − (result empty cycle 3)]} × 100. Linearity was determined according to Emancipator–Kroll [
21] using Analyse-it
®. Stability was reviewed for statistical significance using a paired comparison test and the Friedman test (Analyse-it
®). For method comparison (agreement), a Pearson coefficient of correlation (
r), Deming linear regression analysis determining the intercept and the slope with a 95% confidence interval, and Bland–Altmann difference plot providing information on bias and the 95% limits of agreement were calculated for both species. The concordance of nRBC counts from the two assessed methods was analyzed using Microsoft
® Excel
® at four levels (≤1; 1.1–5; 5.1–20; >20 nRBCs/100 WBCs). The total observed error was calculated according to the ASVCP (American Society for Veterinary Clinical Pathology) guidelines [
22]. The coefficient of correlation was considered excellent if
r ≥ 0.95, very good if
r = 0.90–0.94, good if
r = 0.80–0.89, fair if
r = 0.59–0.79, and poor if
r < 0.59 [
23]. Statistical significance was defined at a
p-value of <0.05.
4. Discussion
The performance of nRBC enumeration by the Sysmex XN series has been previously evaluated with human samples, revealing it to be a precise and accurate method, especially in specimens with less than 200 nRBCs/100 WBCs [
27]. The overall concordance rate between the manual and automated counts by Sysmex XN has been shown to be high (85.1% and 93.2%) [
16,
27,
28]. Previous studies on the veterinary analyzer Sysmex XN-V revealed a low imprecision with an overall CV of 7.1%. Additionally, good agreement has been found for feline and canine specimens with absolute nRBC counts of <1000 nRBCs/µL, excluding samples without circulating nRBCs in data analysis. In cases with higher nRBC counts, blood smear evaluation is recommended for confirmation due to a wider 95% LoA [
29]. Correlation between the Sysmex XN-V and manual nRBC counts was found to be fair (
r = 0.57) in a study using 63 canine specimens with a median automated absolute count of 10 nRBCs/µL (ranging from 0 to 1120 nRBCs/µL). This low correlation may be due to the low nRBC counts assessed and the poor reliability of the manual nRBC counting [
30]. However, the performance of Sysmex XN-V in the enumeration of nRBCs using a wide range of feline and canine specimens, also including samples without normoblastosis, had not yet been evaluated. Therefore, the present method comparison study is unique in that it investigated a high number of patient samples at the full measurement range to detect both false negative and false positive results since they could affect patients’ treatment and outcome. In addition to previous studies, linearity, stability, carryover, and the investigation of interferences of nRBCs with the white blood cell differential in the WDF channel were studied.
The present study investigated a wide range of nRBC counts from 0 to 272 nRBCs/100 WBCs in dogs and 0 to 527 nRBCs/100 WBCs in cats, also including specimens without normoblastosis. A total of 3810 dog and 2844 cat specimens were used for assessing the agreement of the method. By analyzing patient samples collected in routine diagnostics over the course of three years, the study population reflects a representative patient population for veterinary laboratories. Since severe normoblastosis is an uncommon finding in canine and feline species, the majority of samples showed normal or low nRBC counts and samples with high numbers of nRBCs were less frequently observed. Still, in the group with >20 nRBCs/100 WBCs, 52 dog and 18 cat samples could be evaluated for this study. To overcome the limitation that the majority of evaluated specimens had normal or low nRBC counts, the performance of Sysmex XN-V was evaluated at three different severity levels of normoblastosis (low: <5, moderate: 5–20, and high: >20 nRBCs/100 WBCs) providing insight in the performance at different ranges. A fair to excellent correlation ranging from 0.743 to 0.998, with the highest correlation in the group with >20 nRBCs/100 WBCs in both species, was determined by regression analysis, indicating a representative study population and range.
The grading of relative or absolute nRBC counts is not harmonized in the veterinary literature. However, up to 1 nRBC/100 WBCs or absolute counts of <100 nRBCs/µL are most often considered normal. Nucleated RBCs of more than 5/100 WBCs or >400 nRBCs/µL are considered indicative of severe normoblastosis [
4,
31]. A relative count of >5/100 WBCs is also often used as the decision limit for the correction of WBC counts if no automatically corrected WBC count is available. Normoblastosis can be assessed as relative (number of nRBCs per 100 white blood cells) or absolute (number of nRBCs per microliter) counts. However, considering the increasing availability of automated nRBC counts in the future, medical decision limits should be set using absolute counts to eliminate the interference of WBC counts in the decision.
Manual nRBC counting was used as the reference method for method comparison, as it represents the most commonly used method for the enumeration of nRBCs in veterinary medicine. However, manual counting may miss low numbers of nRBCs and shows high imprecision and interobserver variation [
16,
17,
20,
29]. A high imprecision of manual counts was also noted in this study, with a mean CV of 19.6%. Manual imprecision is especially high at low ranges of nRBCs, as also previously reported [
16,
27]. This finding may be associated with the low numbers of cells counted manually, observer bias, and poor distribution of cells. Precision analysis of automated nRBC counts has shown low within- and between-run imprecision, with a mean CV of 4.5% and 5.4%, respectively. The imprecision of Sysmex XN-V determined in this study was found to be lower than previously reported [
29]. Given the higher imprecision of manual counts compared to automated counts, data from method comparisons using manual counting as the reference method need to be interpreted in light of the possible unfavorable performance of the reference method (manual count). Method comparison of nRBC counts using a gold standard method, such as flow cytometric analysis and electron microscopy, has not yet been performed in veterinary medicine, possibly due to the low availability or unavailability of these methods. However, this gap should be filled in future investigations.
Stability testing showed no significant changes over 48 h at room temperature and refrigerated. Linearity was excellent at most investigated levels, especially at high levels of nRBC counts. However, at the lowest investigated level (<300 nRBC/µL), the difference exceeded the manufacturer’s specifications. A dilution error can be excluded since the linearity was excellent in red blood cell and white blood cell measurements in the investigated samples. According to our findings, linearity was confirmed at a range between 300 and 3000 nRBCs/µL. Carry-over was not evident. The limitations of this study include the relatively low number of samples used for the assessment of precision, linearity, and stability. This was due to the low numbers of specimens with increased nRBC counts available and the limited volume of residual samples, especially in cats.
Method comparison for the feline samples revealed a high overall concordance of 93%, with the highest concordance at the lowest ranges (≤1 nRBCs/100 WBCs). The mean bias was small at all levels. Although the 95% LoA was wider in the groups with moderate and high nRBC counts (>5 nRBCs/100 WBCs; 86 feline specimens), a normoblastosis was detected by Sysmex XN-V in all of these specimens. The clinical importance of wider 95% LoA at higher ranges is difficult to assess since no medical decision limits have been defined for nRBC counts in veterinary medicine. It therefore remains unclear if the observed variation would alter medical decisions. Furthermore, since severe normoblastosis was a rare finding in feline samples (3% samples with >5 nRBCs/100 WBCs), the overall agreement of the automated counts by Sysmex XN-V and the manual counts needs to be considered good in feline samples. However, manual counts may be performed to confirm the degree of normoblastosis in cases of high automated nRBC counts, since a wider variation is possible. If done so, the higher imprecision of manual counts must be considered when comparing the results.
Method comparison for the canine samples revealed a high overall concordance of 93%, with the highest concordance at the lowest ranges (≤1 nRBCs/100 WBCs). A negligible bias and narrow 95% LoA were found for specimens below 5 nRBCs/100 WBCs. A negative bias and wider 95% LoA were determined for samples with 5 or more nRBCs/100 WBCs, suggesting either an underestimation of automated counts or an overestimation of manual counts. Despite the better performance of manual counts at higher ranges, the imprecision of the manual counts still exceeded the very low imprecision of the automated counts. Therefore, the contribution of the poorer performance of the manual counts to the bias cannot be excluded. However, the negative proportional bias may be also driven by an underestimation of the automated counts in patients with severe normoblastosis. An underlying reason for this may be a poorer identification of early precursors (e.g., rubriblasts and prorubricytes), as previously hypothesized by Brown et al., where a negative proportional bias was also observed [
29]. This hypothesis was not further investigated in the frame of the present study. In the canine specimens with manual relative nRBC counts above 5 (5.4% of all samples), normoblastosis was detected by Sysmex XN-V in all cases. The importance of wider 95% LoA in the canine specimens at higher nRBC counts must be correlated with medical decision limits, which are not yet defined for nRBC counts. Therefore, the influence of a wider 95% LoA and proportional negative bias on medical decisions remains unclear. However, given these findings, confirmation of nRBC counts by manual counts may be performed for samples with moderate and severe normoblastosis in addition to scattergram evaluation and inspection for flags.
The total observable error (TEobs) was below 9% for both species over the full range of investigation. No total allowable error (TEa) has yet been defined for the enumeration of nRBCs, and no expert opinion values are available to the best of the authors’ knowledge. Therefore, the TEobs was compared to the TEa for other common hematological values (TEa for WBC: 15%; TEa for RBC: 6–10%) defined by ASVCP QALS (American Society for Veterinary Clinical Pathology Quality Assurance and Laboratory Standards Committee) and CLIA (Clinical Laboratory Improvement Amendments) recommendations [
32,
33]. Thus, the TEobs for the enumeration of nRBCs by Sysmex XN-V was found to be within these limits, even for samples with marked normoblastosis, suggesting good performance of the assay investigated.
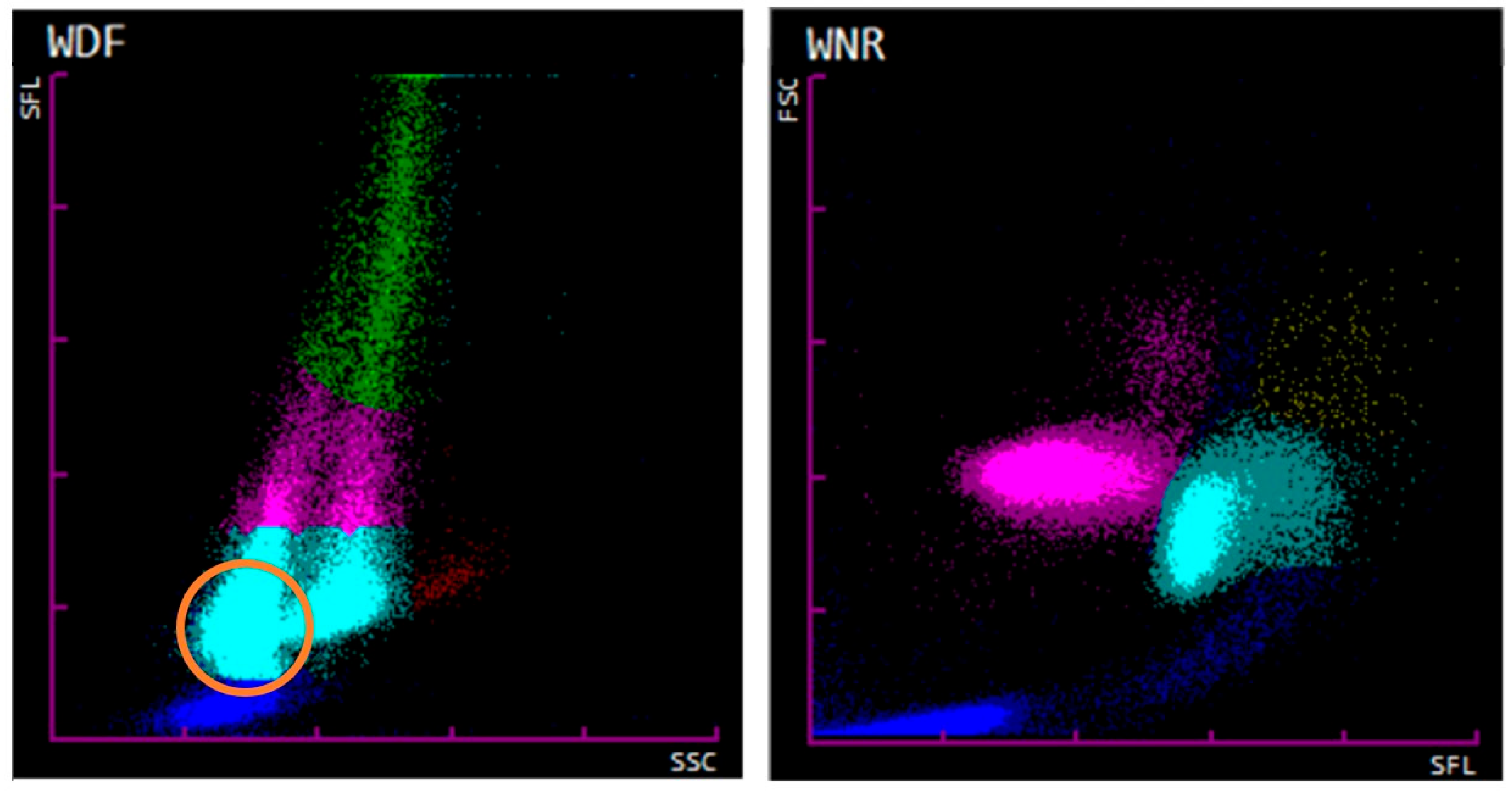
False classification of nRBCs, as shown by the example of Cat 51, probably exceeds medical decision limits for WBC counts and therefore will affect the treatment, test regimen, and, possibly, the outcome of the patient. However, this infrequently observed severe error was noticed by non-statistical quality control procedures, which were performed for each hematological evaluation according to a standard operating procedure, including inspection for flags, scattergram evaluation, and blood smear examination. If such a case was noticed, manual re-gating was performed after confirmation of the absence of a significant normoblastosis by blood smear evaluation. This procedure resulted in a good agreement between manual and automated nRBC counts and reliable values could be obtained. Brown et al. previously reported a correlation between a high or high normal hematocrit and the misclassification of WBC as nRBCs in the WNR channel [
29]. This phenomenon can also be confirmed for our investigated cases including the case of Cat 51. A mean hematocrit of 54% in cats (ten cases) and a mean hematocrit of 56% in dogs (four cases) was found. Further investigation in cases with severe normoblastosis showed that WBC differentiation is also affected by nRBCs entering the WDF channel. This emphasizes the importance of manual WBC differentiation by blood smear evaluation in cases of remarkable normoblastosis. After the course of this study, Sysmex introduced the Sysmex XN-V Series IPU software update version 3.07-00 in July 2022 with improvements on the fractioning of nRBCs and WBCs in the WNR channel by adjusting the algorithm. Since then, significant misclassifications, as described by the case of Cat 51, were subjectively noted to a much lesser extent in our routine diagnostics. However, rare misclassifications of WBCs as nRBCs must still be anticipated, especially in patients showing erythrocytosis. Thus, non-statistical quality control procedures, such as scattergram evaluation, investigation for flags, and blood smear evaluation, should be carried out for patients with severe normoblastosis and concurrent leukopenia to identify possible errors.
Our findings suggest, for both species, that automated and manual nRBC counts can be used interchangeably in samples with low nRBC ranges (<5 nRBCs/100 WBCs). In the higher ranges, Sysmex XV-V performed satisfactorily compared to the manual method. Besides the essential evaluation for flags and scattergram examination, manual counts by blood smear evaluation are recommended for confirmation of the degree of normoblastosis in cases with high nRBC counts. The importance of blood smear evaluation in cases with remarkable normoblastosis is further supported by the observed interference of nRBCs with the WBC differentials and the rare chance of misclassification of WBCs as nRBCs.
The advantages of the automated enumeration of nRBCs include the readily and cost-effective availability of nRBC counts for each patient, the elimination of undetected significant normoblastosis, the reduced hands-on time, and increased precision of counts compared to manual counting. The elimination of undetected normoblastosis is especially important in patients with leukopenia and severe normoblastosis as, e.g., seen in oncological patients receiving chemotherapeutics (e.g., vincristine, doxorubicine, vinblastine, and prednisone), where drug-induced severe normoblastosis may mask concurrent leukopenia due to overestimation of automated leukocyte counts if no WBC correction is performed [
4,
19]. Furthermore, by using automated nRBC counts, this measurement will be readily available for more patients and can provide a foundation to propagate research on circulating nRBCs, providing further insight into, e.g., the regenerative response of anemic patients, the severity of disease, and the risk of mortality in critically ill patients.