Whole-Genome Scanning for Selection Signatures Reveals Candidate Genes Associated with Growth and Tail Length in Sheep
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Information and Sequencing
2.2. Quality Controlling, Reads Mapping, and SNP Calling and Annotating
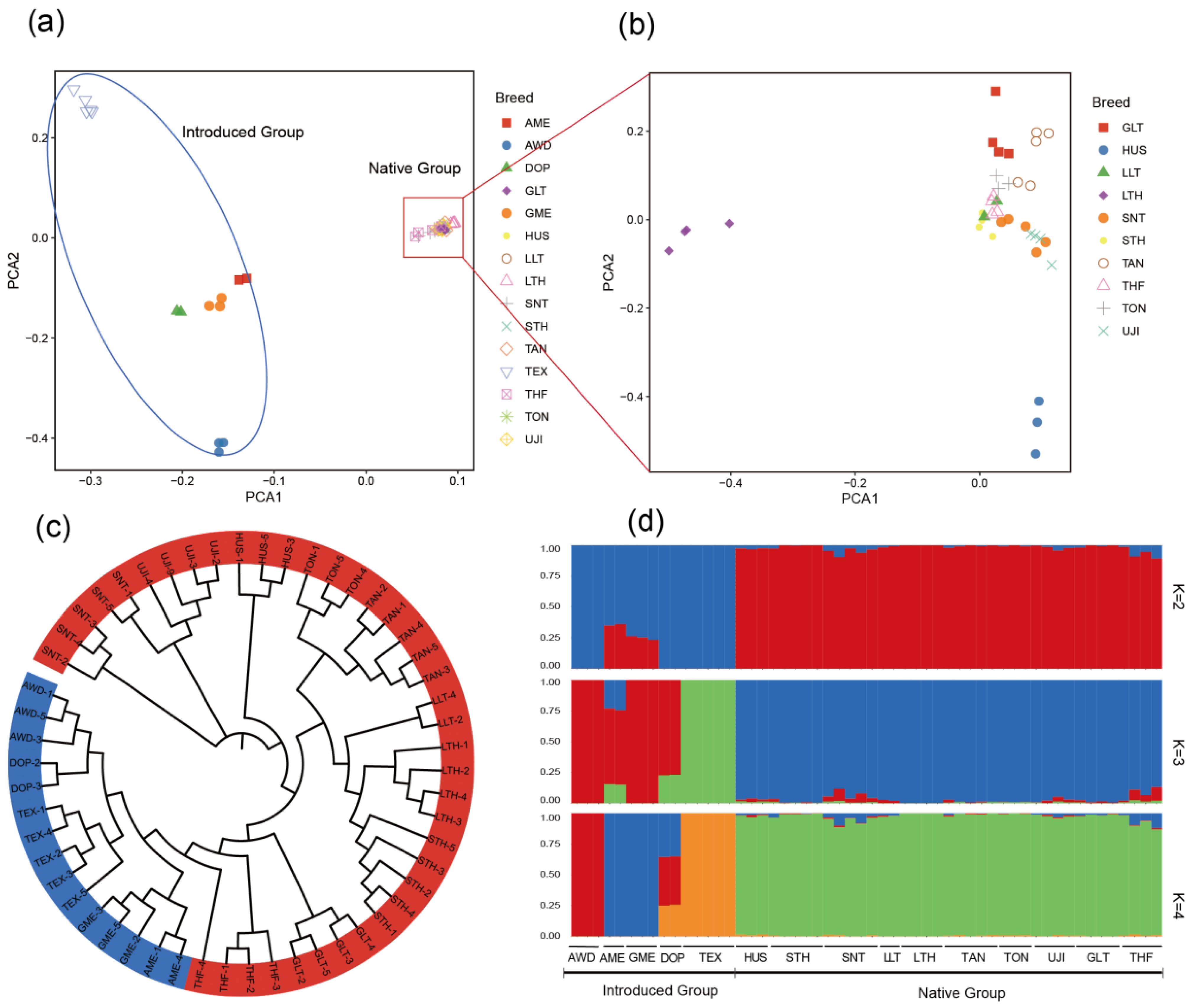
2.3. Population Structure Analysis
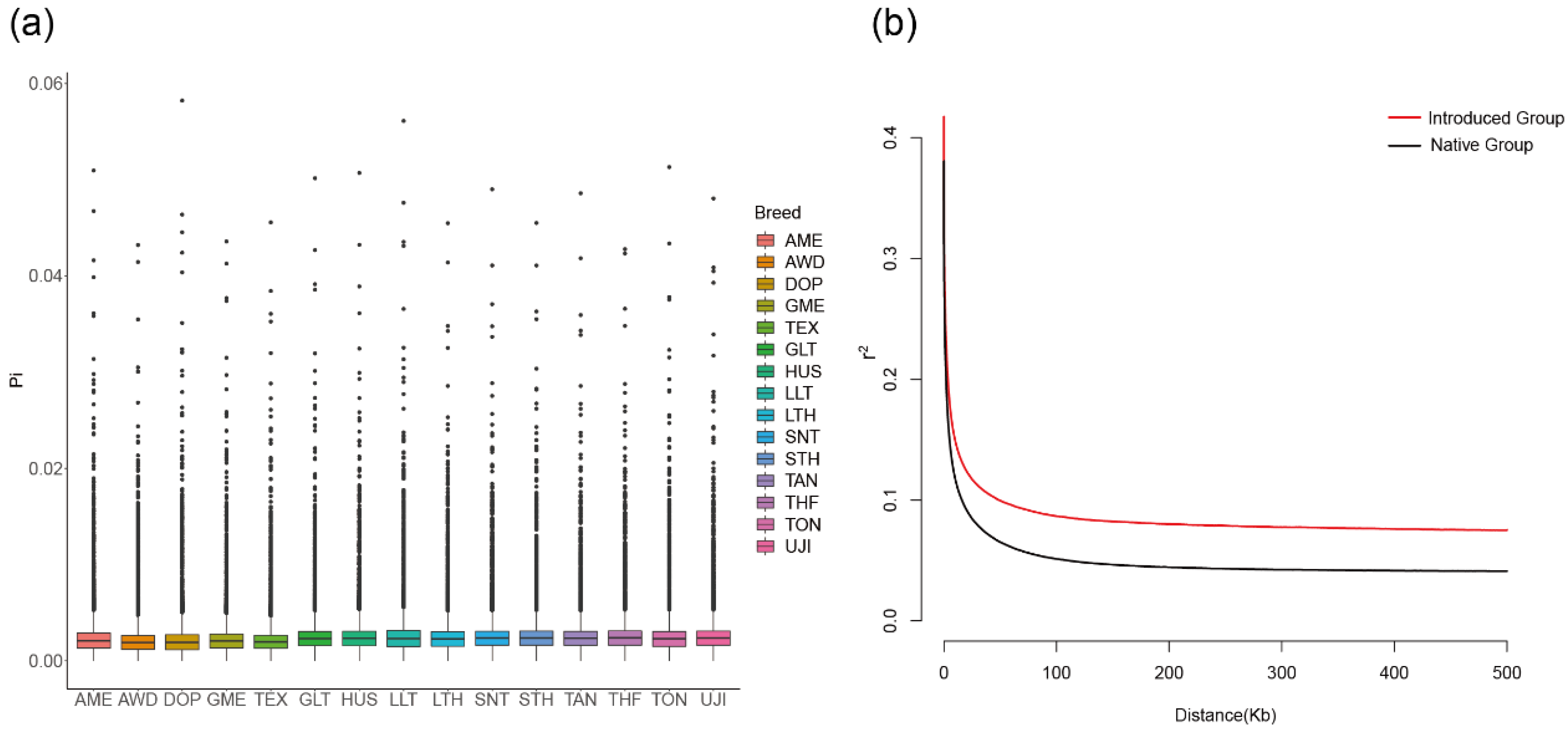
2.4. Nucleotide Diversity and Linkage Disequilibrium
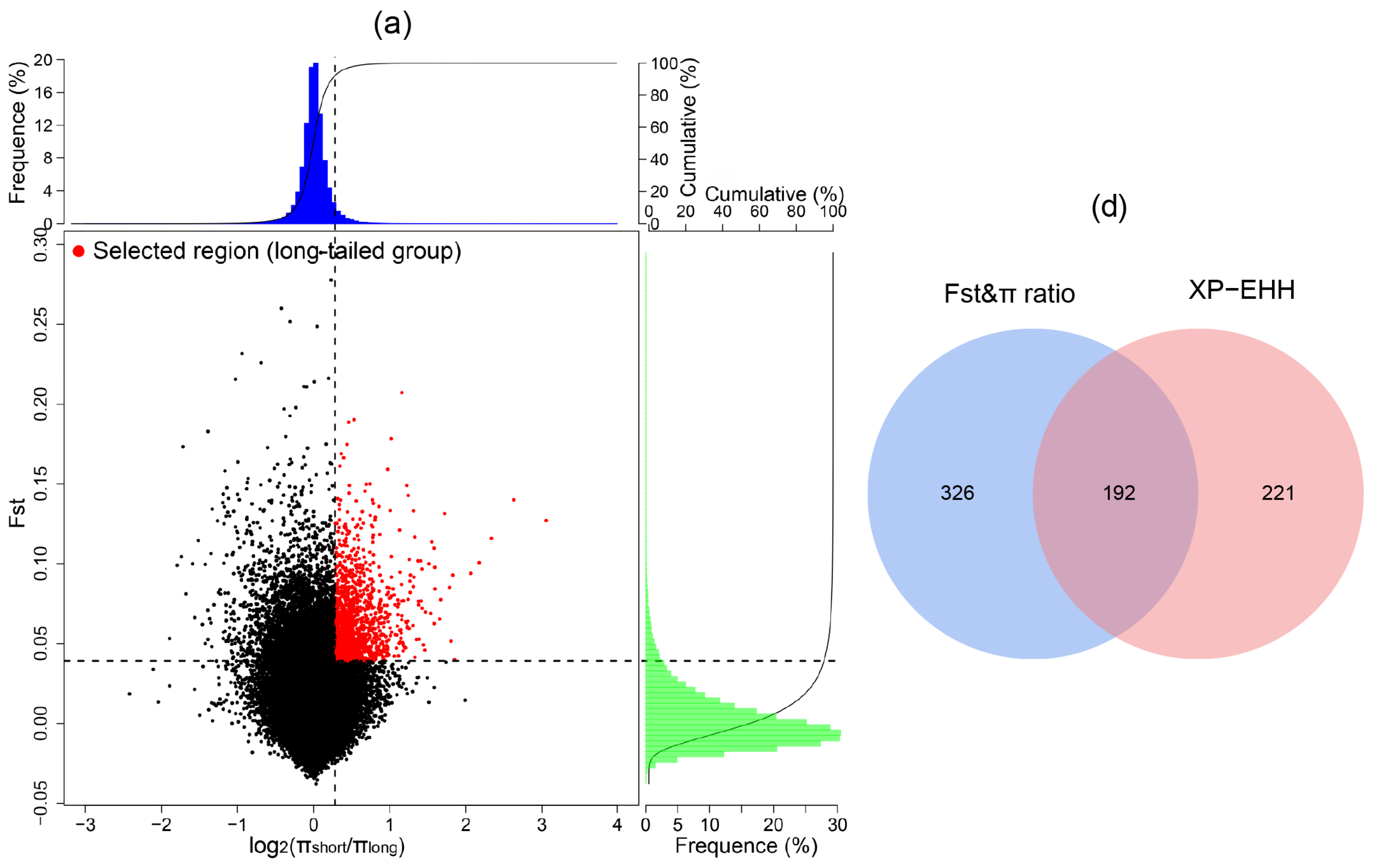
2.5. Selective Sweep Detection
2.6. DCMS Analysis
2.7. Variant Functional Annotations
3. Results
3.1. Sequencing and Variation Calling
3.2. Population Structure Analysis
3.3. Nucleotide Diversity and Linkage Disequilibrium
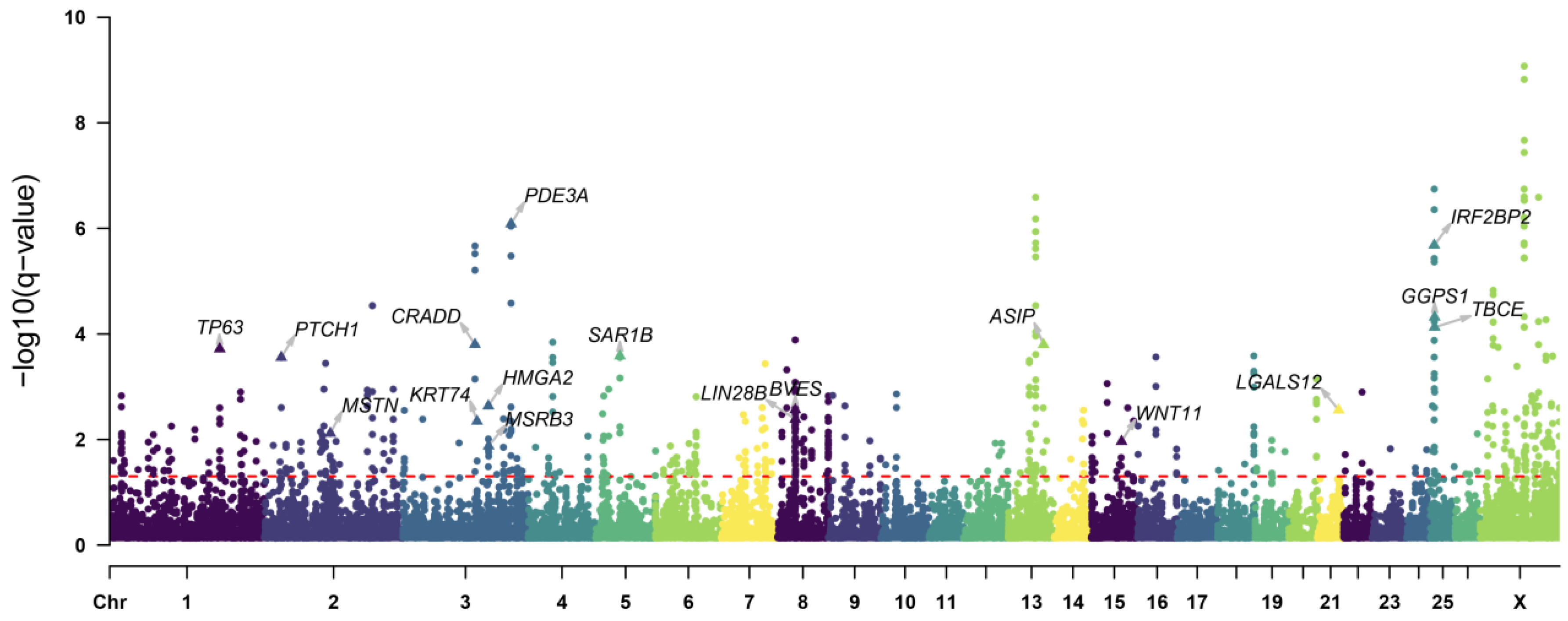
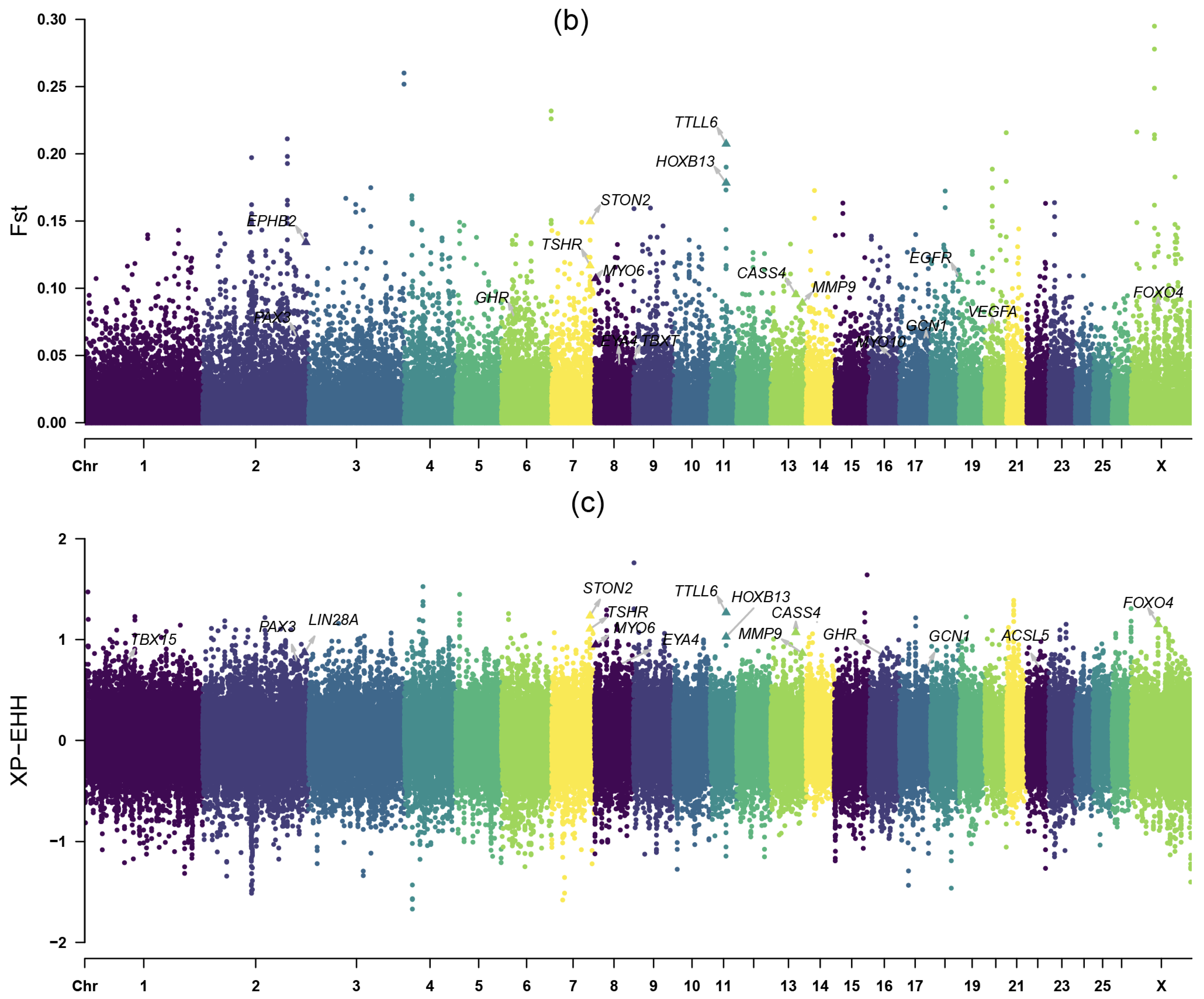
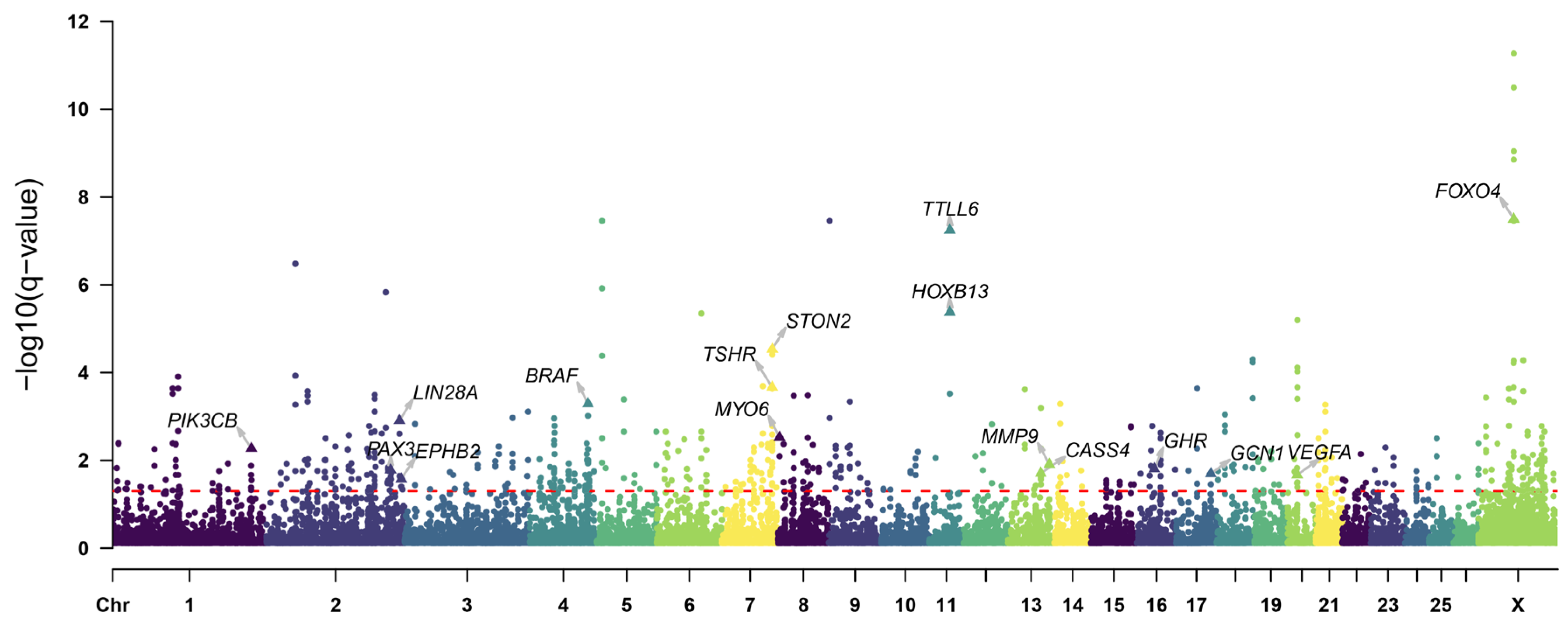
3.4. Positive Selection Signatures in the Introduced Population
3.5. Positive Selection Signatures in the Long-Tailed Population
4. Discussion
4.1. Selection Signatures for Growth Traits
4.2. Selection Signatures for Tail Length Trait
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Song, Y.; Zhang, Q.; Shi, J.; Fu, L.; Cheng, S. Screening of Genes Related to Growth, Development and Meat Quality of Sahan Crossbred F1 Sheep Based on RNA-Seq Technology. Front. Vet. Sci. 2022, 9, 831519. [Google Scholar] [CrossRef]
- Quan, K.; Li, J.; Han, H.; Wei, H.; Zhao, J.; Si, H.A.; Zhang, X.; Zhang, D. Review of Huang-Huai Sheep, a New Multiparous Mutton Sheep Breed First Identified in China. Trop. Anim. Health Prod. 2020, 53, 35. [Google Scholar] [CrossRef]
- Posbergh, C.J.; Huson, H.J. All Sheeps and Sizes: A Genetic Investigation of Mature Body Size across Sheep Breeds Reveals a Polygenic Nature. Anim. Genet. 2021, 52, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Al-Mamun, H.A.; Kwan, P.; Clark, S.A.; Ferdosi, M.H.; Tellam, R.; Gondro, C. Genome-Wide Association Study of Body Weight in Australian Merino Sheep Reveals an Orthologous Region on Oar6 to Human and Bovine Genomic Regions Affecting Height and Weight. Genet. Sel. Evol. 2015, 47, 66. [Google Scholar] [CrossRef] [PubMed]
- Makvandi-Nejad, S.; Hoffman, G.E.; Allen, J.J.; Chu, E.; Gu, E.; Chandler, A.M.; Loredo, A.I.; Bellone, R.R.; Mezey, J.G.; Brooks, S.A.; et al. Four Loci Explain 83% of Size Variation in the Horse. PLoS ONE 2012, 7, e39929. [Google Scholar] [CrossRef] [PubMed]
- Schwaner, M.J.; Hsieh, S.T.; Braasch, I.; Bradley, S.; Campos, C.B.; Collins, C.E.; Donatelli, C.M.; Fish, F.E.; Fitch, O.E.; Flammang, B.E.; et al. Future Tail Tales: A Forward-Looking, Integrative Perspective on Tail Research. Integr. Comp. Biol. 2021, 61, 521–537. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Wang, H.; Liu, G.; Wu, M.; Cao, J.; Liu, Z.; Liu, R.; Zhao, F.; Zhang, L.; Lu, J.; et al. Genome-Wide Analysis Reveals Population Structure and Selection in Chinese Indigenous Sheep Breeds. BMC Genom. 2015, 16, 194. [Google Scholar] [CrossRef] [PubMed]
- Moioli, B.; Pilla, F.; Ciani, E. Signatures of Selection Identify Loci Associated with Fat Tail in Sheep. J. Anim. Sci. 2015, 93, 4660–4669. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Gong, M.; Zhang, X.; Wang, F.; Liu, Z.; Zhang, L.; Xu, M.; Zhang, Y.; Dai, X.; Zhang, Z.; et al. The First Sheep Graph-Based Pan-Genome Reveals the Spectrum of Structural Variations and Their Effects on Tail Phenotypes. bioRxiv 2021. [Google Scholar] [CrossRef]
- Lagler, D.K.; Hannemann, E.; Eck, K.; Klawatsch, J.; Seichter, D.; Russ, I.; Mendel, C.; Lühken, G.; Krebs, S.; Blum, H.; et al. Fine-Mapping and Identification of Candidate Causal Genes for Tail Length in the Merinolandschaf Breed. Commun. Biol. 2022, 5, 918. [Google Scholar] [CrossRef]
- Li, X.; He, S.G.; Li, W.R.; Luo, L.Y.; Yan, Z.; Mo, D.X.; Wan, X.; Lv, F.H.; Yang, J.; Xu, Y.X.; et al. Genomic Analyses of Wild Argali, Domestic Sheep, and Their Hybrids Provide Insights into Chromosome Evolution, Phenotypic Variation, and Germplasm Innovation. Genome Res. 2022, 32, 1669–1684. [Google Scholar] [CrossRef]
- Lotterhos, K.E.; Whitlock, M.C. The Relative Power of Genome Scans to Detect Local Adaptation Depends on Sampling Design and Statistical Method. Mol. Ecol. 2015, 24, 1031–1046. [Google Scholar] [CrossRef]
- Lotterhos, K.E.; Card, D.C.; Schaal, S.M.; Wang, L.; Collins, C.; Verity, B. Composite Measures of Selection Can Improve the Signal-to-Noise Ratio in Genome Scans. Methods Ecol. Evol. 2017, 8, 717–727. [Google Scholar] [CrossRef]
- Ma, Y.; Ding, X.; Qanbari, S.; Weigend, S.; Zhang, Q.; Simianer, H. Properties of Different Selection Signature Statistics and a New Strategy for Combining Them. Heredity 2015, 115, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of Samtools and Bcftools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and Vcftools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. Annovar: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Benyamin, B.; McEvoy, B.P.; Gordon, S.; Henders, A.K.; Nyholt, D.R.; Madden, P.A.; Heath, A.C.; Martin, N.G.; Montgomery, G.W.; et al. Common Snps Explain a Large Proportion of the Heritability for Human Height. Nat. Genet. 2010, 42, 565–569. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast Model-Based Estimation of Ancestry in Unrelated Individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Verity, R.; Collins, C.; Card, D.C.; Schaal, S.M.; Wang, L.; Lotterhos, K.E. Minotaur: A Platform for the Analysis and Visualization of Multivariate Results from Genome Scans with R Shiny. Mol. Ecol. Resour. 2017, 17, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Boitard, S.; Boussaha, M.; Capitan, A.; Rocha, D.; Servin, B. Uncovering Adaptation from Sequence Data: Lessons from Genome Resequencing of Four Cattle Breeds. Genetics 2016, 203, 433–450. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical Significance for Genomewide Studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, J.; Shen, M.; Xie, X.L.; Liu, G.J.; Xu, Y.X.; Lv, F.H.; Yang, H.; Yang, Y.L.; Liu, C.B.; et al. Whole-Genome Resequencing of Wild and Domestic Sheep Identifies Genes Associated with Morphological and Agronomic Traits. Nat. Commun. 2020, 11, 2815. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, X.; Zhou, X.; Zhang, Y.; La, Y.; Zhang, Y.; Li, C.; Zhao, Y.; Li, F.; Liu, B.; et al. Deep Genome Resequencing Reveals Artificial and Natural Selection for Visual Deterioration, Plateau Adaptability and High Prolificacy in Chinese Domestic Sheep. Front. Genet. 2019, 10, 300. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Zhang, X.X.; Li, F.D.; Yuan, L.F.; Li, X.L.; Zhang, Y.K.; Zhao, Y.; Zhao, L.M.; Wang, J.H.; Xu, D.; et al. Whole-Genome Resequencing Reveals Molecular Imprints of Anthropogenic and Natural Selection in Wild and Domesticated Sheep. Zool. Res. 2022, 43, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Srinivasula, S.M.; Wang, L.; Talanian, R.V.; Litwack, G.; Fernandes-Alnemri, T.; Alnemri, E.S. Cradd, a Novel Human Apoptotic Adaptor Molecule for Caspase-2, and Fasl/Tumor Necrosis Factor Receptor-Interacting Protein Rip. Cancer Res. 1997, 57, 615–619. [Google Scholar]
- Di Donato, N.; Jean, Y.Y.; Maga, A.M.; Krewson, B.D.; Shupp, A.B.; Avrutsky, M.I.; Roy, A.; Collins, S.; Olds, C.; Willert, R.A.; et al. Mutations in Cradd Result in Reduced Caspase-2-Mediated Neuronal Apoptosis and Cause Megalencephaly with a Rare Lissencephaly Variant. Am. J. Hum. Genet. 2016, 99, 1117–1129. [Google Scholar] [CrossRef]
- Horvat, S.; Medrano, J.F. A 500-Kb Yac and Bac Contig Encompassing the High-Growth Deletion in Mouse Chromosome 10 and Identification of the Murine Raidd/Cradd Gene in the Candidate Region. Genomics 1998, 54, 159–164. [Google Scholar] [CrossRef]
- Famula, T.R.; Calvert, C.C.; Luna, E.; Bradford, G.E. Organ and Skeletal Growth in Mice with a Major Gene for Rapid Postweaning Growth. Growth Dev. Aging 1988, 52, 145–150. [Google Scholar] [PubMed]
- Easa, A.A.; Selionova, M.; Aibazov, M.; Mamontova, T.; Sermyagin, A.; Belous, A.; Abdelmanova, A.; Deniskova, T.; Zinovieva, N. Identification of Genomic Regions and Candidate Genes Associated with Body Weight and Body Conformation Traits in Karachai Goats. Genes 2022, 13, 1773. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Bai, X.; Liu, H.; Zhao, B.; Yan, Z.; Hou, Y.; Chu, Q. Population Genomic Sequencing Delineates Global Landscape of Copy Number Variations That Drive Domestication and Breed Formation of in Chicken. Front. Genet. 2022, 13, 830393. [Google Scholar] [CrossRef] [PubMed]
- Zhi, Y.; Wang, D.; Zhang, K.; Wang, Y.; Geng, W.; Chen, B.; Li, H.; Li, Z.; Tian, Y.; Kang, X.; et al. Genome-Wide Genetic Structure of Henan Indigenous Chicken Breeds. Animals 2023, 13, 753. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Zhao, D.; Bownes, M. Blood Vessel/Epicardial Substance (Bves) Expression, Essential for Embryonic Development, Is Down Regulated by Grk/Efgr Signalling. Int. J. Dev. Biol. 2007, 51, 37–44. [Google Scholar] [CrossRef]
- Osler, M.E.; Smith, T.K.; Bader, D.M. Bves, a Member of the Popeye Domain-Containing Gene Family. Dev. Dyn. 2006, 235, 586–593. [Google Scholar] [CrossRef]
- Beecher, G.; Tang, C.; Liewluck, T. Severe Adolescent-Onset Limb-Girdle Muscular Dystrophy Due to a Novel Homozygous Nonsense Bves Variant. J. Neurol. Sci. 2021, 420, 117259. [Google Scholar] [CrossRef]
- Graf, R.; Munschauer, M.; Mastrobuoni, G.; Mayr, F.; Heinemann, U.; Kempa, S.; Rajewsky, N.; Landthaler, M. Identification of Lin28b-Bound Mrnas Reveals Features of Target Recognition and Regulation. RNA Biol. 2013, 10, 1146–1159. [Google Scholar] [CrossRef]
- Lettre, G.; Jackson, A.U.; Gieger, C.; Schumacher, F.R.; Berndt, S.I.; Sanna, S.; Eyheramendy, S.; Voight, B.F.; Butler, J.L.; Guiducci, C.; et al. Identification of Ten Loci Associated with Height Highlights New Biological Pathways in Human Growth. Nat. Genet. 2008, 40, 584–591. [Google Scholar] [CrossRef]
- Widén, E.; Ripatti, S.; Cousminer, D.L.; Surakka, I.; Lappalainen, T.; Järvelin, M.R.; Eriksson, J.G.; Raitakari, O.; Salomaa, V.; Sovio, U.; et al. Distinct Variants at Lin28b Influence Growth in Height from Birth to Adulthood. Am. J. Hum. Genet. 2010, 86, 773–782. [Google Scholar] [CrossRef]
- Cousminer, D.L.; Berry, D.J.; Timpson, N.J.; Ang, W.; Thiering, E.; Byrne, E.M.; Taal, H.R.; Huikari, V.; Bradfield, J.P.; Kerkhof, M.; et al. Genome-Wide Association and Longitudinal Analyses Reveal Genetic Loci Linking Pubertal Height Growth, Pubertal Timing and Childhood Adiposity. Hum. Mol. Genet. 2013, 22, 2735–2747. [Google Scholar] [CrossRef] [PubMed]
- Sinha, T.; Lin, L.; Li, D.; Davis, J.; Evans, S.; Wynshaw-Boris, A.; Wang, J. Mapping the Dynamic Expression of Wnt11 and the Lineage Contribution of Wnt11-Expressing Cells During Early Mouse Development. Dev. Biol. 2015, 398, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Rankin, S.A.; Sinner, D.; Kenny, A.P.; Krieg, P.A.; Zorn, A.M. Sfrp5 Coordinates Foregut Specification and Morphogenesis by Antagonizing Both Canonical and Noncanonical Wnt11 Signaling. Genes Dev. 2008, 22, 3050–3063. [Google Scholar] [CrossRef] [PubMed]
- Caetano da Silva, C.; Edouard, T.; Fradin, M.; Aubert-Mucca, M.; Ricquebourg, M.; Raman, R.; Salles, J.P.; Charon, V.; Guggenbuhl, P.; Muller, M.; et al. Wnt11, a New Gene Associated with Early Onset Osteoporosis, Is Required for Osteoblastogenesis. Hum. Mol. Genet. 2022, 31, 1622–1634. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y. The Methionine Sulfoxide Reduction System: Selenium Utilization and Methionine Sulfoxide Reductase Enzymes and Their Functions. Antioxid. Redox Signal. 2013, 19, 958–969. [Google Scholar] [CrossRef]
- Ahmed, Z.M.; Yousaf, R.; Lee, B.C.; Khan, S.N.; Lee, S.; Lee, K.; Husnain, T.; Rehman, A.U.; Bonneux, S.; Ansar, M.; et al. Functional Null Mutations of Msrb3 Encoding Methionine Sulfoxide Reductase Are Associated with Human Deafness Dfnb74. Am. J. Hum. Genet. 2011, 88, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Vaysse, A.; Ratnakumar, A.; Derrien, T.; Axelsson, E.; Rosengren Pielberg, G.; Sigurdsson, S.; Fall, T.; Seppälä, E.H.; Hansen, M.S.; Lawley, C.T.; et al. Identification of Genomic Regions Associated with Phenotypic Variation between Dog Breeds Using Selection Mapping. PLoS Genet. 2011, 7, e1002316. [Google Scholar] [CrossRef]
- Boyko, A.R.; Quignon, P.; Li, L.; Schoenebeck, J.J.; Degenhardt, J.D.; Lohmueller, K.E.; Zhao, K.; Brisbin, A.; Parker, H.G.; vonHoldt, B.M.; et al. A Simple Genetic Architecture Underlies Morphological Variation in Dogs. PLoS Biol. 2010, 8, e1000451. [Google Scholar] [CrossRef]
- Chen, C.; Liu, C.; Xiong, X.; Fang, S.; Yang, H.; Zhang, Z.; Ren, J.; Guo, Y.; Huang, L. Copy Number Variation in the Msrb3 Gene Enlarges Porcine Ear Size through a Mechanism Involving Mir-584-5p. Genet. Sel. Evol. 2018, 50, 72. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, J.; Zhang, L.; Wang, L.; Liu, X.; Yan, H.; Zhao, K.; Shi, H.; Zhang, T.; Li, N.; et al. Porcine Methionine Sulfoxide Reductase B3: Molecular Cloning, Tissue-Specific Expression Profiles, and Polymorphisms Associated with Ear Size in Sus Scrofa. J. Anim. Sci. Biotechnol. 2015, 6, 60. [Google Scholar] [CrossRef]
- Saatchi, M.; Schnabel, R.D.; Taylor, J.F.; Garrick, D.J. Large-Effect Pleiotropic or Closely Linked Qtl Segregate within and across Ten Us Cattle Breeds. BMC Genom. 2014, 15, 442. [Google Scholar] [CrossRef]
- Zapata, I.; Serpell, J.A.; Alvarez, C.E. Genetic Mapping of Canine Fear and Aggression. BMC Genom. 2016, 17, 572. [Google Scholar] [CrossRef]
- Kader, A.; Li, Y.; Dong, K.; Irwin, D.M.; Zhao, Q.; He, X.; Liu, J.; Pu, Y.; Gorkhali, N.A.; Liu, X.; et al. Population Variation Reveals Independent Selection toward Small Body Size in Chinese Debao Pony. Genome Biol. Evol. 2015, 8, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Zhang, X.; Collins, B.; Sper, R.B.; Gleason, K.; Simpson, S.; Koh, S.; Sommer, J.; Flowers, W.L.; Petters, R.M.; et al. High Mobility Group A2 (Hmga2) Deficiency in Pigs Leads to Dwarfism, Abnormal Fetal Resource Allocation, and Cryptorchidism. Proc. Natl. Acad. Sci. USA 2018, 115, 5420–5425. [Google Scholar] [CrossRef] [PubMed]
- Norton, E.M.; Avila, F.; Schultz, N.E.; Mickelson, J.R.; Geor, R.J.; McCue, M.E. Evaluation of an Hmga2 Variant for Pleiotropic Effects on Height and Metabolic Traits in Ponies. J. Vet. Intern. Med. 2019, 33, 942–952. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, A.C.; Daetwyler, H.D.; Chamberlain, A.J.; Ponce, C.H.; Sargolzaei, M.; Schenkel, F.S.; Sahana, G.; Govignon-Gion, A.; Boitard, S.; Dolezal, M.; et al. Meta-Analysis of Genome-Wide Association Studies for Cattle Stature Identifies Common Genes That Regulate Body Size in Mammals. Nat. Genet. 2018, 50, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Gianferante, D.M.; Rotunno, M.; Dean, M.; Zhou, W.; Hicks, B.D.; Wyatt, K.; Jones, K.; Wang, M.; Zhu, B.; Goldstein, A.M.; et al. Whole-Exome Sequencing of Nevoid Basal Cell Carcinoma Syndrome Families and Review of Human Gene Mutation Database Ptch1 Mutation Data. Mol. Genet. Genom. Med. 2018, 6, 1168–1180. [Google Scholar] [CrossRef] [PubMed]
- Derwińska, K.; Smyk, M.; Cooper, M.L.; Bader, P.; Cheung, S.W.; Stankiewicz, P. Ptch1 Duplication in a Family with Microcephaly and Mild Developmental Delay. Eur. J. Hum. Genet. 2009, 17, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.S.; Gao, L.; Shen, M.; Lyu, F. Whole-Genome Selective Scans Detect Genes Associated with Important Phenotypic Traits in Sheep (Ovis Aries). Front. Genet. 2021, 12, 738879. [Google Scholar] [CrossRef] [PubMed]
- Clop, A.; Marcq, F.; Takeda, H.; Pirottin, D.; Tordoir, X.; Bibé, B.; Bouix, J.; Caiment, F.; Elsen, J.M.; Eychenne, F.; et al. A Mutation Creating a Potential Illegitimate Microrna Target Site in the Myostatin Gene Affects Muscularity in Sheep. Nat. Genet. 2006, 38, 813–818. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lee, S.J. Double Muscling in Cattle Due to Mutations in the Myostatin Gene. Proc. Natl. Acad. Sci. USA 1997, 94, 12457–12461. [Google Scholar] [CrossRef]
- Löbbert, R.W.; Winterpacht, A.; Seipel, B.; Zabel, B.U. Molecular Cloning and Chromosomal Assignment of the Human Homologue of the Rat Cgmp-Inhibited Phosphodiesterase 1 (Pde3a)—A Gene Involved in Fat Metabolism Located at 11p15.1. Genomics 1996, 37, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Yin, Y.; Xu, K.; Peng, Y.; Zhang, J. Knockdown of Lgals12 Inhibits Porcine Adipocyte Adipogenesis Via Pka-Erk1/2 Signaling Pathway. Acta Biochim. Biophys. Sin. 2018, 50, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Foley, A.R.; Zou, Y.; Dunford, J.E.; Rooney, J.; Chandra, G.; Xiong, H.; Straub, V.; Voit, T.; Romero, N.; Donkervoort, S.; et al. Ggps1 Mutations Cause Muscular Dystrophy/Hearing Loss/Ovarian Insufficiency Syndrome. Ann. Neurol. 2020, 88, 332–347. [Google Scholar] [CrossRef] [PubMed]
- Kaiyrzhanov, R.; Perry, L.; Rocca, C.; Zaki, M.S.; Hosny, H.; Araujo Martins Moreno, C.; Phadke, R.; Zaharieva, I.; Camelo Gontijo, C.; Beetz, C.; et al. Ggps1-Associated Muscular Dystrophy with and without Hearing Loss. Ann. Clin. Transl. Neurol. 2022, 9, 1465–1474. [Google Scholar] [CrossRef] [PubMed]
- Bertolio, R.; Napoletano, F.; Mano, M.; Maurer-Stroh, S.; Fantuz, M.; Zannini, A.; Bicciato, S.; Sorrentino, G.; Del Sal, G. Sterol Regulatory Element Binding Protein 1 Couples Mechanical Cues and Lipid Metabolism. Nat. Commun. 2019, 10, 1326. [Google Scholar] [CrossRef] [PubMed]
- Moreira, G.C.; Godoy, T.F.; Boschiero, C.; Gheyas, A.; Gasparin, G.; Andrade, S.C.; Paduan, M.; Montenegro, H.; Burt, D.W.; Ledur, M.C.; et al. Variant Discovery in a Qtl Region on Chromosome 3 Associated with Fatness in Chickens. Anim. Genet. 2015, 46, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Goyache, F.; Fernández, I.; Tapsoba, A.S.R.; Traoré, A.; Menéndez-Arias, N.A.; Álvarez, I. Functional Characterization of Copy Number Variations Regions in Djallonké Sheep. J. Anim. Breed. Genet. 2021, 138, 600–612. [Google Scholar] [CrossRef]
- Xu, L.; Bickhart, D.M.; Cole, J.B.; Schroeder, S.G.; Song, J.; Tassell, C.P.; Sonstegard, T.S.; Liu, G.E. Genomic Signatures Reveal New Evidences for Selection of Important Traits in Domestic Cattle. Mol. Biol. Evol. 2015, 32, 711–725. [Google Scholar] [CrossRef]
- Norris, B.J.; Whan, V.A. A Gene Duplication Affecting Expression of the Ovine Asip Gene Is Responsible for White and Black Sheep. Genome Res. 2008, 18, 1282–1293. [Google Scholar] [CrossRef]
- Gong, G.; Fan, Y.; Li, W.; Yan, X.; Yan, X.; Zhang, L.; Wang, N.; Chen, O.; Zhang, Y.; Wang, R.; et al. Identification of the Key Genes Associated with Different Hair Types in the Inner Mongolia Cashmere Goat. Animals 2022, 12, 1456. [Google Scholar] [CrossRef]
- Demars, J.; Cano, M.; Drouilhet, L.; Plisson-Petit, F.; Bardou, P.; Fabre, S.; Servin, B.; Sarry, J.; Woloszyn, F.; Mulsant, P.; et al. Genome-Wide Identification of the Mutation Underlying Fleece Variation and Discriminating Ancestral Hairy Species from Modern Woolly Sheep. Mol. Biol. Evol. 2017, 34, 1722–1729. [Google Scholar] [CrossRef]
- Shimomura, Y.; Wajid, M.; Petukhova, L.; Kurban, M.; Christiano, A.M. Autosomal-Dominant Woolly Hair Resulting from Disruption of Keratin 74 (Krt74), a Potential Determinant of Human Hair Texture. Am. J. Hum. Genet. 2010, 86, 632–638. [Google Scholar] [CrossRef]
- Cain, B.; Gebelein, B. Mechanisms Underlying Hox-Mediated Transcriptional Outcomes. Front. Cell Dev. Biol. 2021, 9, 787339. [Google Scholar] [CrossRef]
- Economides, K.D.; Zeltser, L.; Capecchi, M.R. Hoxb13 Mutations Cause Overgrowth of Caudal Spinal Cordand Tail Vertebrae. Dev. Biol. 2003, 256, 317–330. [Google Scholar] [CrossRef]
- Mallo, M. Reassessing the Role of Hox Genes During Vertebrate Development and Evolution. Trends Genet. 2018, 34, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Aires, R.; de Lemos, L.; Nóvoa, A.; Jurberg, A.D.; Mascrez, B.; Duboule, D.; Mallo, M. Tail Bud Progenitor Activity Relies on a Network Comprising Gdf11, Lin28, and Hox13 Genes. Dev. Cell 2019, 48, 383–395.e8. [Google Scholar] [CrossRef] [PubMed]
- Ahbara, A.M.; Musa, H.H.; Robert, C.; Abebe, A.; Al-Jumaili, A.S.; Kebede, A.; Latairish, S.; Agoub, M.O.; Clark, E.; Hanotte, O.; et al. Natural Adaptation and Human Selection of Northeast African Sheep Genomes. Genomics 2022, 114, 110448. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Yang, S.L.; Herrlinger, S.; Liang, C.; Dzieciatkowska, M.; Hansen, K.C.; Desai, R.; Nagy, A.; Niswander, L.; Moss, E.G.; et al. Lin28 Promotes the Proliferative Capacity of Neural Progenitor Cells in Brain Development. Development 2015, 142, 1616–1627. [Google Scholar] [CrossRef] [PubMed]
- Robinton, D.A.; Chal, J.; Lummertz da Rocha, E.; Han, A.; Yermalovich, A.V.; Oginuma, M.; Schlaeger, T.M.; Sousa, P.; Rodriguez, A.; Urbach, A.; et al. The Lin28/Let-7 Pathway Regulates the Mammalian Caudal Body Axis Elongation Program. Dev. Cell 2019, 48, 396–405.e393. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Kataoka, K.; Ito, Y.; Yokoyama, S.; Inui, M.; Mori, M.; Takahashi, S.; Akita, K.; Takada, S.; Ueno-Kudoh, H.; et al. Lin28a/Let-7 Pathway Modulates the Hox Code Via Polycomb Regulation During Axial Patterning in Vertebrates. eLife 2020, 9, e53608. [Google Scholar] [CrossRef]
- Viswanathan, S.R.; Daley, G.Q. Lin28: A Microrna Regulator with a Macro Role. Cell 2010, 140, 445–449. [Google Scholar] [CrossRef]
- Epstein, J.A. Pax3 and Vertebrate Development. Methods Mol. Biol. 2000, 137, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Orvis, J.; Stolfi, A. Pax3/7 Regulates Neural Tube Closure and Patterning in a Non-Vertebrate Chordate. Front. Cell Dev. Biol. 2022, 10, 999511. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Gan, Q.; Stokes, A.; Lassiter, R.N.; Wang, Y.; Chan, J.; Han, J.X.; Pleasure, D.E.; Epstein, J.A.; Zhou, C.J. Β-Catenin Regulates Pax3 and Cdx2 for Caudal Neural Tube Closure and Elongation. Development 2014, 141, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Fatima, L.A.; Campello, R.S.; Santos, R.S.; Freitas, H.S.; Frank, A.P.; Machado, U.F.; Clegg, D.J. Estrogen Receptor 1 (Esr1) Regulates Vegfa in Adipose Tissue. Sci. Rep. 2017, 7, 16716. [Google Scholar] [CrossRef] [PubMed]
- Yonekura, S.; Tokutake, Y.; Hirota, S.; Rose, M.T.; Katoh, K.; Aso, H. Proliferating Bovine Intramuscular Preadipocyte Cells Synthesize Leptin. Domest. Anim. Endocrinol. 2013, 45, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; van de Wall, E.; Laplante, M.; Azzara, A.; Trujillo, M.E.; Hofmann, S.M.; Schraw, T.; Durand, J.L.; Li, H.; Li, G.; et al. Obesity-Associated Improvements in Metabolic Profile through Expansion of Adipose Tissue. J. Clin. Investig. 2007, 117, 2621–2637. [Google Scholar] [CrossRef]

| Breed | Abbr. | Sample Size | Category | Tail Type |
|---|---|---|---|---|
| Hu sheep | HUS | 3 | Chinese | Short fat tail |
| Sunit sheep | SNT | 5 | Chinese | Short fat tail |
| Ujimqin sheep | UJI | 4 | Chinese | Short fat tail |
| Taihang fur sheep | THF | 4 | Chinese | Short fat tail |
| Small-tailed Han sheep | STH | 5 | Chinese | Short fat tail |
| Tan sheep | TAN | 5 | Chinese | Long fat tail |
| Tong sheep | TON | 3 | Chinese | Long fat tail |
| Large-tailed Han sheep | LTH | 4 | Chinese | Long fat tail |
| Guangling Large-tailed sheep | GLT | 4 | Chinese | Long fat tail |
| Lanzhou Large-tailed sheep | LLT | 2 | Chinese | Long fat tail |
| Dorper sheep | AWD | 3 | Western | Thin tail |
| Texel sheep | TEX | 5 | Western | Thin tail |
| Hornless Poll Dorset sheep | DOP | 2 | Western | Thin tail |
| Australian Merino sheep | AME | 2 | Western | Thin tail |
| German Mutton Merino sheep | GME | 3 | Western | Thin tail |
| Catalogue | SNP Numbers |
|---|---|
| Upstream | 87,385 |
| Stop gain (Exonic) | 649 |
| Stop loss (Exonic) | 178 |
| Synonymous (Exonic) | 63,547 |
| Non-synonymous (Exonic) | 75,476 |
| Intronic | 7,633,513 |
| Splicing | 3867 |
| Downstream | 117,367 |
| Upstream/downstream | 2363 |
| Intergenic | 12,638,742 |
| ts | 13,684,673 |
| tv | 7,141,784 |
| ts/tv | 1.916 |
| Total | 20,826,457 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, T.; Jin, M.; Wang, H.; Zhang, W.; Yuan, Z.; Wei, C. Whole-Genome Scanning for Selection Signatures Reveals Candidate Genes Associated with Growth and Tail Length in Sheep. Animals 2024, 14, 687. https://doi.org/10.3390/ani14050687
Li T, Jin M, Wang H, Zhang W, Yuan Z, Wei C. Whole-Genome Scanning for Selection Signatures Reveals Candidate Genes Associated with Growth and Tail Length in Sheep. Animals. 2024; 14(5):687. https://doi.org/10.3390/ani14050687
Chicago/Turabian StyleLi, Taotao, Meilin Jin, Huihua Wang, Wentao Zhang, Zehu Yuan, and Caihong Wei. 2024. "Whole-Genome Scanning for Selection Signatures Reveals Candidate Genes Associated with Growth and Tail Length in Sheep" Animals 14, no. 5: 687. https://doi.org/10.3390/ani14050687
APA StyleLi, T., Jin, M., Wang, H., Zhang, W., Yuan, Z., & Wei, C. (2024). Whole-Genome Scanning for Selection Signatures Reveals Candidate Genes Associated with Growth and Tail Length in Sheep. Animals, 14(5), 687. https://doi.org/10.3390/ani14050687






