Functional Analysis and Tissue-Specific Expression of Calcitonin and CGRP with RAMP-Modulated Receptors CTR and CLR in Chickens
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Primers, Peptides, and Antibodies
2.2. Extraction of Total RNA and Reverse Transcription
2.3. Cloning the cDNAs of Chicken CTR, CLR, RAMP1, RAMP2, and RAMP3
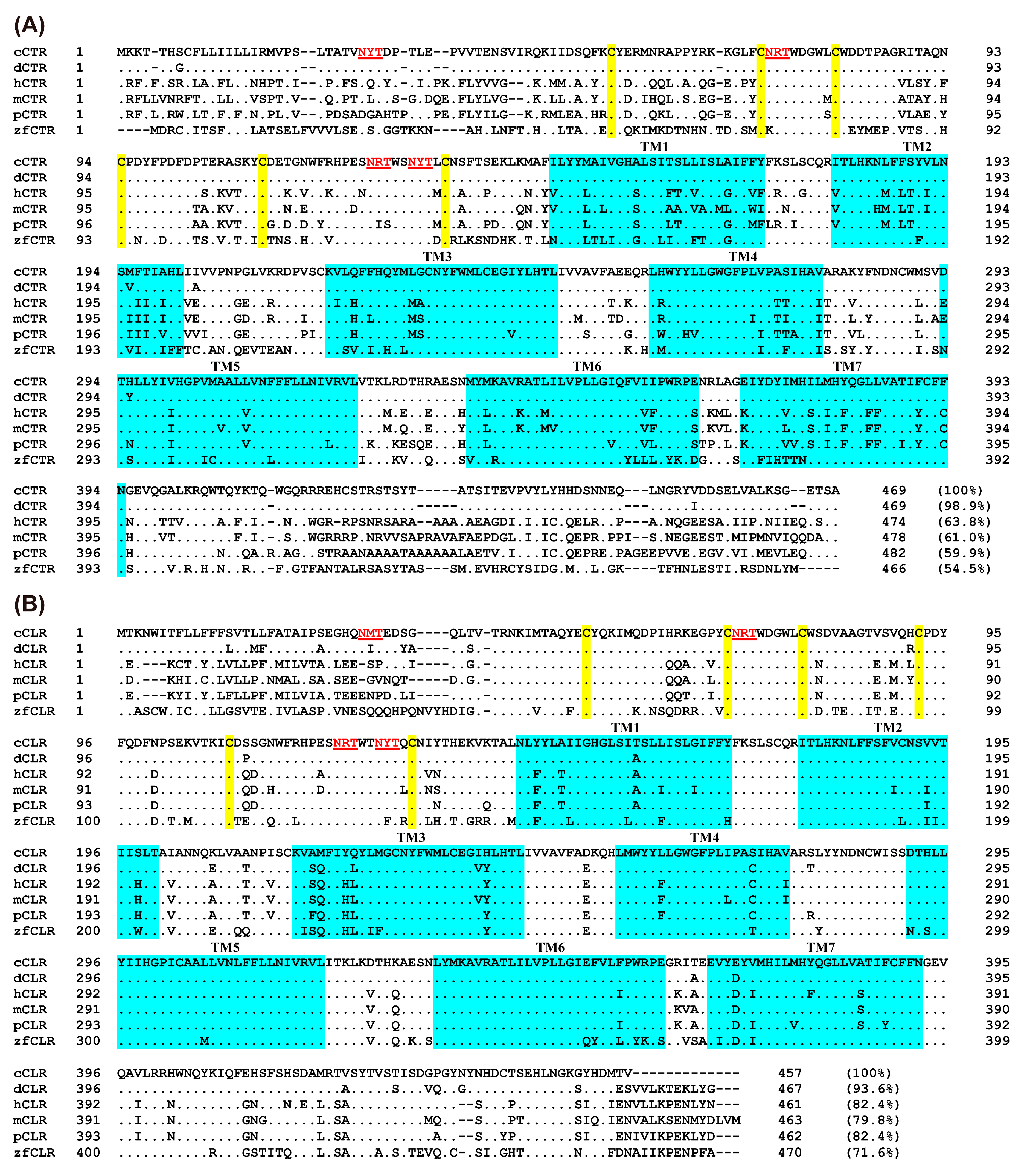
2.4. Sequence Alignment and Analysis
2.5. Functional Characterization of the Chicken Calcitonin Family Receptors (CTR and CLR)
2.6. Western Blot
2.7. Tissue-Specific Expression Analysis of Chicken CTR, CLR, RAMP1, RAMP2, and RAMP3 Using RNA-Seq Data
2.8. Data Analysis
3. Results
3.1. Characterization of the Coding Regions of Chicken CT, CGRP, CTR, and CLR
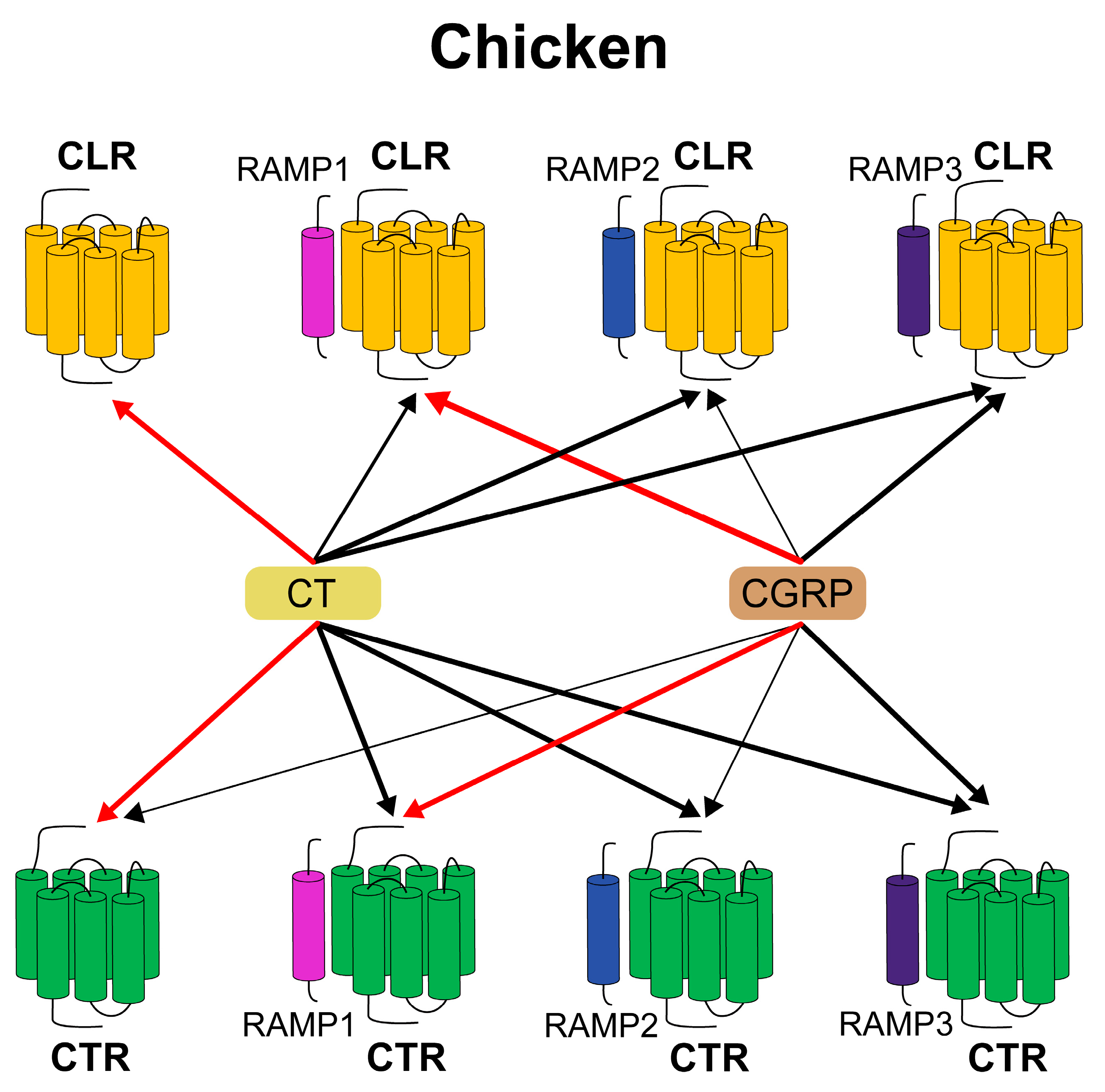
3.2. Overview of CTR/CLR Receptor and RAMP Interaction in Chickens
3.3. Effect of CT/CGRP on CTR/CLR–RAMP Complexes in cAMP/PKA Signaling Pathway
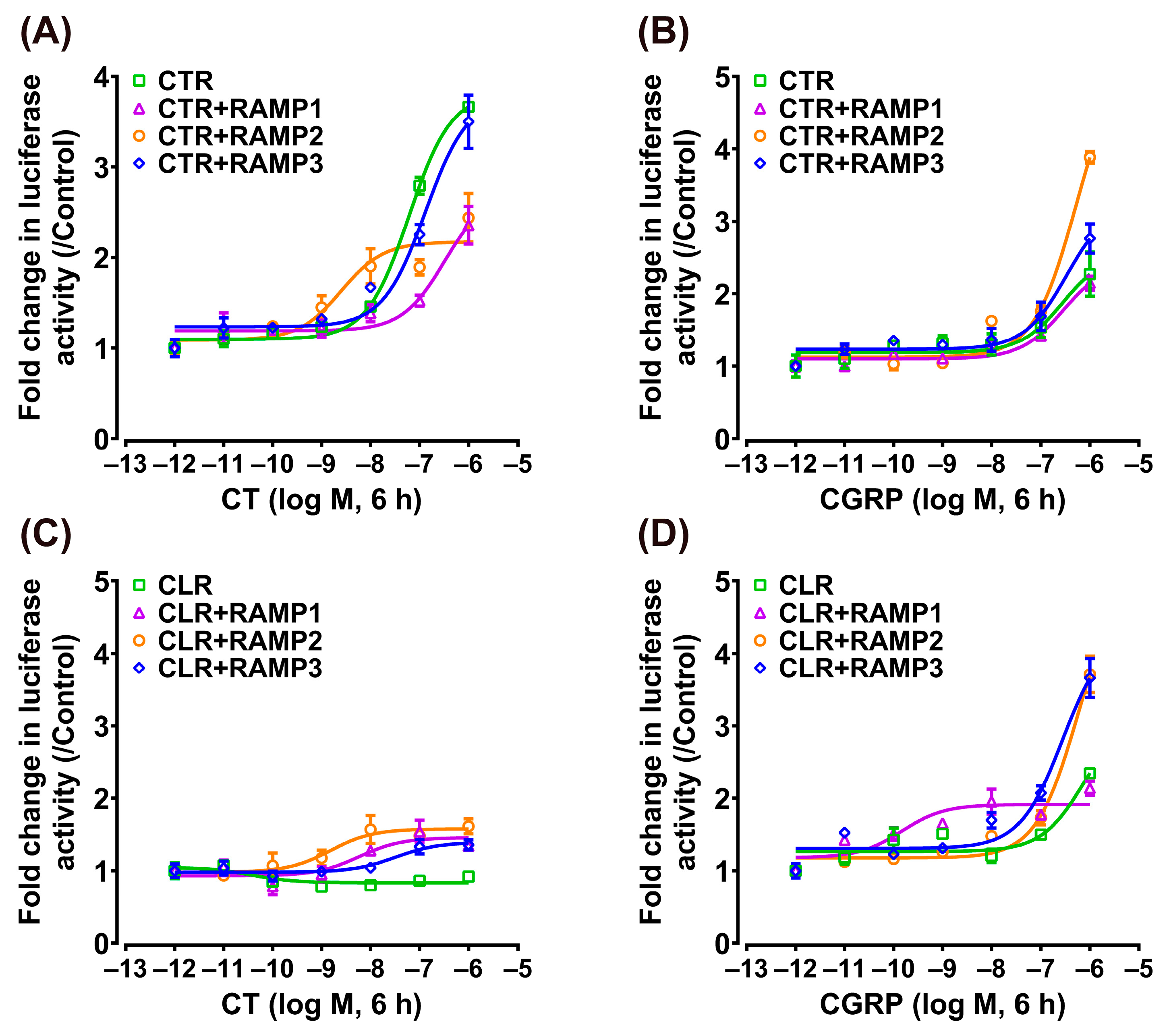
3.4. Effect of CT/CGRP on CTR/CLR–RAMP Complexes in MAPK/ERK Signaling Pathway
3.5. Effect of CT/CGRP on CTR/CLR–RAMP Complexes in Calcium Signaling Pathway
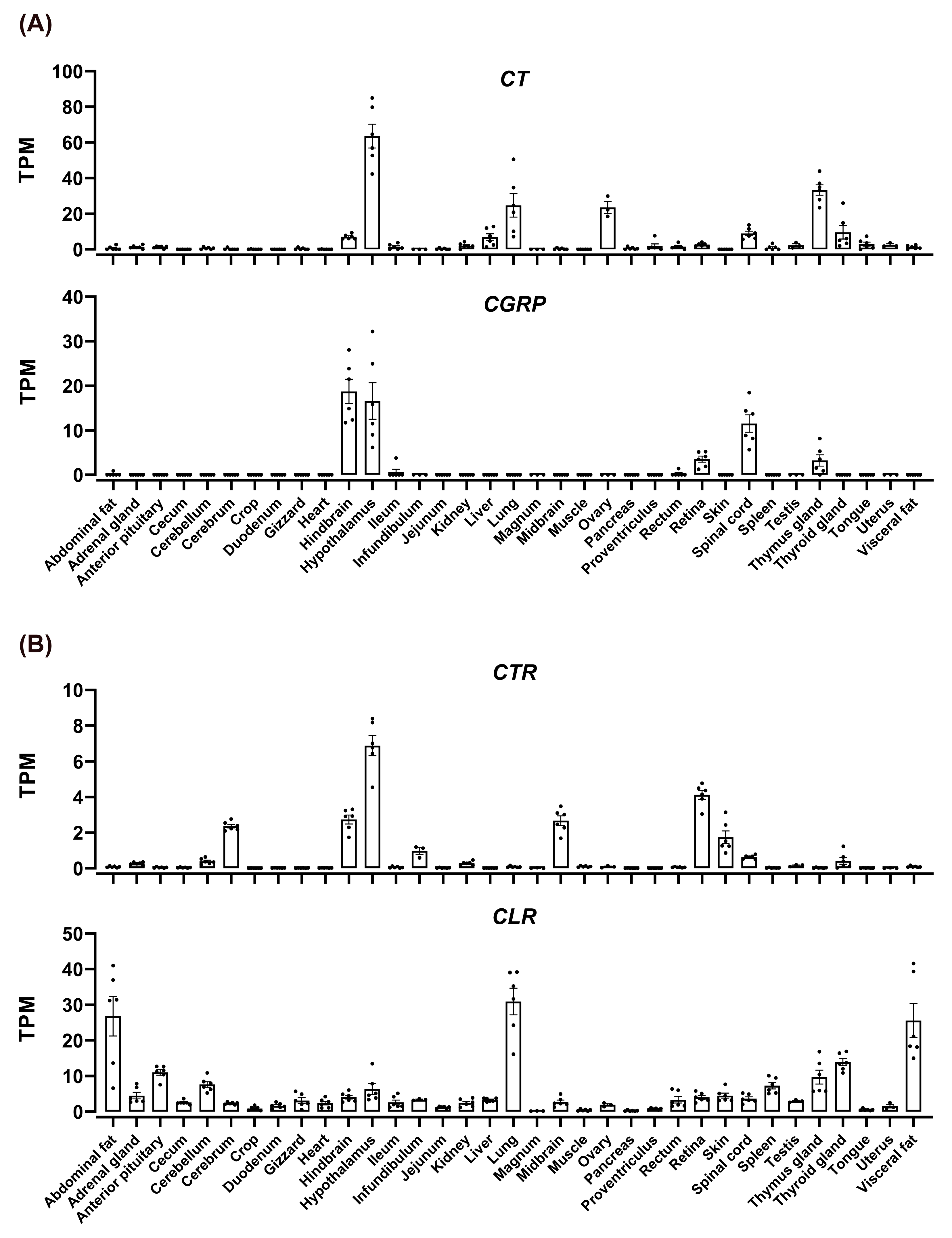
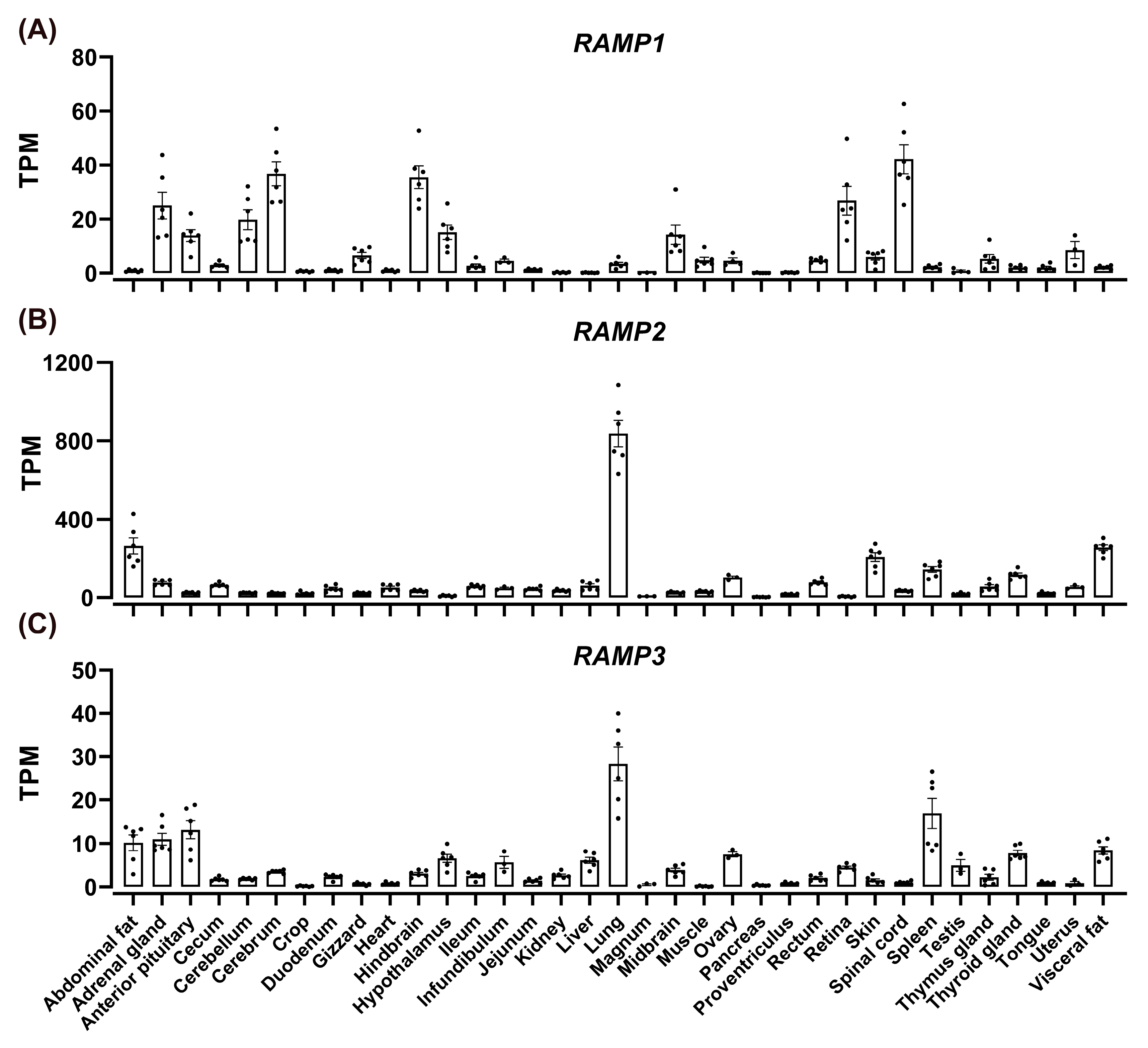
3.6. Tissue Distribution of CT, CGTRP, CTR, CLR, and RAMPs in Chickens
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Pearse, A.G.E. The cytochemistry of the thyroid C cells and their relationship to calcitonin. Proc. R. Soc. Lond. Ser. B. Biol. Sci. 1966, 164, 478–487. [Google Scholar]
- Amara, S.G.; Jonas, V.; Rosenfeld, M.G.; Ong, E.S.; Evans, R.M. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature 1982, 298, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Copp, D.H.; Cockcroft, D.W.; Kueh, Y. Ultimobranchial origin of calcitonin. Hypocalcemic effect of extracts from chicken glands. Can. J. Physiol. Pharmacol. 1967, 45, 1095–1099. [Google Scholar] [CrossRef] [PubMed]
- Sexton, P.M.; Findlay, D.M.; Martin, T.J. Calcitonin. Curr. Med. Chem. 1999, 6, 1067–1093. [Google Scholar] [CrossRef] [PubMed]
- Brain, S.D.; Tippins, J.R.; Morris, H.R.; MacIntyre, I.; Williams, T.J. Potent vasodilator activity of calcitonin gene-related peptide in human skin. J. Investig. Dermatol. 1986, 87, 533–536. [Google Scholar] [CrossRef] [PubMed]
- Russell, F.A.; King, R.; Smillie, S.J.; Kodji, X.; Brain, S.D. Calcitonin gene-related peptide: Physiology and pathophysiology. Physiol. Rev. 2014, 94, 1099–1142. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.; Gagel, R.F.; Berget, S.M. An intron enhancer recognized by splicing factors activates polyadenylation. Genes Dev. 1996, 10, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Born, W.; Muff, R.; Fischer, J.A. Functional interaction of G protein-coupled receptors of the adrenomedullin peptide family with accessory receptor-activity-modifying proteins (RAMP). Microsc. Res. Tech. 2002, 57, 14–22. [Google Scholar] [CrossRef]
- Poyner, D.R.; Sexton, P.M.; Marshall, I.; Smith, D.M.; Quirion, R.; Born, W.; Muff, R.; Fischer, J.A.; Foord, S.M. International Union of Pharmacology. XXXII. The mammalian calcitonin gene-related peptides, adrenomedullin, amylin, and calcitonin receptors. Pharmacol. Rev. 2002, 54, 233–246. [Google Scholar] [CrossRef]
- Poyner, D.R. Molecular pharmacology of receptors for calcitonin-gene-related peptide, amylin and adrenomedullin. Biochem. Soc. Trans. 1997, 25, 1032–1036. [Google Scholar] [CrossRef]
- Poyner, D.R. Calcitonin gene-related peptide: Multiple actions, multiple receptors. Pharmacol. Ther. 1992, 56, 23–51. [Google Scholar] [CrossRef] [PubMed]
- Roh, J.; Chang, C.L.; Bhalla, A.; Klein, C.; Hsu, S.Y. Intermedin is a calcitonin/calcitonin gene-related peptide family peptide acting through the calcitonin receptor-like receptor/receptor activity-modifying protein receptor complexes. J. Biol. Chem. 2004, 279, 7264–7274. [Google Scholar] [CrossRef] [PubMed]
- Alexander, S.P.H.; Christopoulos, A.; Davenport, A.P.; Kelly, E.; Mathie, A.A.; Peters, J.A.; Veale, E.L.; Armstrong, J.F.; Faccenda, E.; Harding, S.D.; et al. The Concise Guide to PHARMACOLOGY 2023/24: G protein-coupled receptors. Br. J. Pharmacol. 2023, 180 (Suppl. S2), S23–S144. [Google Scholar] [CrossRef] [PubMed]
- Conner, A.C.; Simms, J.; Barwell, J.; Wheatley, M.; Poyner, D.R. Ligand binding and activation of the CGRP receptor. Biochem. Soc. Trans. 2007, 35, 729–732. [Google Scholar] [CrossRef] [PubMed]
- Bortolato, A.; Doré, A.S.; Hollenstein, K.; Tehan, B.G.; Mason, J.S.; Marshall, F.H. Structure of C lass B GPCRs: New horizons for drug discovery. Br. J. Pharmacol. 2014, 171, 3132–3145. [Google Scholar] [CrossRef] [PubMed]
- Purdue, B.W.; Tilakaratne, N.; Sexton, P.M. Molecular pharmacology of the calcitonin receptor. Recept. Channels 2002, 8, 243–255. [Google Scholar] [CrossRef] [PubMed]
- McLatchie, L.M.; Fraser, N.J.; Main, M.J.; Wise, A.; Brown, J.; Thompson, N.; Solari, R.; Lee, M.G.; Foord, S.M. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature 1998, 393, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Gingell, J.J.; Simms, J.; Barwell, J.; Poyner, D.R.; Watkins, H.A.; Pioszak, A.A.; Sexton, P.M.; Hay, D.L. An allosteric role for receptor activity-modifying proteins in defining GPCR pharmacology. Cell Discov. 2016, 2, 16012. [Google Scholar] [CrossRef]
- Goltzman, D.; Mitchell, J. Interaction of calcitonin and calcitonin gene-related peptide at receptor sites in target tissues. Science 1985, 227, 1343–1345. [Google Scholar] [CrossRef]
- Garelja, M.L.; Bower, R.L.; Brimble, M.A.; Chand, S.; Harris, P.W.R.; Jamaluddin, M.A.; Petersen, J.; Siow, A.; Walker, C.S.; Hay, D.L. Pharmacological characterisation of mouse calcitonin and calcitonin receptor-like receptors reveals differences compared with human receptors. Br. J. Pharmacol. 2022, 179, 416–434. [Google Scholar] [CrossRef]
- Bailey, R.J.; Walker, C.S.; Ferner, A.H.; Loomes, K.M.; Prijic, G.; Halim, A.; Whiting, L.; Phillips, A.R.; Hay, D.L. Pharmacological characterization of rat amylin receptors: Implications for the identification of amylin receptor subtypes. Br. J. Pharmacol. 2012, 166, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Wootten, D.L.; Simms, J.; Hay, D.L.; Christopoulos, A.; Sexton, P.M. Receptor activity modifying proteins and their potential as drug targets. Prog. Mol. Biol. Transl. Sci. 2010, 91, 53–79. [Google Scholar] [CrossRef] [PubMed]
- Hagner, S.; Haberberger, R.V.; Overkamp, D.; Hoffmann, R.; Voigt, K.H.; McGregor, G.P. Expression and distribution of calcitonin receptor-like receptor in human hairy skin. Peptides 2002, 23, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Minvielle, S.; Cressent, M.; Lasmoles, F.; Jullienne, A.; Milhaud, G.; Moukhtar, M.J.F.l. Isolation and partial characterization of the calcitonin gene in a lower vertebrate: Predicted structure of avian calcitonin gene-related peptide. FEBS Lett. 1986, 203, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Scanes, C.G.; Dridi, S. Sturkie’s Avian Physiology; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar]
- Nicholson, G.; Moseley, J.; Sexton, P.; Martin, T. Chicken osteoclasts do not possess calcitonin receptors. J. Bone Miner. Res. 1987, 2, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Ieda, T.; Takahashi, T.; Saito, N.; Yasuoka, T.; Kawashima, M.; Izumi, T.; Shimada, K. Changes in calcitonin receptor binding in the shell gland of laying hens (Gallus domesticus) during the oviposition cycle. J. Poult. Sci. 2001, 38, 203–212. [Google Scholar] [CrossRef]
- Ogawa, H.; Takahashi, T.; Kuwayama, T.; Kawashima, M. Presence of calcitonin receptors in shell gland of the guineafowl and changes in binding property during an oviposition cycle. Poult. Sci. 2003, 82, 1302–1306. [Google Scholar] [CrossRef]
- Yasuoka, T.; Kawashima, M.; Takahashi, T.; Tatematsu, N.; Tanaka, K. Calcitonin receptor binding properties in bone and kidney of the chicken during the oviposition cycle. J. Bone Miner. Res. 1998, 13, 1412–1419. [Google Scholar] [CrossRef]
- Lanuza, E.; Davies, D.C.; Landete, J.M.; Novejarque, A.; Martínez-García, F. Distribution of CGRP-like immunoreactivity in the chick and quail brain. J. Comp. Neurol. 2000, 421, 515–532. [Google Scholar] [CrossRef]
- Laufer, R.; Changeux, J.-P. Calcitonin gene-related peptide elevates cyclic AMP levels in chick skeletal muscle: Possible neurotrophic role for a coexisting neuronal messenger. EMBO J. 1987, 6, 901–906. [Google Scholar] [CrossRef]
- Cline, M.A.; Calchary, W.A.; Nandar, W. Effect of calcitonin gene-related peptide (CGRP) on avian appetite-related processes. Behav. Brain Res. 2009, 196, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, X.; Zhou, Y.; Cui, L.; Li, J.; Wu, C.; Wan, Y.; Li, J.; Wang, Y. The interaction of MC3R and MC4R with MRAP2, ACTH, alpha-MSH and AgRP in chickens. J. Endocrinol. 2017, 234, 155–174. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Zhang, J.; Wan, Y.; Li, Z.; Qi, F.; Dang, Y.; Li, J.; Wang, Y. Neuropeptide S (NPS) and its receptor (NPSR1) in chickens: Cloning, tissue expression, and functional analysis. Poult. Sci. 2021, 100, 101445. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Su, J.; Huang, T.; Wang, X.; Wu, C.; Li, J.; Li, J.; Zhang, J.; Wang, Y. Characterization of the chicken melanocortin 5 receptor and its potential role in regulating hepatic glucolipid metabolism. Front. Physiol. 2022, 13, 917712. [Google Scholar] [CrossRef]
- Wan, Y.; Deng, Q.; Zhou, Z.; Deng, Y.; Zhang, J.; Li, J.; Wang, Y. Cholecystokinin (CCK) and its receptors (CCK1R and CCK2R) in chickens: Functional analysis and tissue expression. Poult. Sci. 2023, 102, 102273. [Google Scholar] [CrossRef]
- Wu, C.; Lv, C.; Wan, Y.; Li, X.; Zhang, J.; Li, J.; Wang, Y. Arginine vasotocin (AVT)/mesotocin (MT) receptors in chickens: Evidence for the possible involvement of AVT-AVPR1 signaling in the regulation of oviposition and pituitary prolactin expression. Gen. Comp. Endocrinol. 2019, 281, 91–104. [Google Scholar] [CrossRef]
- Guo, X.; Chen, F.; Gao, F.; Li, L.; Liu, K.; You, L.; Hua, C.; Yang, F.; Liu, W.; Peng, C.; et al. CNSA: A data repository for archiving omics data. Database 2020, baaa055. [Google Scholar] [CrossRef]
- Chen, F.Z.; You, L.J.; Yang, F.; Wang, L.N.; Guo, X.Q.; Gao, F.; Hua, C.; Tan, C.; Fang, L.; Shan, R.Q.; et al. CNGBdb: China National GeneBank DataBase. Yi Chuan 2020, 42, 799–809. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, X.; Lv, C.; Wan, Y.; Zhang, X.; Li, J.; Wang, Y. A gene expression atlas of Lohmann white chickens. bioRxiv 2022. [Google Scholar] [CrossRef]
- Sun, C.; Qiu, Y.; Ren, Q.; Zhang, X.; Cao, B.; Zou, Y.; Li, J.; Zhang, J.; Wang, Y. Molecular Cloning and Functional Characterization of Three 5-HT Receptor Genes (HTR1B, HTR1E, and HTR1F) in Chickens. Genes 2021, 12, 891. [Google Scholar] [CrossRef]
- Zhang, J.; Li, J.; Wu, C.; Hu, Z.; An, L.; Wan, Y.; Fang, C.; Zhang, X.; Li, J.; Wang, Y. The Asp298Asn polymorphism of melanocortin-4 receptor (MC4R) in pigs: Evidence for its potential effects on MC4R constitutive activity and cell surface expression. Anim. Genet. 2020, 51, 694–706. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Lv, C.; Zhang, J.; Li, J.; Wang, Y. Characterization of four urotensin II receptors (UTS2Rs) in chickens. Peptides 2021, 138, 170482. [Google Scholar] [CrossRef] [PubMed]
- Martins, R.; Vieira, F.A.; Power, D.M. Calcitonin receptor family evolution and fishing for function using in silico promoter analysis. Gen. Comp. Endocrinol. 2014, 209, 61–73. [Google Scholar] [CrossRef]
- Buhlmann, N.; Leuthauser, K.; Muff, R.; Fischer, J.A.; Born, W. A receptor activity modifying protein (RAMP)2-dependent adrenomedullin receptor is a calcitonin gene-related peptide receptor when coexpressed with human RAMP1. Endocrinology 1999, 140, 2883–2890. [Google Scholar] [CrossRef]
- Muff, R.; Buhlmann, N.; Fischer, J.A.; Born, W. An amylin receptor is revealed following co-transfection of a calcitonin receptor with receptor activity modifying proteins-1 or -3. Endocrinology 1999, 140, 2924–2927. [Google Scholar] [CrossRef]
- Aldecoa, A.; Gujer, R.; Fischer, J.A.; Born, W. Mammalian calcitonin receptor-like receptor/receptor activity modifying protein complexes define calcitonin gene-related peptide and adrenomedullin receptors in Drosophila Schneider 2 cells. FEBS Lett. 2000, 471, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Husmann, K.; Sexton, P.M.; Fischer, J.A.; Born, W. Mouse receptor-activity-modifying proteins 1, -2 and -3: Amino acid sequence, expression and function. Mol. Cell. Endocrinol. 2000, 162, 35–43. [Google Scholar] [CrossRef]
- Pondel, M. Calcitonin and calcitonin receptors: Bone and beyond. Int. J. Exp. Pathol. 2000, 81, 405–422. [Google Scholar] [CrossRef]
- Russo, A.F.; Hay, D.L. CGRP physiology, pharmacology, and therapeutic targets: Migraine and beyond. Physiol. Rev. 2023, 103, 1565–1644. [Google Scholar] [CrossRef]
- Hay, D.L.; Poyner, D.R.; Sexton, P.M. GPCR modulation by RAMPs. Pharmacol. Ther. 2006, 109, 173–197. [Google Scholar] [CrossRef]
- Booe, J.M.; Walker, C.S.; Barwell, J.; Kuteyi, G.; Simms, J.; Jamaluddin, M.A.; Warner, M.L.; Bill, R.M.; Harris, P.W.; Brimble, M.A.; et al. Structural Basis for Receptor Activity-Modifying Protein-Dependent Selective Peptide Recognition by a G Protein-Coupled Receptor. Mol. Cell 2015, 58, 1040–1052. [Google Scholar] [CrossRef] [PubMed]
- Hay, D.L.; Christopoulos, G.; Christopoulos, A.; Poyner, D.R.; Sexton, P.M. Pharmacological discrimination of calcitonin receptor: Receptor activity-modifying protein complexes. Mol. Pharmacol. 2005, 67, 1655–1665. [Google Scholar] [CrossRef] [PubMed]
- Hay, D.L.; Chen, S.; Lutz, T.A.; Parkes, D.G.; Roth, J.D. Amylin: Pharmacology, Physiology, and Clinical Potential. Pharmacol. Rev. 2015, 67, 564–600. [Google Scholar] [CrossRef] [PubMed]
- Christopoulos, G.; Perry, K.J.; Morfis, M.; Tilakaratne, N.; Gao, Y.; Fraser, N.J.; Main, M.J.; Foord, S.M.; Sexton, P.M. Multiple amylin receptors arise from receptor activity-modifying protein interaction with the calcitonin receptor gene product. Mol. Pharmacol. 1999, 56, 235–242. [Google Scholar] [CrossRef]
- Walker, C.S.; Eftekhari, S.; Bower, R.L.; Wilderman, A.; Insel, P.A.; Edvinsson, L.; Waldvogel, H.J.; Jamaluddin, M.A.; Russo, A.F.; Hay, D.L. A second trigeminal CGRP receptor: Function and expression of the AMY1 receptor. Ann. Clin. Transl. Neurol. 2015, 2, 595–608. [Google Scholar] [CrossRef]
- Ogoshi, M.; Takahashi, M.; Aoyagi, K.; Ukena, K.; Aizawa, S.; Takeuchi, H.; Takahashi, S.; Takeuchi, S. Adrenomedullin 2 and 5 activate the calcitonin receptor-like receptor (clr)—Receptor activity-modifying protein 3 (ramp3) receptor complex in Xenopus tropicalis. Gen. Comp. Endocrinol. 2021, 306, 113752. [Google Scholar] [CrossRef] [PubMed]
- Nag, K.; Kato, A.; Nakada, T.; Hoshijima, K.; Mistry, A.C.; Takei, Y.; Hirose, S. Molecular and functional characterization of adrenomedullin receptors in pufferfish. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 290, R467–R478. [Google Scholar] [CrossRef]
- Nag, K.; Kato, A.; Sultana, N.; Ogoshi, M.; Takei, Y.; Hirose, S. Fish calcitonin receptor has novel features. Gen. Comp. Endocrinol. 2007, 154, 48–58. [Google Scholar] [CrossRef]
- Galan Galan, F.; Rogers, R.M.; Girgis, S.I.; MacIntyre, I. Immunoreactive calcitonin in the central nervous system of the pigeon. Brain Res. 1981, 212, 59–66. [Google Scholar] [CrossRef]
- Krzysik-Walker, S.M.; Ocon-Grove, O.M.; Maddineni, S.B.; Hendricks, G.L., 3rd; Ramachandran, R. Identification of calcitonin expression in the chicken ovary: Influence of follicular maturation and ovarian steroids. Biol. Reprod. 2007, 77, 626–635. [Google Scholar] [CrossRef]
- Cline, M.A.; Siders, R.; Newmyer, B.A.; Smith, M.L.; Siegel, P.B. Both calcitonin and calcitonin gene-related peptides’ thresholds of hypophagia are considerably lower in chicks selected for high rather than low juvenile body weight. Physiol. Behav. 2010, 101, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.L.; Snider, R.H.; Moore, C.F.; Monoghan, K.G.; Silva, O.L. Calcitonin in extrathyroidal tissues of man. Acta Endocrinol. 1979, 92, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.L.; Geelhoed, G.; O’Neill, W.; Monaghan, K.G.; Snider, R.H.; Moore, C.F.; Silva, O.L. Calcitonin in tissues of thyroidectomized monkey. Experientia 1980, 36, 609–610. [Google Scholar] [CrossRef]
- Cohen, R.; Becker, K.; Jullienne, A. Calcitonin and related peptides. EMC-Endocrinol. 2004, 1, 200–213. [Google Scholar] [CrossRef]
- Nakagawa-Mizuyachi, K.; Takahashi, T.; Kasai, S.; Nakayama, H.; Kawashima, M. Calcitonin Directly Inhibits Luteinizing Hormone-Stimulated Progesterone Production in Granulosa Cells of the Largest Follicle of Hen. J. Poult. Sci. 2010, 47, 170–175. [Google Scholar] [CrossRef]
- Nakayama, H.; Takahashi, T.; Funaki, W.; Nakagawa-Mizuyachi, K.; Kawashima, M. Calcitonin receptor bindings in the hen hypothalamus before and after oviposition. Poult. Sci. 2011, 90, 642–647. [Google Scholar] [CrossRef]
- Nakayama, H.; Takahashi, T.; Nakagawa-Mizuyachi, K.; Kawashima, M. Effect of calcitonin on adrenocorticotropic hormone secretion stimulated by corticotropin-releasing hormone in the hen anterior pituitary. Anim. Sci. J. 2011, 82, 475–480. [Google Scholar] [CrossRef]
- Hagner, S.; Haberberger, R.; Hay, D.L.; Facer, P.; Reiners, K.; Voigt, K.; McGregor, G.P. Immunohistochemical detection of the calcitonin receptor-like receptor protein in the microvasculature of rat endothelium. Eur. J. Pharmacol. 2003, 481, 147–151. [Google Scholar] [CrossRef]
- Hagner, S.; Stahl, U.; Knoblauch, B.; McGregor, G.P.; Lang, R.E. Calcitonin receptor-like receptor: Identification and distribution in human peripheral tissues. Cell Tissue Res. 2002, 310, 41–50. [Google Scholar] [CrossRef]
- Dakhama, A.; Larsen, G.L.; Gelfand, E.W. Calcitonin gene-related peptide: Role in airway homeostasis. Curr. Opin. Pharmacol. 2004, 4, 215–220. [Google Scholar] [CrossRef]
- Aguilera, C.M.; Gomez-Llorente, C.; Tofe, I.; Gil-Campos, M.; Canete, R.; Gil, A. Genome-wide expression in visceral adipose tissue from obese prepubertal children. Int. J. Mol. Sci. 2015, 16, 7723–7737. [Google Scholar] [CrossRef] [PubMed]
- Na, W.; Yu, J.Q.; Xu, Z.C.; Zhang, X.Y.; Yang, L.L.; Cao, Z.P.; Li, H.; Zhang, H. Important candidate genes for abdominal fat content identified by linkage disequilibrium and fixation index information. Poult. Sci. 2019, 98, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.B.; Ding, S.; Nielsen, N.R.; Pawlak, J.B.; Blakeney, E.S.; Caron, K.M. Calcitonin-Receptor-Like Receptor Signaling Governs Intestinal Lymphatic Innervation and Lipid Uptake. ACS Pharmacol. Transl. Sci. 2019, 2, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Yang, Y.; Deng, Q.-W.; Zhang, B.-B.; Ruan, J.-W.; Jin, H.; Wang, J.-H.; Ren, J.; Jiang, B.; Sun, J.-H. Governor vessel electro-acupuncture promotes the intrinsic growth ability of spinal neurons through activating calcitonin gene-related peptide/α-calcium/calmodulin-dependent protein kinase/neurotrophin-3 pathway after spinal cord injury. J. Neurotrauma 2021, 38, 734–745. [Google Scholar] [CrossRef] [PubMed]
- Brain, S.D.; Grant, A.D. Vascular actions of calcitonin gene-related peptide and adrenomedullin. Physiol. Rev. 2004, 84, 903–934. [Google Scholar] [CrossRef]
- Amisten, S.; Neville, M.; Hawkes, R.; Persaud, S.J.; Karpe, F.; Salehi, A. An atlas of G-protein coupled receptor expression and function in human subcutaneous adipose tissue. Pharmacol. Ther. 2015, 146, 61–93. [Google Scholar] [CrossRef]


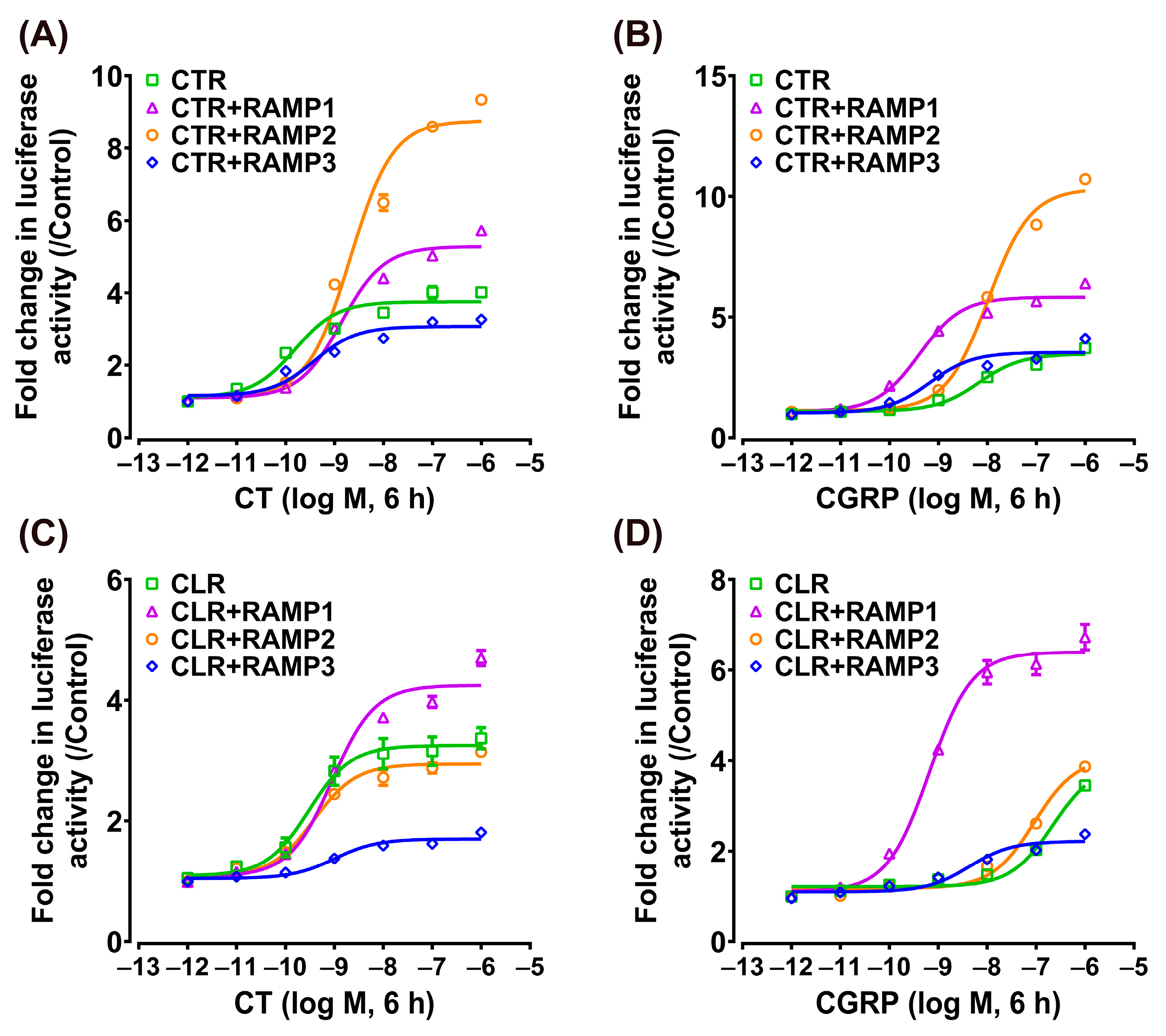
| EC50 Values (nM) | |||||
|---|---|---|---|---|---|
| cAMP/PKA signaling pathway | |||||
| CT | CGRP | CT | CGRP | ||
| CTR + pcDNA | 0.15 ± 0.04 | 2.35 ± 0.63 | CLR + pcDNA | 0.75 ± 0.21 | — |
| CTR + RAMP1 | 0.77 ± 0.07 | 0.10 ± 0.01 | CLR + RAMP1 | 3.30 ± 0.96 | 0.26 ± 0.03 |
| CTR + RAMP2 | 0.67 ± 0.10 | 1.63 ± 0.19 | CLR + RAMP2 | 0.93 ± 0.15 | 40.86 ± 6.76 |
| CTR + RAMP3 | 0.44 ± 0.10 | 0.24 ± 0.07 | CLR + RAMP3 | 1.27 ± 0.83 | 1.84 ± 0.73 |
| MAPK/ERK signaling pathway | |||||
| CT | CGRP | CT | CGRP | ||
| CTR + pcDNA | 0.15 ± 0.05 | 6.73 ± 1.78 | CLR + pcDNA | 0.30 ± 0.12 | — |
| CTR + RAMP1 | 1.38 ± 0.24 | 0.41 ± 0.08 | CLR + RAMP1 | 0.89 ± 0.20 | 0.66 ± 0.11 |
| CTR + RAMP2 | 2.09 ± 0.42 | 10.38 ± 1.12 | CLR + RAMP2 | 0.38 ± 0.09 | 90.93 ± 17.36 |
| CTR + RAMP3 | 0.38 ± 0.13 | 0.67 ± 0.24 | CLR + RAMP3 | 1.00 ± 0.45 | 4.31 ± 1.73 |
| Calcium signaling pathway | |||||
| CT | CGRP | CT | CGRP | ||
| CTR + pcDNA | 61.10 ± 8.55 | — | CLR + pcDNA | NA a | — |
| CTR + RAMP1 | — | — | CLR + RAMP1 | 5.81 ± 5.75 | 0.15 ± 0.11 |
| CTR + RAMP2 | 2.50 ± 1.81 | — | CLR + RAMP2 | 1.52 ± 1.32 | — |
| CTR + RAMP3 | — | — | CLR + RAMP3 | 32.3 ± 30.3 | — |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, T.; Su, J.; Wang, X.; Shi, N.; Zhang, X.; He, J.; Li, J.; Zhang, J.; Wang, Y. Functional Analysis and Tissue-Specific Expression of Calcitonin and CGRP with RAMP-Modulated Receptors CTR and CLR in Chickens. Animals 2024, 14, 1058. https://doi.org/10.3390/ani14071058
Huang T, Su J, Wang X, Shi N, Zhang X, He J, Li J, Zhang J, Wang Y. Functional Analysis and Tissue-Specific Expression of Calcitonin and CGRP with RAMP-Modulated Receptors CTR and CLR in Chickens. Animals. 2024; 14(7):1058. https://doi.org/10.3390/ani14071058
Chicago/Turabian StyleHuang, Tianjiao, Jiancheng Su, Xinglong Wang, Ningkun Shi, Xiao Zhang, Jiliang He, Juan Li, Jiannan Zhang, and Yajun Wang. 2024. "Functional Analysis and Tissue-Specific Expression of Calcitonin and CGRP with RAMP-Modulated Receptors CTR and CLR in Chickens" Animals 14, no. 7: 1058. https://doi.org/10.3390/ani14071058
APA StyleHuang, T., Su, J., Wang, X., Shi, N., Zhang, X., He, J., Li, J., Zhang, J., & Wang, Y. (2024). Functional Analysis and Tissue-Specific Expression of Calcitonin and CGRP with RAMP-Modulated Receptors CTR and CLR in Chickens. Animals, 14(7), 1058. https://doi.org/10.3390/ani14071058






