Exploring the Effects of Robertsonian Translocation 1/29 (Rob (1;29)) on Genetic Diversity in Minor Breeds of Spanish Berrenda Cattle via Genome-Wide Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
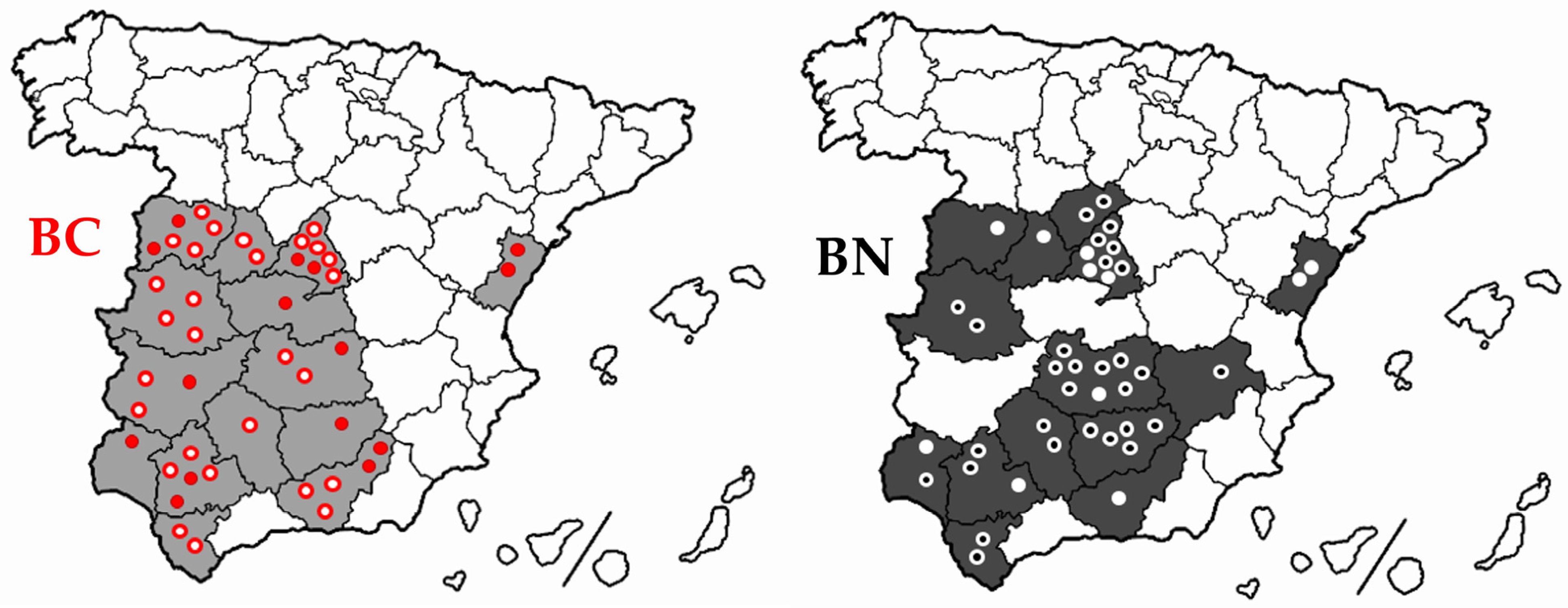
2.1. Sampling for the Identification of the Robertsonian Translocation Rob (1;29)
2.2. Chromosomal Analyses
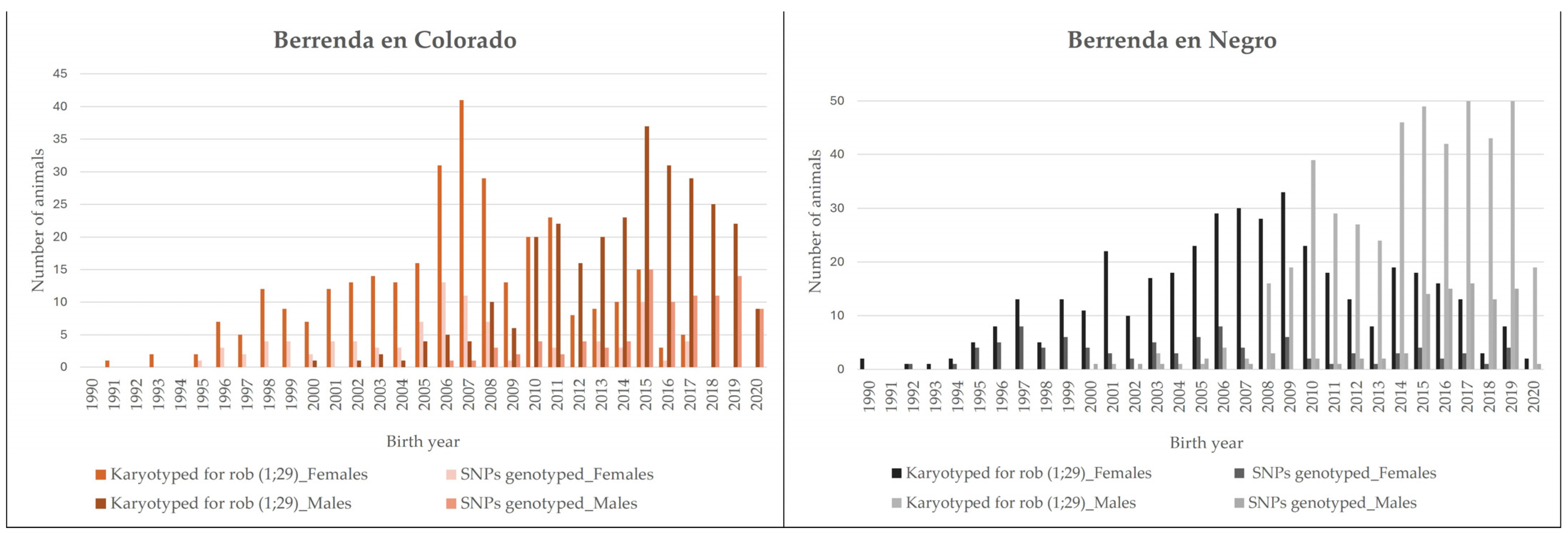
2.3. Animals Tested for Genetic Diversity Studies via SNP Genotyping
2.4. SNP Genotyping and Quality Control
2.5. Genetic Diversity Estimation and Population Structure
3. Results and Discussion
3.1. Genetic Diversity Analyses
3.2. Analyses of Inbreeding and Runs of Homozygosity (ROH)
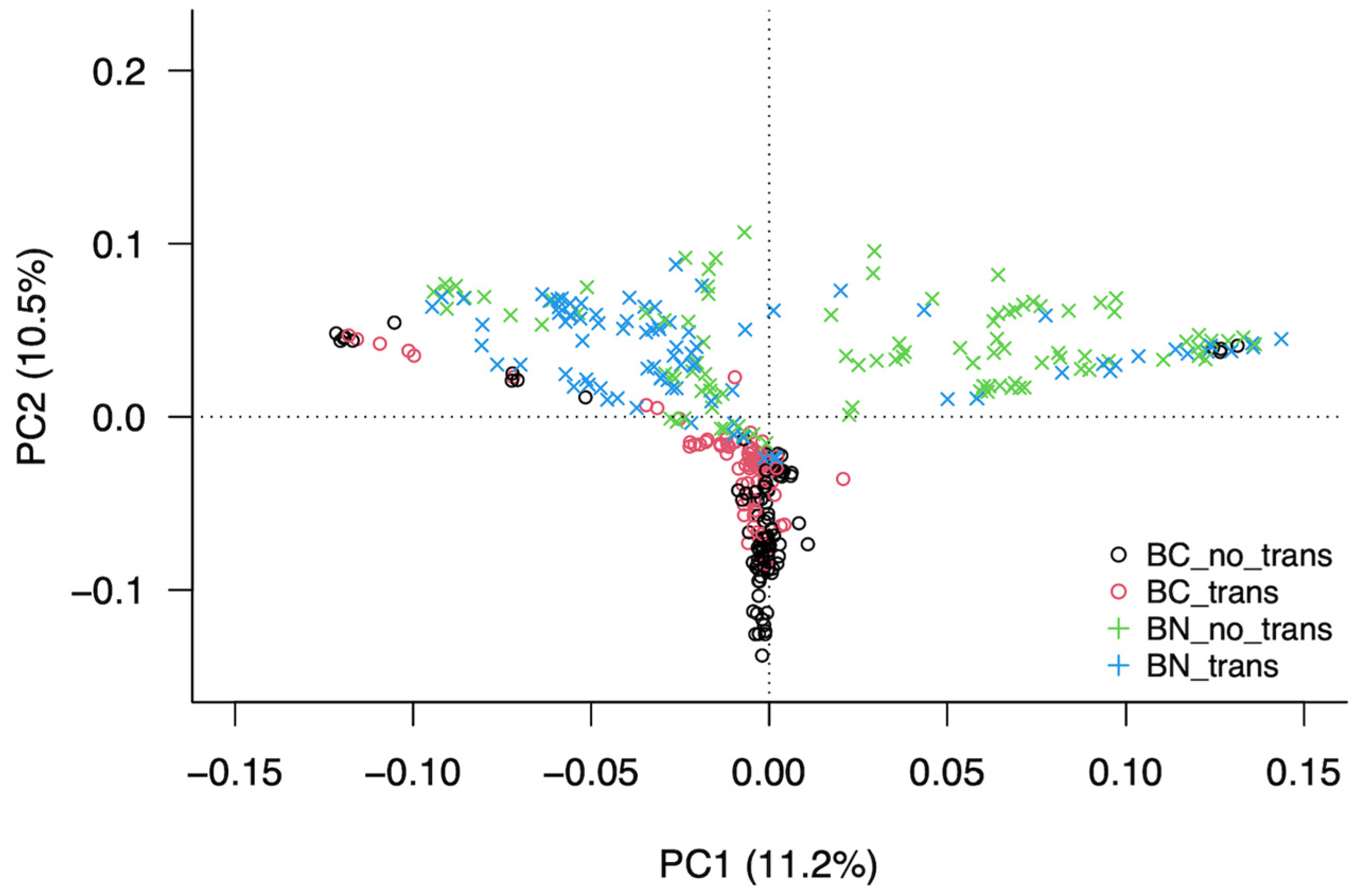
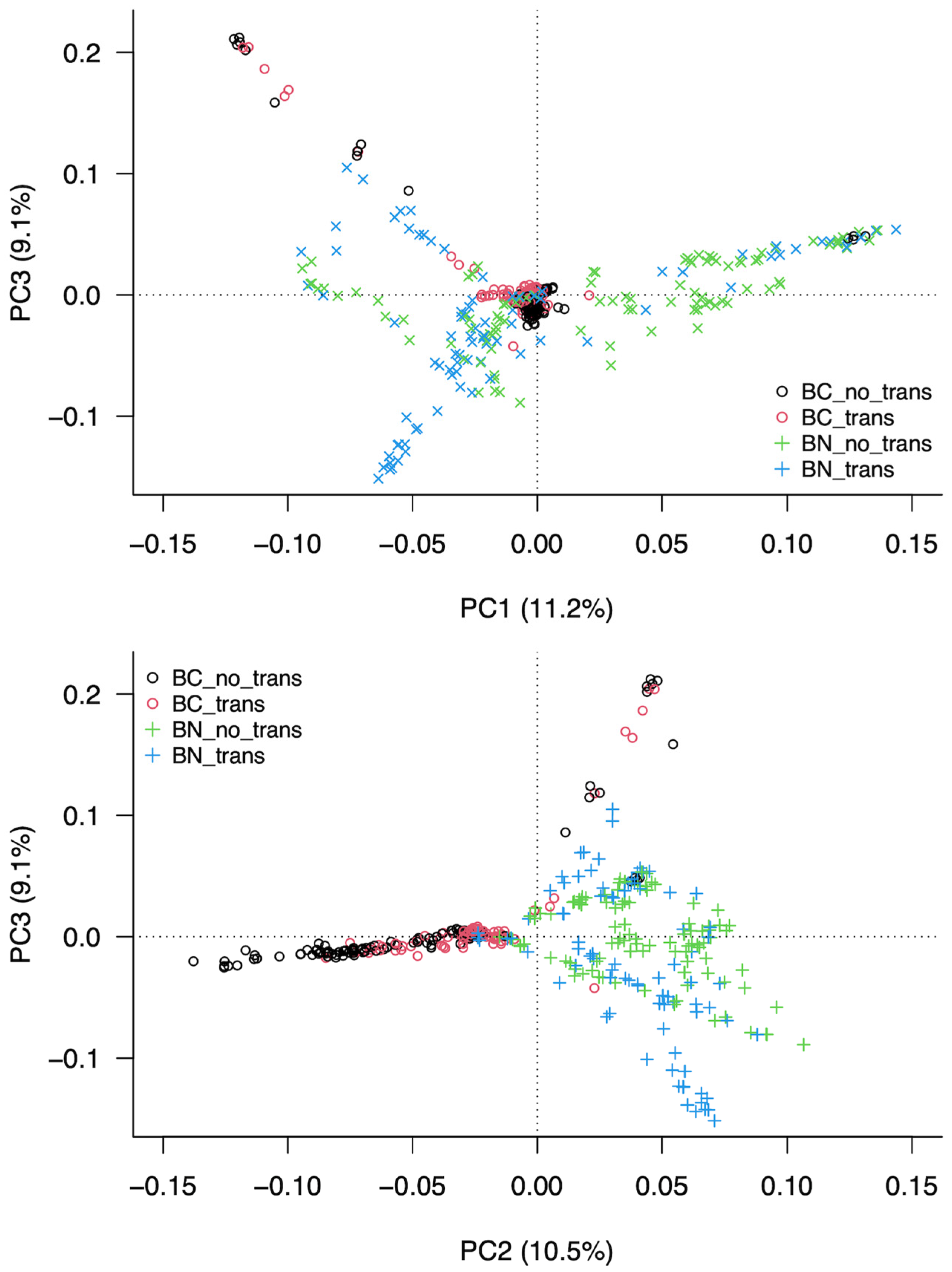
3.3. Analysis of the Structure of the Population
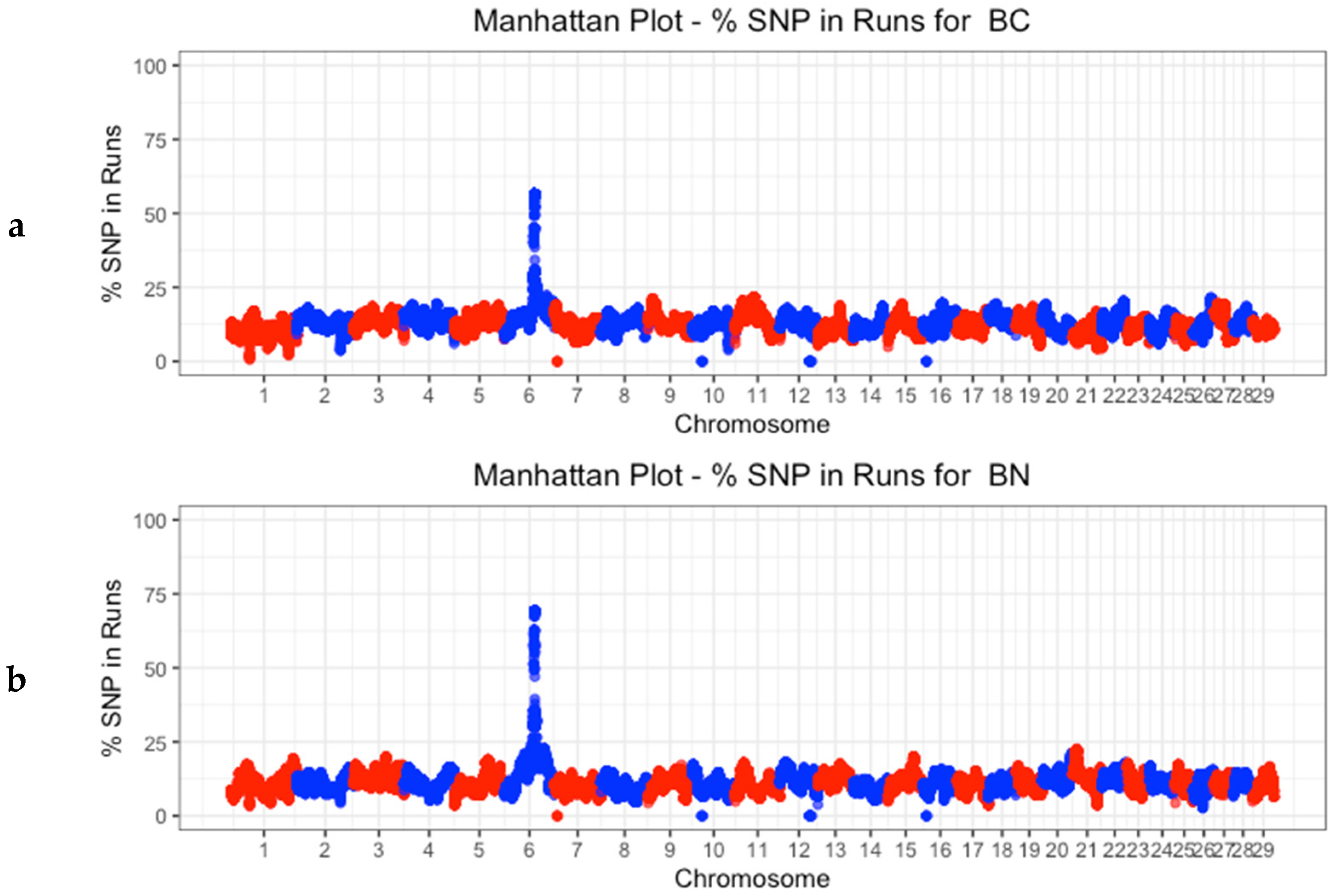
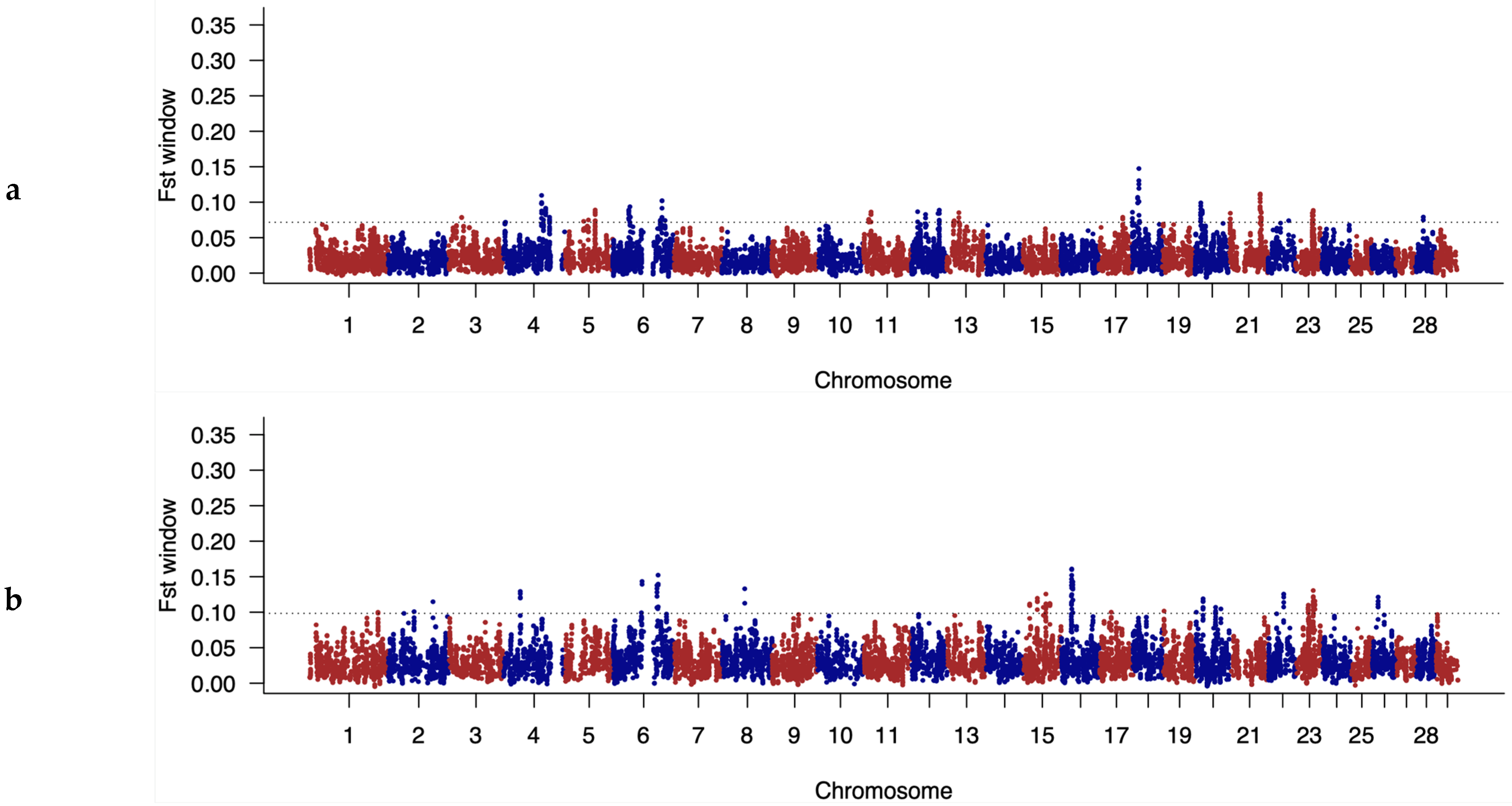
3.4. Identification of Discriminatory Regions between Populations along the Genome
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Real Decreto 45/2019, de 8 de Febrero, por el que se Establecen las Normas Zootécnicas Aplicables a los Animales Reproductores de raza pura, Porcinos Reproductores híbridos y su Material Reproductivo, se Actualiza el Programa Nacional de Conservación, Mejora y Fomento de las razas Ganaderas y se Modifican los Reales Decretos 558/2001, de 25 de mayo; 1316/1992, de 30 de Octubre; 1438/1992, de 27 de Noviembre; y 1625/2011, de 14 de Noviembre. Boletín Oficial del Estado, núm. 52, de 01/03/2019. Available online: https://www.boe.es/eli/es/rd/2019/02/08/45/con (accessed on 12 January 2024).
- Spanish Breed Information System of the Ministry of Agriculture, Fisheries and Food (ARCA, by Its Initials in Spanish). 2023. Available online: https://www.mapa.gob.es/es/ganaderia/temas/zootecnia/razas-ganaderas/razas/catalogo-razas/bovino/berrenda-Colorado/default.aspx (accessed on 12 January 2024).
- González-Cano, R.; González-Martínez, A.; Muñoz-Mejías, M.E.; Valera, P.; Rodero, E. Analyses of Genetic Diversity in the Endangered Berrenda Spanish Cattle Breeds Using Pedigree Data. Animals 2022, 12, 249. [Google Scholar] [CrossRef]
- Rodero Serrano, E.; González, A.; Luque, A. Las razas bovinas andaluzas de protección especial: Berrendo en Negro, Berrendo en Colorado, Cárdena Andaluza, Negra andaluza de las Campiñas, Pajuna y Marismeña. In Patrimonio Ganadero Andaluz: Las Razas Ganaderas de Andalucía (Volumen II); Consejería de Agricultura y Pesca. Junta de Andalucía: Sevilla, Spain, 2007; pp. 53–120. [Google Scholar]
- Ferraz-de-Oliveira, M.I.; Azeda, C.; Pinto-Correia, T. Management of Montados and Dehesas for High Nature Value: An interdisciplinary pathway. Agrofor. Syst. 2016, 90, 1–6. [Google Scholar] [CrossRef]
- Cañón, J.; García, D.; Delgado, J.V.; Dunner, S.; Da Gama, L.T.; Landi, V.; Martin-Burriel, I.; Martinez, A.; Penedo, C.; Rodellar, C.; et al. Relative breed contributions to neutral genetic diversity of a comprehensive representation of Iberian native cattle. Animal 2011, 5, 1323–1334. [Google Scholar] [CrossRef]
- Gustavsson, I.; Rockborn, G. Chromosome abnormality in three cases of lymphatic leukaemia in cattle. Nature 1964, 203, 990. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, I. Cytogenetics, distribution and phenotypic effects of a translocation in Swedish cattle. Hereditas 1969, 63, 68–169. [Google Scholar] [CrossRef] [PubMed]
- Popescu, C.P.; Pech, A. Une bibliographie sur la translocation 1/29 de bovins dans le monde (1964–1990). Ann. Zootech. 1991, 40, 271–305. [Google Scholar] [CrossRef]
- Switonski, M. 50 years of studies on bovine 1/29 Robertsonian translocation-from Giemsa staining to genomic analysis. Chromosome Res. 2014, 22, 395. [Google Scholar]
- Holečková, B.; Schwarzbacherová, V.; Galdíková, M.; Koleničová, S.; Halušková, J.; Staničová, J.; Jutková, A. Chromosomal Aberrations in Cattle. Genes 2021, 12, 1330. [Google Scholar] [CrossRef]
- Jiménez, J.M.; Morales, R.; Molina, A.; Moreno-Millán, M.; Demyda-Peyrás, S. Effect of the rob (1;29) translocation on the fertility of beef cattle reared under extensive conditions: A 30-year retrospective study. Reprod. Domest. Anim. 2022, 57, 349–356. [Google Scholar] [CrossRef]
- Rodero-Serrano, E.; Demyda-Peyrás, S.; González-Martinez, A.; Rodero-Franganillo, A.; Moreno-Millán, M. The rob (1;29) chromosome translocation in endangered Andalusian cattle breeds. Livest. Sci. 2013, 158, 32–39. [Google Scholar] [CrossRef]
- Refsdal, A.O. Fertiliteten hos ketosekyr. (Fertility of ketotic cows). Norsk Vet. Tidsskr. 1977, 89, 219–221. [Google Scholar]
- Popescu, C.P.; Tixier, M. The frequency of chromosome abnormalities in farm animals and their economic consequences. Ann. Genet. 1984, 27, 69–72. [Google Scholar]
- ANABE. Agrupación Nacional de Asociaciones de Criadores de Raza Berrenda en Negro y Berrenda en Colorado. Available online: https://anabe.webgescan.com/ (accessed on 12 January 2024).
- Rodero Serrano, E.; González Cano, R.; Moreno Millán, M.; Muñoz-Mejías, E.; Valera, P.; González, A.; González, R.; González, A.; Muñoz, E.; Valera, P.; et al. Evolución de la translocación Robertsoniana t 1;29 en las razas bovinas Berrendas. In Proceedings of the XII Congreso de la Federación Iberoamericana de Razas Criollas y Autóctonas, Veracruz, Mexico, 9–12 October 2019. [Google Scholar]
- FAO. Surveying and Monitoring of Animal Genetic Resources. FAO Animal Production and Health Guidelines. No. 7. Rome. 2011. Available online: https://www.fao.org/3/ba0055e/ba0055e00.htm (accessed on 12 January 2024).
- Cortés, O.; Eusebi, P.; Dunner, S.; Sevane, N.; Cañón, J. Comparison of diversity parameters from SNP, microsatellites and pedigree records in the Lidia cattle breed. Livest. Sci. 2019, 219, 80–85. [Google Scholar] [CrossRef]
- Álvarez, I.; Fernández, I.; Traoré, A.; Menéndez-Arias, N.A.; Goyache, F. Population structure assessed using microsatellite and SNP data: An empirical comparison in West African cattle. Animals 2021, 11, 151. [Google Scholar] [CrossRef]
- Ajmone-Marsan, P.; Boettcher, P.J.; Ginja, C.; Kantanen, J.; Lenstra, J.A. Genomic Characterization of Animal Genetic Resources: Practical Guide; Food & Agriculture Org.: Rome, Italy, 2023.
- Santure, A.W.; Stapley, J.; Ball, A.D.; Birkhead, T.R.; Burke, T.; Slate, J.O.N. On the use of large marker panels to estimate inbreeding and relatedness: Empirical and simulation studies of a pedigreed zebra finch population typed at 771 SNPs. Mol. Ecol. 2010, 19, 1439–1451. [Google Scholar] [CrossRef]
- Sun, C.; VanRaden, P.M.; O’connell, J.R.; Weigel, K.A.; Gianola, D. Mating programs including genomic relationships and dominance effects. J. Dairy Sci. 2013, 96, 8014–8023. [Google Scholar] [CrossRef] [PubMed]
- Ruiter, J.; Plá, J.L.V.; Franganillo, A.R.; Zamorano, M.J. Análisis genético de marcadores microsatélites en dos poblaciones de la raza bovina Berrenda en Negro. Arch. Zootec. 1998, 47, 195–200. [Google Scholar]
- González, R.; González, A.; Muñoz, E.; Valera, P.; Rodero, E. Evolución de la variabilidad genética de las dos razas bovinas berrendas autóctonas españolas (Periodo 2010–2018). In Proceedings of the XII Congreso de la Federación Iberoamericana de Razas Criollas y Autóctonas, Veracruz, Mexico, 9–12 October 2019. [Google Scholar]
- Iannuzzi, A.; Pistucci, R.; Perucatti, A.; Zannotti, M.; Iannuzzi, L.; Parma, P. Characterization of Robertsonian and Reciprocal Translocations in Cattle through NGS. Animals 2023, 13, 3018. [Google Scholar] [CrossRef]
- de Grouchy, J.R. Deletion partielle des bars longs du chromosome 18. Path. Biol. 1964, 12, 579–582. [Google Scholar]
- Iannuzzi, L. G-and R-banded prometaphase karyotypes in cattle (Bos taurus L.). Chromosome Res. 1996, 4, 448–456. [Google Scholar] [CrossRef]
- Purcell, S.; Chang, C. PLINK 1.9. 2015. Available online: https://www.cog-genomics.org/plink/1.9/ (accessed on 12 January 2024).
- Biscarini, F.; Cozzi, P.; Gaspa, G.; Marras, G. DetectRUNS: An R Package to Detect Runs of Homozygosity and Heterozygosity in Diploid Genomes; CRAN (The Comprehensive R Archive Network): Windhoek, Namibia, 2018. [Google Scholar]
- Chitneedi, P.K.; Arranz, J.J.; Suarez-Vega, A.; García-Gámez, E.; Gutiérrez-Gil, B. Estimations of linkage disequilibrium, effective population size and ROH-based inbreeding coefficients in Spanish Churra sheep using imputed high-density SNP genotypes. Anim. Genet. 2017, 48, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ramilo, S.T.; Elsen, J.M.; Legarra, A. Inbreeding and effective population size in French dairy sheep: Comparison between genomic and pedigree estimates. J. Dairy Sci. 2019, 102, 4227–4237. [Google Scholar] [CrossRef]
- Mastrangelo, S.; Sardina, M.T.; Tolone, M.; Di Gerlando, R.; Sutera, A.M.; Fontanesi, L.; Portolano, B. Genome-wide identification of runs of homozygosity islands and associated genes in local dairy cattle breeds. Animal 2018, 12, 2480–2488. [Google Scholar] [CrossRef]
- Michailidou, S.; Tsangaris, G.; Fthenakis, G.C.; Tzora, A.; Skoufos, I.; Karkabounas, S.C.; Banos, G.; Argiriou, A.; Arsenos, G. Genomic diversity and population structure of three autochthonous Greek sheep breeds assessed with genome-wide DNA arrays. Mol. Genet. Genom. 2018, 293, 753–768. [Google Scholar] [CrossRef]
- Granado-Tajada, I.; Rodríguez-Ramilo, S.T.; Legarra, A.; Ugarte, E. Inbreeding, effective population size, and coancestry in the Latxa dairy sheep breed. J. Dairy Sci. 2020, 103, 5215–5226. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Kyselová, J.; Gurgul, A.; Jasielczuk, I.; Bugno-Poniewierska, M.; Mátlová, V.; Pikousová, J.; Musková, M.; Jochová, K.; Tichý, L.; Sztankóová, Z.; et al. Investigation of genetic diversity and selection signatures in Czech cattle genetic resources revealed by genome-wide analysis. Livest. Sci. 2022, 256, 104825. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Schmidtmann, C.; Schönherz, A.; Guldbrandtsen, B.; Marjanovic, J.; Calus, M.; Hinrichs, D.; Thaller, G. Assessing the genetic background and genomic relatedness of red cattle populations originating from Northern Europe. Genet. Sel. Evol. 2021, 53, 23. [Google Scholar] [CrossRef]
- Mastrangelo, S.; Ciani, E.; Ajmone Marsan, P.; Bagnato, A.; Battaglini, L.; Bozzi, R.; Carta, A.; Catillo, G.; Cassandro, M.; Casu, S.; et al. Conservation status and historical relatedness of Italian cattle breeds. Genet. Sel. Evol. 2018, 50, 35. [Google Scholar] [CrossRef]
- Matukumalli, L.K.; Lawley, C.T.; Schnabel, R.D.; Taylor, J.F.; Allan, M.F.; Heaton, M.P.; O’Connell, J.; Moore, S.S.; Smith, T.P.L.; Sonstegard, T.S.; et al. Development and characterization of a high-density SNP genotyping assay for cattle. PLoS ONE 2009, 4, e5350. [Google Scholar] [CrossRef]
- Cañas-Álvarez, J.J.; González-Rodríguez, A.; Munilla, S.; Varona, L.; Díaz, C.; Baro, J.A.; Altarriba, J.; Molina, A.; Piedrafita, J. Genetic diversity and divergence among Spanish beef cattle breeds assessed by a bovine high-density SNP chip. J. Anim. Sci. 2015, 93, 5164–5174. [Google Scholar] [CrossRef]
- Alvariño Martínez, P. Evaluación de la Diversidad Genética y de Parentesco en Poblaciones de Rubia Gallega (Bos Taurus). Trabajo Fin de Grado. Universidad de la Coruña, Spain. 2019. Available online: https://ruc.udc.es/dspace/handle/2183/24504 (accessed on 12 January 2024).
- Gautier, M.; Faraut, T.; Moazami-Goudarzi, K.; Navratil, V.; Foglio, M.; Grohs, C.; Boland, A.; Garnier, J.-G.; Boichard, D.; Lathrop, G.M.; et al. Genetic and haplotypic structure in 14 European and African cattle breeds. Genetics 2007, 177, 1059–1070. [Google Scholar] [CrossRef]
- Lenstra, J.A.; Groeneveld, L.F.; Eding, H.; Kantanen, J.; Williams, J.L.; Taberlet, P.; Nicolazzi, E.L.; Sölkner, J.; Simianer, H.; Ciani, E.; et al. Molecular tools and analytical approaches for the characterization of farm animal genetic diversity. Anim. Genet. 2012, 43, 483–502. [Google Scholar] [CrossRef]
- Laoun, A.; Harkat, S.; Lafri, M.; Gaouar, S.B.S.; Belabdi, I.; Ciani, E.; De Groot, M.; Blanquet, V.; Leroy, G.; Rognon, X.; et al. Inference of breed structure in farm animals: Empirical comparison between SNP and microsatellite performance. Genes 2020, 11, 57. [Google Scholar] [CrossRef]
- Peripolli, E.; Munari, D.P.; Silva, M.V.G.B.; Lima, A.L.F.; Irgang, R.; Baldi, F. Runs of homozygosity: Current knowledge and applications in livestock. Anim. Genet. 2017, 48, 255–271. [Google Scholar] [CrossRef]
- Purfield, D.C.; Berry, D.P.; McParland, S.; Bradley, D.G. Runs of homozygosity and population history in cattle. BMC Genom. 2012, 13, 70. [Google Scholar] [CrossRef]
- Howrigan, D.P.; Simonson, M.A.; Keller, M.C. Detecting autozygosity through runs of homozygosity: A comparison of three autozygosity detection algorithms. BMC Genom. 2011, 12, 460. [Google Scholar] [CrossRef]
- Detera-Wadleigh, S.D.; Yoon, S.W.; Berrettini, W.H.; Goldin, L.R.; Turner, G.; Yoshikawa, T.; Rollins, D.Y.; Muniec, D.; Nurnberger, J.I., Jr.; Gershon, E.S. Adrenocorticotropin receptor/melanocortin receptor-2 maps within a reported susceptibility region for bipolar illness on chromosome 18. Am. J. Med. Genet. 1995, 60, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Perdomo-González, D.I.; Laseca, N.; Demyda-Peyrás, S.; Valera, M.; Cervantes, I.; Molina, A. Fine-tuning genomic and pedigree inbreeding rates in equine population with a deep and reliable stud book: The case of the Pura Raza Espanola horse. J. Anim. Sci. Biotechnol. 2022, 13, 127. [Google Scholar] [CrossRef]
- Iannuzzi, A.; Demyda-Peyrás, S.; Pistucci, R.; Morales, R.; Zannotti, M.; Sbarra, F.; Quaglia, A.; Parma, P. A genomic biomarker for the rapid identification of the rob (1;29) translocation in beef cattle breeds. Sci. Rep. 2024, 14, 2951. [Google Scholar] [CrossRef] [PubMed]
- Grosz, M.D.; MacNeil, M.D. The” spotted” locus maps to bovine chromosome 6 in a Hereford-Cross population. J. Hered. 1999, 90, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Fontanesi, L.; Scotti, E.; Russo, V. Haplotype variability in the bovine MITF gene and association with piebaldism in Holstein and Simmental cattle breeds. Anim. Genet. 2012, 43, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Boitard, S.; Rodríguez, W.; Jay, F.; Mona, S.; Austerlitz, F. Inferring population size history from large samples of genome-wide molecular data—An approximate Bayesian computation approach. PLoS Genet. 2016, 12, e1005877. [Google Scholar]
- Hayes, B.J.; Daetwyler, H.D. 1000 bull genomes project to map simple and complex genetic traits in cattle: Applications and outcomes. Annu. Rev. Anim. 2019, 7, 89–102. [Google Scholar] [CrossRef]
- Moreno-Millán, M.; Demyda Peyrás, S. Causas cromosómicas de infertilidad y mortalidad embrionaria en mamíferos domésticos. Spernova 2014, 4, 145–152. [Google Scholar]

| Berrenda en Colorado | Berrenda en Negro | |||||
|---|---|---|---|---|---|---|
| Region | Males | Females | Farms | Males | Females | Farms |
| Castilla y León | 6 | 29 | 8 | 13 | 7 | 4 |
| Madrid | 14 | 3 | 7 | 11 | 3 | 8 |
| Extremadura | 33 | 2 | 7 | 25 | 1 | 3 |
| Castilla La Mancha | 2 | 15 | 4 | 7 | 31 | 10 |
| Andalucía | 39 | 42 | 15 | 36 | 51 | 16 |
| Comunidad Valenciana | 0 | 2 | 1 | 1 | 2 | 2 |
| Animals | BC | BN | ||||||
|---|---|---|---|---|---|---|---|---|
| Total | Free | HeC | HmC | Total | Free | HeC | HmC | |
| Females | 93 | 51 | 38 | 4 | 94 | 30 | 63 | 1 |
| Males | 94 | 53 | 37 | 4 | 92 | 70 | 22 | |
| Total | 187 (184) | 104 (101) | 75 (75) | 8 (8) | 186 (185) | 100 (99) | 85 (85) | 1 (1) |
| Farms | 42 | 14 | 28 | 7 | 43 | 11 | 32 | 1 |
| Group | n | MAF | %MAF05 | Np | Np% | FIT (±SE) | Ho (±SE) | He (±SE) |
|---|---|---|---|---|---|---|---|---|
| BC | 184 | 0.205 | 71.5% | 44,129 | 81.5 | 0.09 (0.01) A | 0.306 (0.01) | 0.334 (0.00) A |
| BN | 185 | 0.203 | 70.9% | 44,951 | 83.0 | 0.09 (0.01) A | 0.308 (0.01) | 0.328 (0.00) B |
| BCF | 101 | 0.201 | 70.3% | 43,096 | 79.6 | 0.12 (0.01) B | 0.302 (0.01) | 0.333 (0.00) B |
| BCC | 83 | 0.206 | 71.7% | 43,720 | 80.7 | 0.08 (0.01) A | 0.311 (0. 01) | 0.336 (0.00) A |
| BNF | 99 | 0.200 | 70.4% | 43,678 | 80.7 | 0.09 (0.01) A | 0.313 (0.01) | 0.328 (0.00) D |
| BNC | 86 | 0.202 | 70.8% | 43,990 | 81.2 | 0.09 (0.01) A | 0.304 (0.01) | 0.329 (0.00) C |
| Group | n | FIS (±SD) | ΔF | FROH (±SD) | FROH < 4 Mb (±SD) | FROH < 10 Mb (±SD) | FROH 10–20 Mb (±SD) | FROH > 20 Mb (±SD) |
|---|---|---|---|---|---|---|---|---|
| BC | 184 | 0.09 (0.13) | 0.101 | 0.13 (0.12) | 0.01 (0.008) | 0.03 (0.02) | 0.04 (0.03) | 0.09 (0.079) |
| BCF | 101 | 0.09 (0.13) | 0.100 | 0.14 (0.12) | 0.01 (0.009) | 0.03 (0.02) | 0.05 (0.03) | 0.010 (0.08) |
| BCC | 83 | 0.08 (0.13) | 0.082 | 0.11 (0.12) | 0.01 (0.008) | 0.02 (0.02) | 0.04 (0.03) | 0.09 (0.08) |
| BN | 185 | 0.07 (0.12) | 0.077 | 0.12 (0.13) | 0.01 (0.009) | 0.03 (0.02) | 0.04 (0.03) | 0.08 (0.06) |
| BNF | 99 | 0.05 (0.13) | 0.053 | 0.10 (0.11) | 0.01 (0.01) | 0.03 (0.02) | 0.04 (0.03) | 0.08 (0.07) |
| BNC | 86 | 0.08 (0.12) | 0.084 | 0.13 (0.11) | 0.01 (0.01) | 0.03 (0.02) | 0.05 (0.03) | 0.08 (0.06) |
| Group | BCC | BNC | BNF |
|---|---|---|---|
| BCF | 0.013 | 0.028 | 0.031 |
| BCC | 0.021 | 0.026 | |
| BNC | 0.012 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Cano, R.; González-Martínez, A.; Ramón, M.; González Serrano, M.; Moreno Millán, M.; Rubio de Juan, A.; Rodero Serrano, E. Exploring the Effects of Robertsonian Translocation 1/29 (Rob (1;29)) on Genetic Diversity in Minor Breeds of Spanish Berrenda Cattle via Genome-Wide Analysis. Animals 2024, 14, 793. https://doi.org/10.3390/ani14050793
González-Cano R, González-Martínez A, Ramón M, González Serrano M, Moreno Millán M, Rubio de Juan A, Rodero Serrano E. Exploring the Effects of Robertsonian Translocation 1/29 (Rob (1;29)) on Genetic Diversity in Minor Breeds of Spanish Berrenda Cattle via Genome-Wide Analysis. Animals. 2024; 14(5):793. https://doi.org/10.3390/ani14050793
Chicago/Turabian StyleGonzález-Cano, Rafael, Ana González-Martínez, Manuel Ramón, Miriam González Serrano, Miguel Moreno Millán, Alejandro Rubio de Juan, and Evangelina Rodero Serrano. 2024. "Exploring the Effects of Robertsonian Translocation 1/29 (Rob (1;29)) on Genetic Diversity in Minor Breeds of Spanish Berrenda Cattle via Genome-Wide Analysis" Animals 14, no. 5: 793. https://doi.org/10.3390/ani14050793
APA StyleGonzález-Cano, R., González-Martínez, A., Ramón, M., González Serrano, M., Moreno Millán, M., Rubio de Juan, A., & Rodero Serrano, E. (2024). Exploring the Effects of Robertsonian Translocation 1/29 (Rob (1;29)) on Genetic Diversity in Minor Breeds of Spanish Berrenda Cattle via Genome-Wide Analysis. Animals, 14(5), 793. https://doi.org/10.3390/ani14050793





