New Insights into Geometric Morphometry Applied to Fish Scales for Species Identification
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Image Acquisition
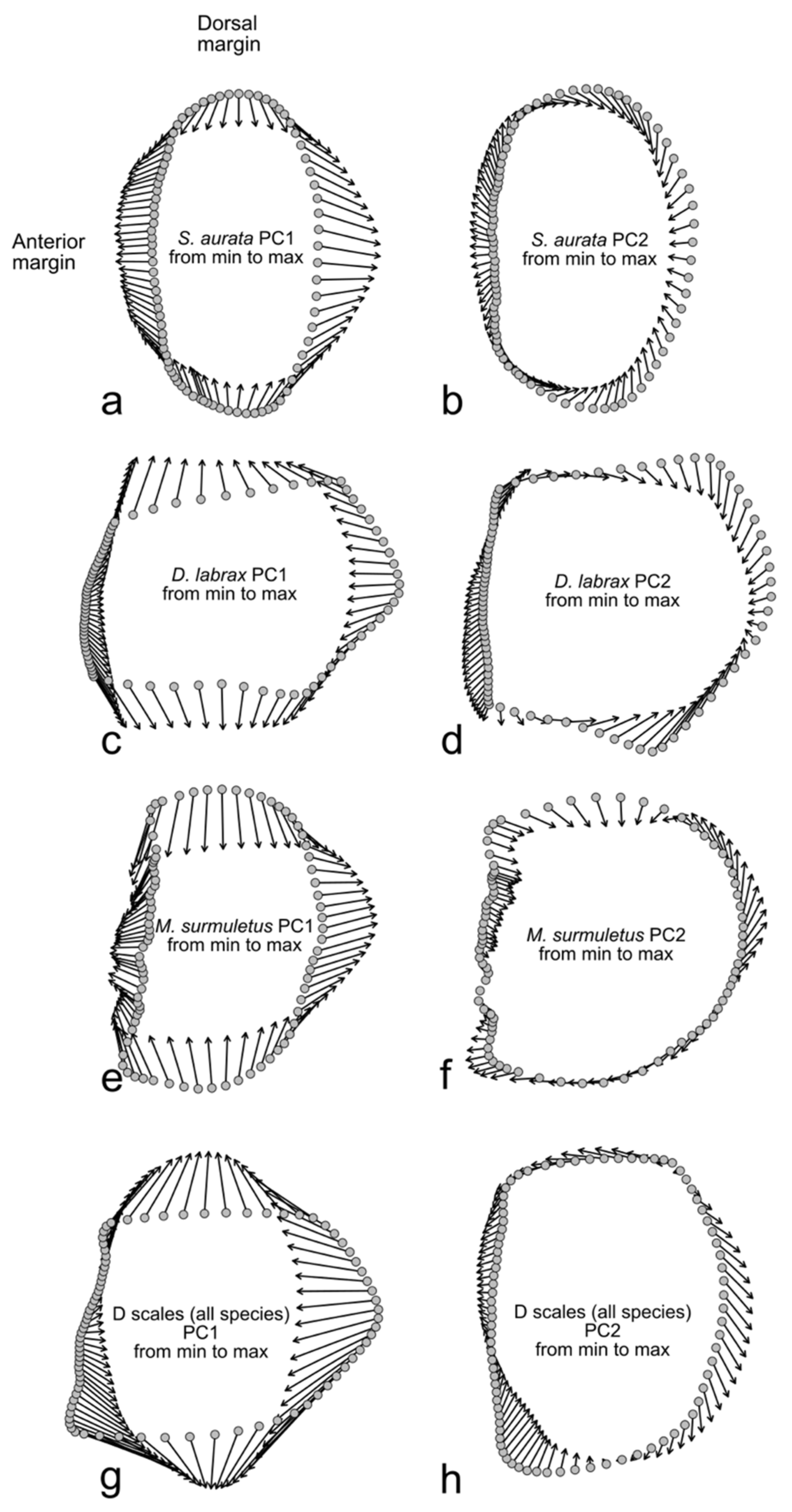
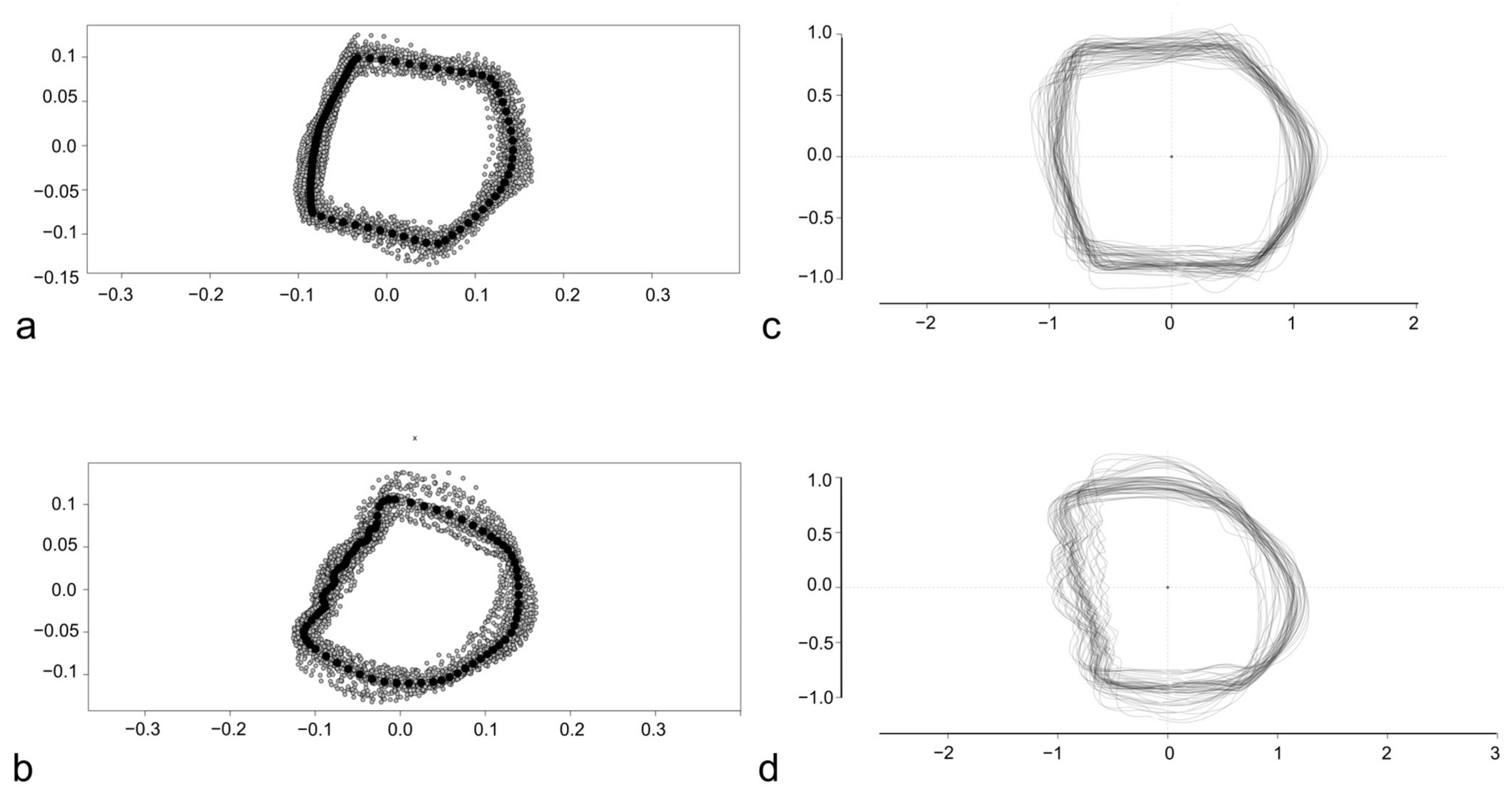
2.3. Geometric Morphometry Based on Landmarks and Semilandmarks (LM-Based GM)
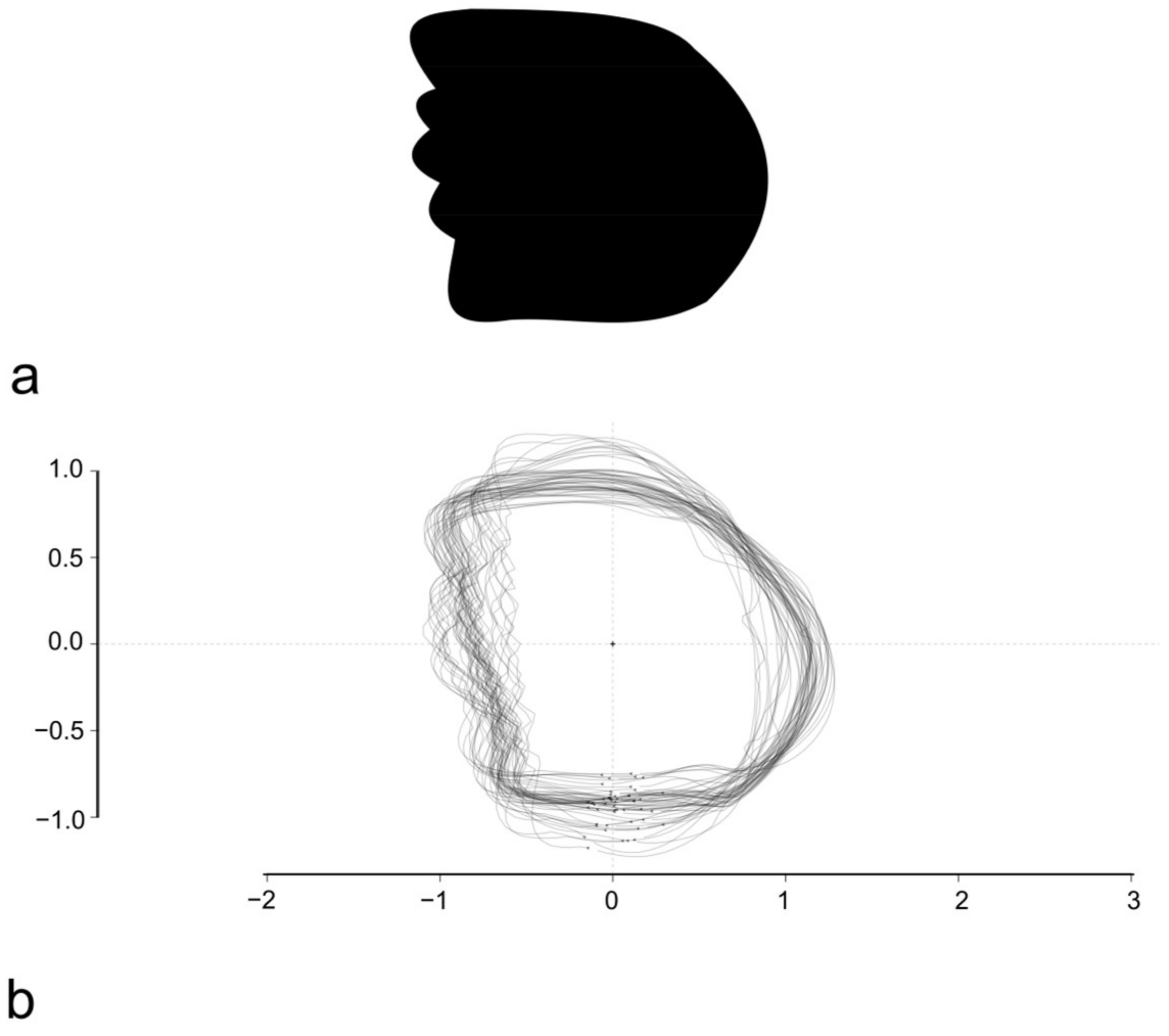
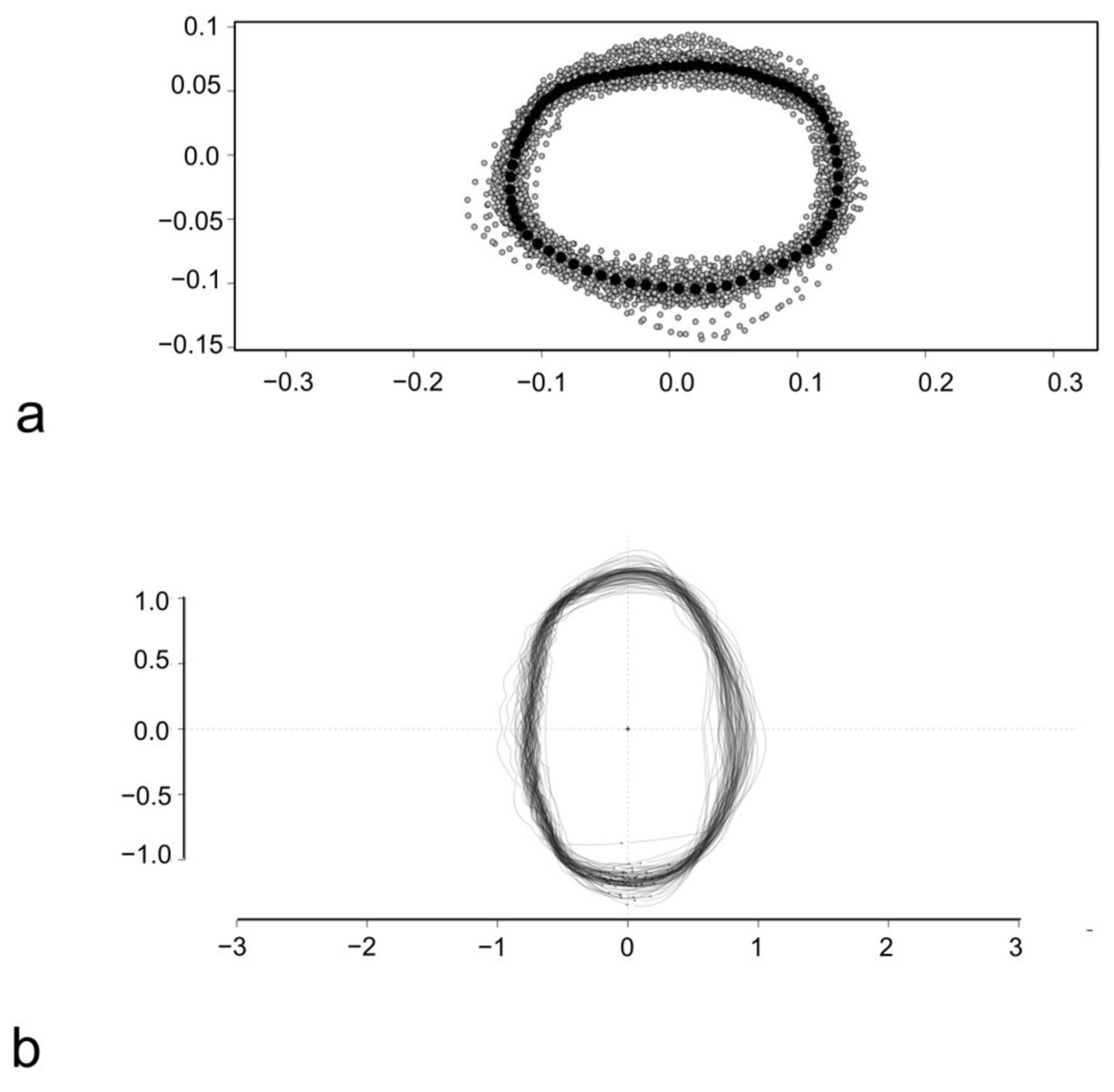
2.4. Outline-Based Geometric Morphometrics (OL-Based GM)
3. Results
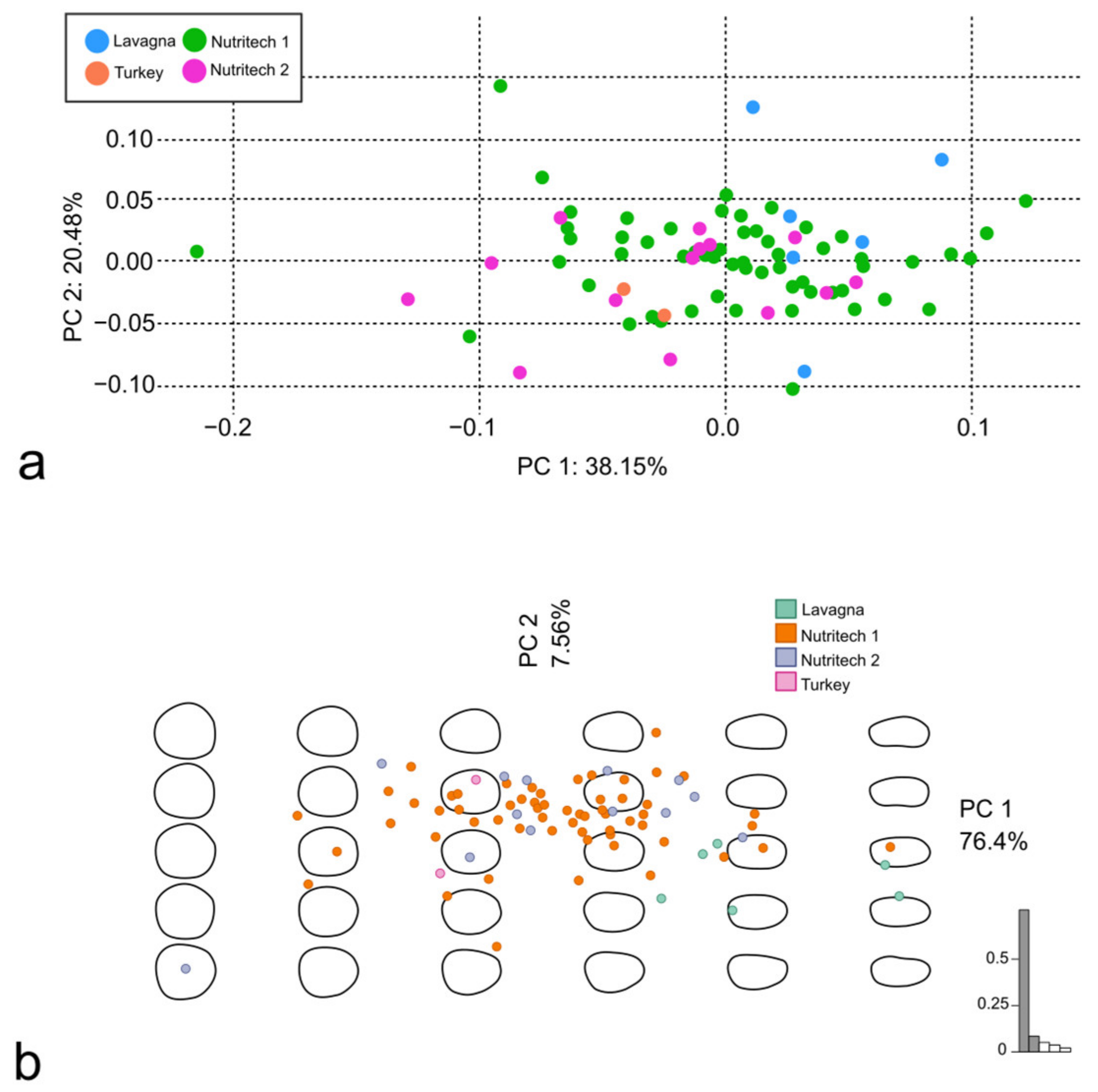
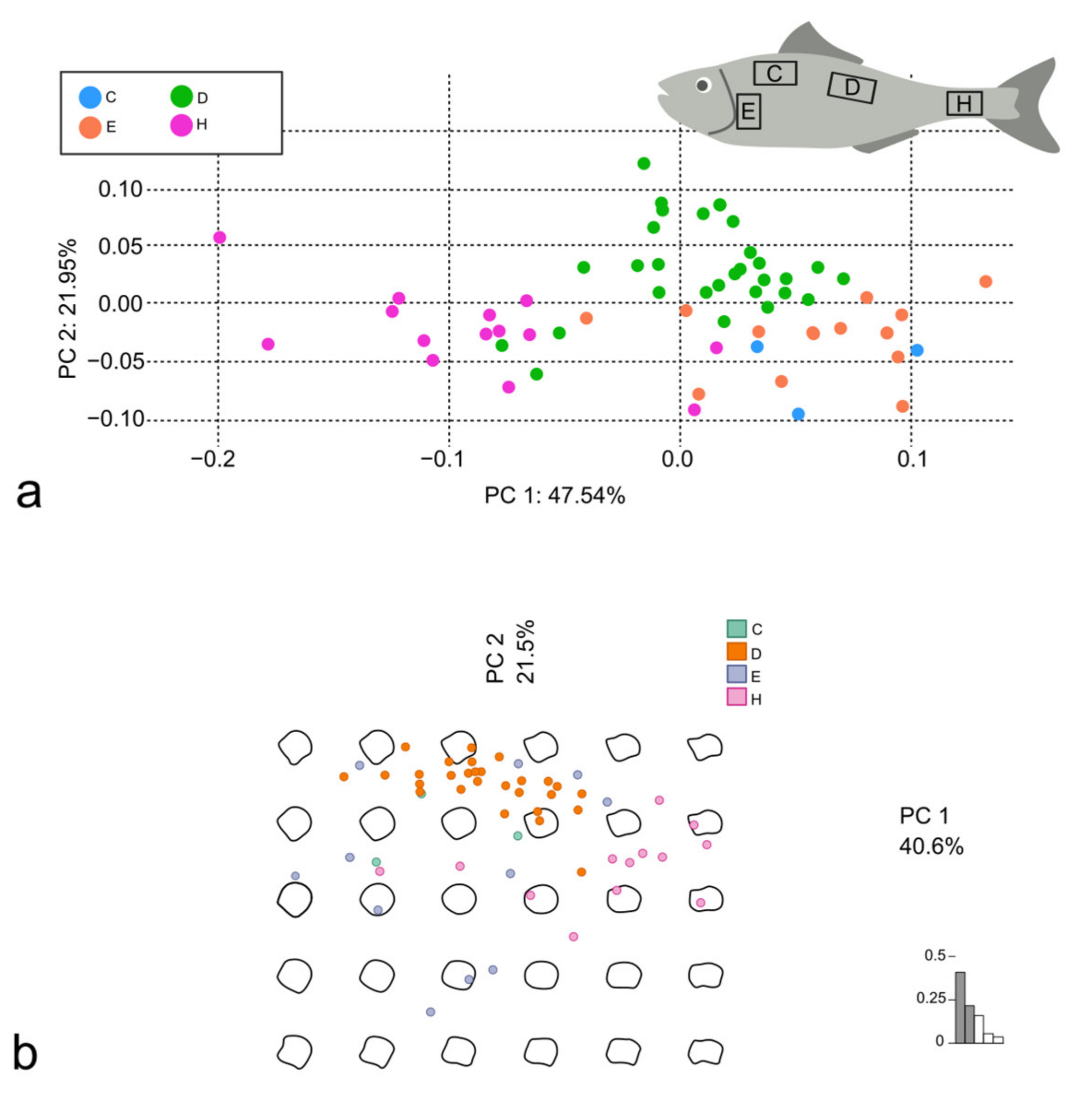
3.1. Discrimination of S. aurata from Different Aquaculture Facilities
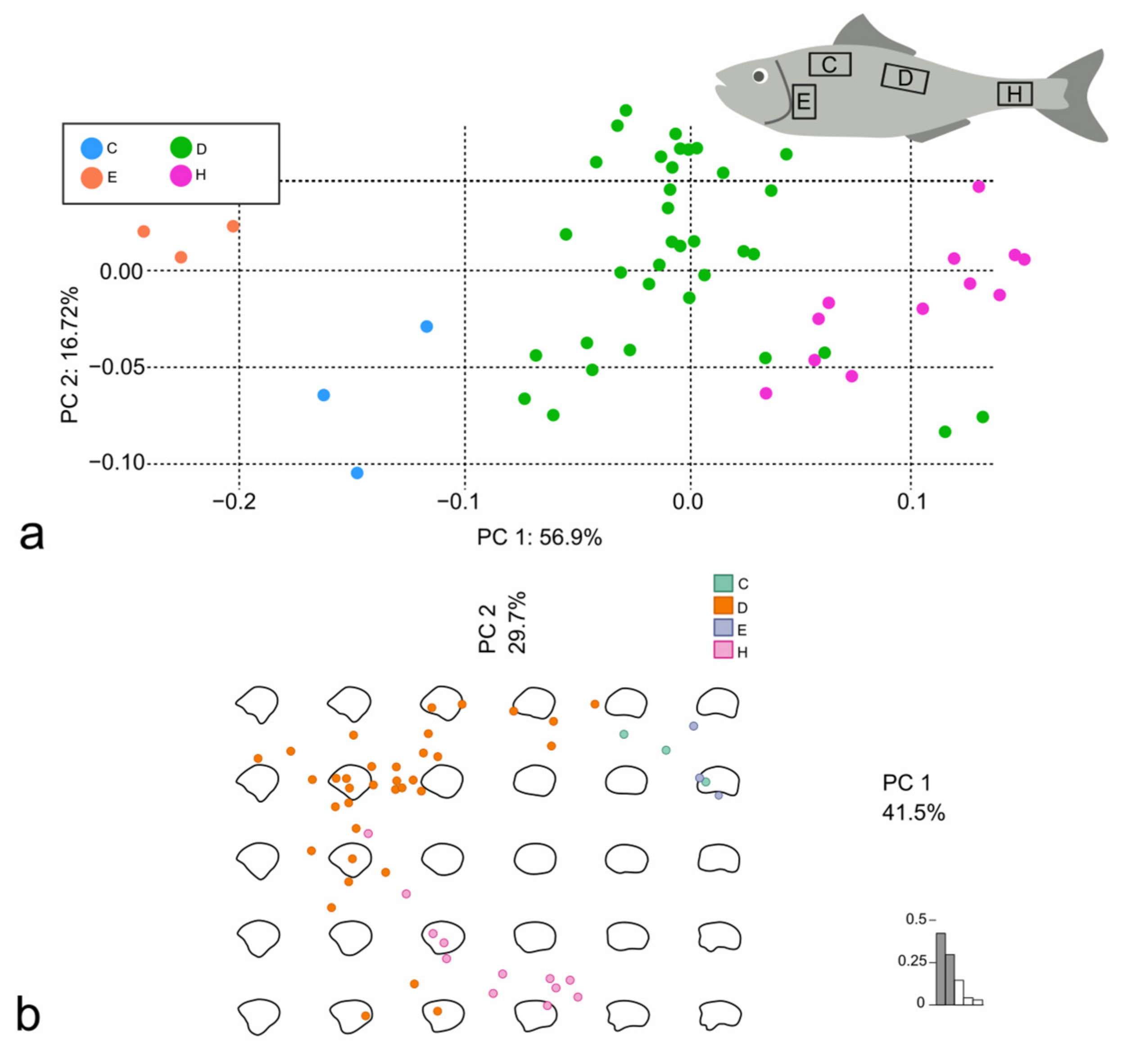
3.2. Discrimination of Different Body Areas in D. labrax and M. surmuletus
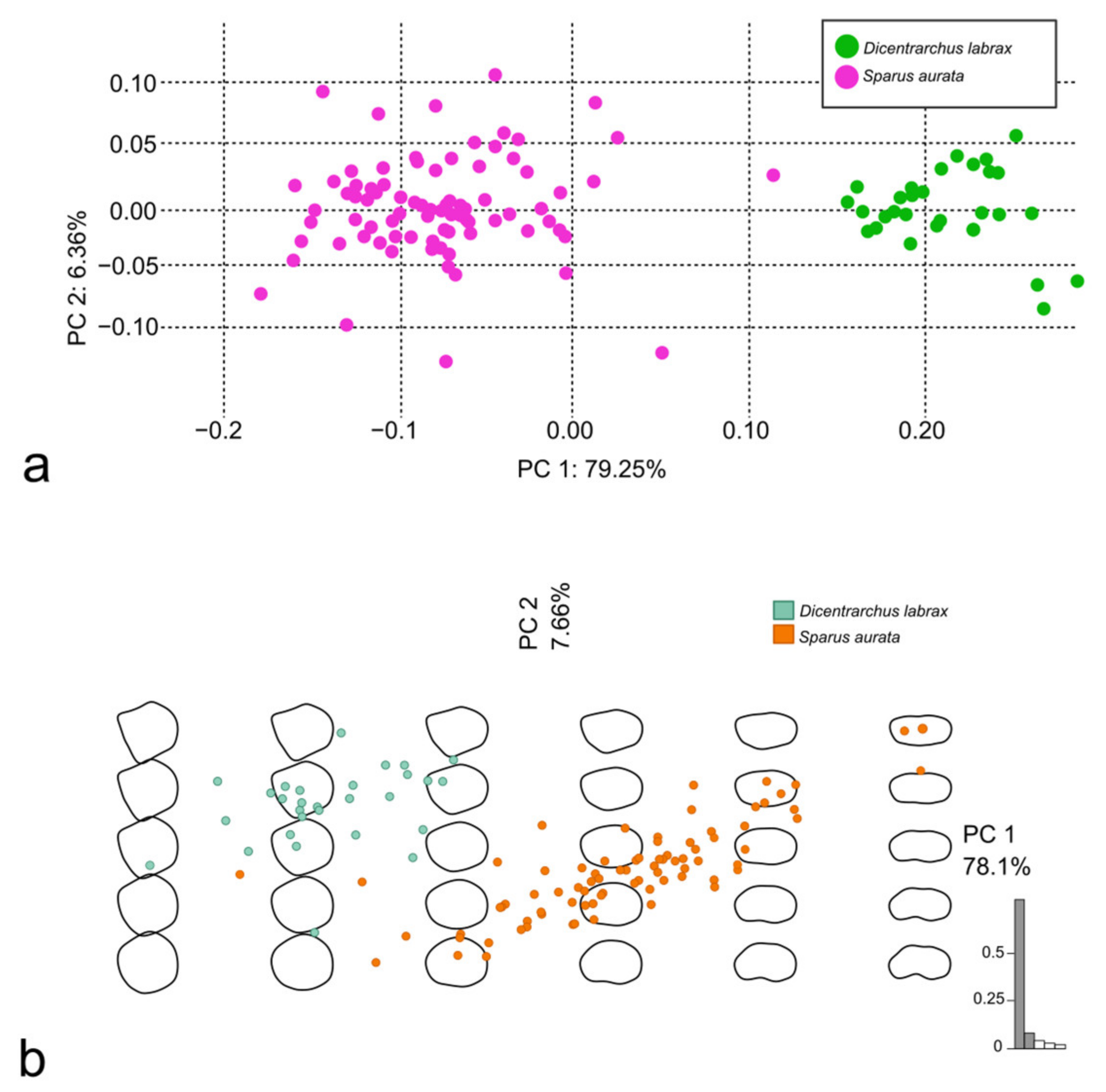
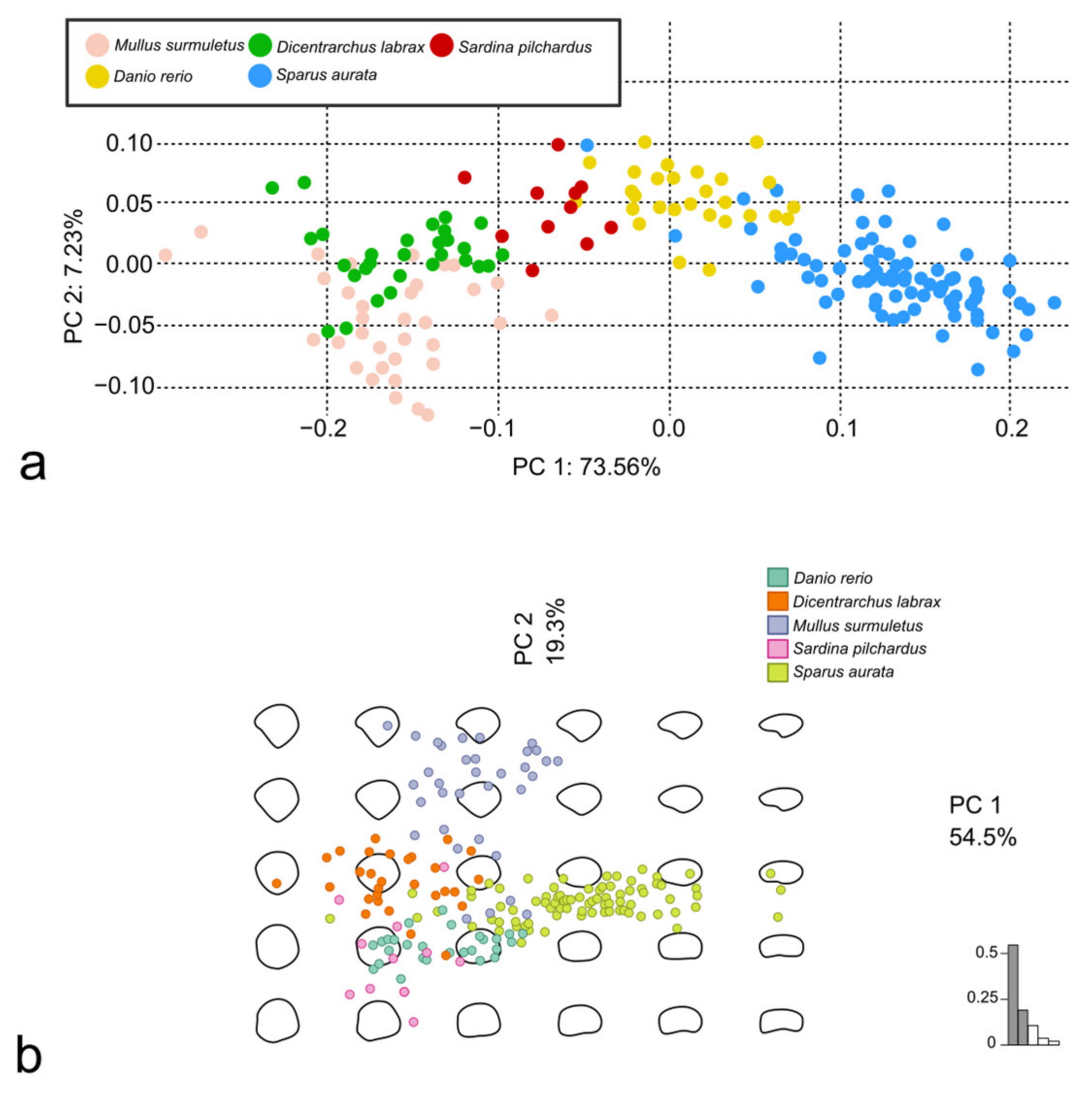
3.3. Discrimination of the Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture; FAO: Rome, Italy, 2022. [Google Scholar]
- De Carvalho, S.C.; Sampaio, I.; Santos, S. DNA Barcoding Reveals Mislabeling and Commercial Fraud in the Marketing of Fillets of the Genus Brachyplatystoma Bleeker, 1862, the Amazonian Freshwater Catfishes Economically Important in Brazil. Heliyon 2020, 6, e04888. [Google Scholar] [CrossRef]
- Dornburg, A.; Near, T.J. The Emerging Phylogenetic Perspective on the Evolution of Actinopterygian Fishes. Annu. Rev. Ecol. Evol. Syst. 2021, 52, 427–452. [Google Scholar] [CrossRef]
- Garman, G.C. Identification of Ingested Prey Fish Based on Scale Characteristics. N. Am. J. Fish. Manag. 1982, 2, 201–203. [Google Scholar] [CrossRef]
- Park, J.M.; Kwak, S.N.; Lee, W.C. Dietary Study Using Set-Nets Produces Bias in Prey Choice of Fish: A Case of Three Coastal Fishes Inhabiting Southern Korean Waters. J. Sea Res. 2020, 157, 101846. [Google Scholar] [CrossRef]
- Da Silveira, E.L.; Semmar, N.; Cartes, J.E.; Tuset, V.M.; Lombarte, A.; Ballester, E.L.C.; Vaz-dos-Santos, A.M. Methods for Trophic Ecology Assessment in Fishes: A Critical Review of Stomach Analyses. Rev. Fish. Sci. Aquac. 2020, 28, 71–106. [Google Scholar] [CrossRef]
- Colley, S.M. The Analysis and Interpretation of Archaeological Fish Remains. Archaeol. Method Theory 1990, 2, 207–253. [Google Scholar]
- Ibáñez, A.L. Fish Traceability: Guessing the Origin of Fish from a Seafood Market Using Fish Scale Shape. Fish. Res. 2015, 170, 82–88. [Google Scholar] [CrossRef]
- Ibáñez, A.L.; Jawad, L.A. Morphometric Variation of Fish Scales among Some Species of Rattail Fish from New Zealand Waters. J. Mar. Biol. Ass. 2018, 98, 1991–1998. [Google Scholar] [CrossRef]
- Çiçek, T.; Kaya, A.; Bilici, S.; Dörtbudak, M.Y. Discrimination of Capoeta Trutta (Heckel, 1843) and Capoeta Umbla (Heckel, 1843) from Scales by Geometric Morphometric Methods. J. Surv. Fish. Sci. 2017, 4, 8–17. [Google Scholar] [CrossRef]
- Bräger, Z.; Staszny, Á.; Mertzen, M.; Moritz, T.; Horváth, G. Fish Scale Identification: From Individual to Species-Specific Shape Variability. Acta Ichthyol. Piscat. 2017, 47, 331–338. [Google Scholar] [CrossRef]
- Šegvić-Bubić, T.; Talijančić, I.; Vulić, L.; Šegvić, B.; Žužul, I.; Radonić, I.; Grubišić, L. Assignment of Gilthead Seabream Sparus aurata to Its Origin through Scale Shape and Microchemistry Composition: Management Implications for Aquaculture Escapees. Water 2020, 12, 3186. [Google Scholar] [CrossRef]
- Staszny, Á.; Ferincz, Á.; Weiperth, A.; Havas, E.; Urbányi, B.; Paulovits, G. Scale-Morphometry Study to Discriminate Gibel Carp (Carassius gibelio) Populations in the Balaton-Catchment (Hungary). Acta Zool. Acad. Sci. Hung. 2012, 58, 19–27. [Google Scholar]
- Garduño-Paz, M.V.; Demetriou, M.; Adams, C.E. Variation in Scale Shape among Alternative Sympatric Phenotypes of Arctic Charr Salvelinus alpinus from Two Lakes in Scotland. J. Fish Biol. 2010, 76, 1491–1497. [Google Scholar] [CrossRef] [PubMed]
- De Pontual, H.; Prouzet, P. Numerical Analysis of Scale Morphology to Discriminate between Atlantic Salmon Stocks. Aquat. Living Resour. 1988, 1, 17–27. [Google Scholar] [CrossRef]
- Dürrani, Ö.; Ateşşahin, T.; Eroğlu, M.; Düşükcan, M. Morphological Variations of an Invasive Cyprinid Fish (Carassius gibelio) in Lentic and Lotic Environments Inferred from the Body, Otolith, and Scale Shapes. Acta Zool. 2023, 104, 458–472. [Google Scholar] [CrossRef]
- Poulet, N.; Reyjol, Y.; Collier, H.; Lek, S. Does Fish Scale Morphology Allow the Identification of Populations at a Local Scale? A Case Study for Rostrum Dace Leuciscus Leuciscus Burdigalensis in River Viaur (SW France). Aquat. Sci. 2005, 67, 122–127. [Google Scholar] [CrossRef]
- Moneva, C.S.O.; Torres, M.A.J.; Tabugo, S.R.; Demayo, C.G. Qualitative and Geometric Morphometric Description of the Shapes of Scales of Viridescent Parrotfish, Calotomus viridescens (Rüppell, 1835). In Proceedings of the Second International Congress on Interdisciplinary Research and Development, Bangkok, Thailand, 31 June 2012; pp. 7.1–7.7. [Google Scholar]
- Albutra, Q.B.; Torres, M.A.J.; Demayo, C.G. Qualitative and Quantitative Analysis of Scale Shape Variation between Different Body Regions of Readbreast Wrasse (Cheilinus fasciatus). Ann. Biol. Sci. 2014, 2, 1–10. [Google Scholar]
- Braeger, Z.; Moritz, T. A Scale Atlas for Common Mediterranean Teleost Fishes. Vertebr. Zool. 2016, 66, 275–386. [Google Scholar] [CrossRef]
- Ibáñez, A.L.; O’Higgins, P. Identifying Fish Scales: The Influence of Allometry on Scale Shape and Classification. Fish. Res. 2011, 109, 54–60. [Google Scholar] [CrossRef]
- Staszny, Á.; Havas, E.; Kovács, R.; Urbányi, B.; Paulovits, G.; Bencsik, D.; Ferincz, Á.; Müller, T.; Specziár, A.; Bakos, K.; et al. Impact of Environmental and Genetic Factors on the Scale Shape of Zebrafish, Danio rerio (Hamilton 1822): A Geometric Morphometric Study. Acta Biol. Hung. 2013, 64, 462–475. [Google Scholar] [CrossRef] [PubMed]
- Bılıcı, S.; Kaya, A.; Cıcek, T.; Dörtbudak, M.Y. Investigation of Size and Shape Differences Depend to Sex, Age and Season on Scales of Smallmouth Lotak (Cyprinion kais). J. Surv. Fish. Sci. 2016, 3, 37–45. [Google Scholar] [CrossRef]
- Pacheco-Almanzar, E.; Loza-Estrada, N.; Ibáñez, A.L. Do the Fish Scales Shape of Mugil Curema Reflect the Genetic Structure Using Microsatellites Markers and the Mexican Marine Ecoregions Classification? Front. Mar. Sci. 2020, 7, 166. [Google Scholar] [CrossRef]
- Șerban, C.; Grigoraş, G. Structural and morphometric study of scales in petzea rudd (Scardinius racovitzai Müller 1958). Appl. Ecol. Environ. Res. 2018, 16, 6063–6076. [Google Scholar] [CrossRef]
- Staszny, Á.; Paulovits, G.; Takács, P.; Juhász, V.; Prigl, S.; Sziráki, B.; Urbányi, B.; Ferincz, Á. The role of ontogenetic development in fish scale shape changes. Appl. Ecol. Environ. Res. 2019, 17, 3535–3544. [Google Scholar] [CrossRef]
- Staszny, A.; Dobosy, P.; Maasz, G.; Szalai, Z.; Jakab, G.; Pirger, Z.; Szeberenyi, J.; Molnar, E.; Pap, L.O.; Juhasz, V.; et al. Effects of Pharmaceutically Active Compounds (PhACs) on Fish Body and Scale Shape in Natural Waters. PeerJ 2021, 9, e10642. [Google Scholar] [CrossRef] [PubMed]
- Takács, P.; Vitál, Z.; Ferincz, Á.; Staszny, Á. Repeatability, Reproducibility, Separative Power and Subjectivity of Different Fish Morphometric Analysis Methods. PLoS ONE 2016, 11, e0157890. [Google Scholar] [CrossRef] [PubMed]
- Albertson, R.C.; Kawasaki, K.C.; Tetrault, E.R.; Powder, K.E. Genetic Analyses in Lake Malawi Cichlids Identify New Roles for Fgf Signaling in Scale Shape Variation. Commun. Biol. 2018, 1, 55. [Google Scholar] [CrossRef] [PubMed]
- Arechavala-Lopez, P.; Sanchez-Jerez, P.; Bayle-Sempere, J.T.; Sfakianakis, D.G.; Somarakis, S. Discriminating Farmed Gilthead Sea Bream Sparus aurata and European Sea Bass Dicentrarchus labrax from Wild Stocks through Scales and Otoliths. J. Fish Biol. 2012, 80, 2159–2175. [Google Scholar] [CrossRef] [PubMed]
- Baken, E.K.; Collyer, M.L.; Kaliontzopoulou, A.; Adams, D.C. Geomorph v4.0 and gmShiny: Enhanced Analytics and a New Graphical Interface for a Comprehensive Morphometric Experience. Methods Ecol. Evol. 2021, 12, 2355–2363. [Google Scholar] [CrossRef]
- Adams, D.C.; Collyer, M.L.; Kaliontzopoulou, A.; Baken, E.K. Geomorph: Software for Geometric Morphometric Analyses, version 4.0.1. 2022. Available online: https://cran.r-project.org/package=geomorph (accessed on 30 March 2024).
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package, version 2.5-7. 2022. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 30 March 2024).
- Harrington, B.; Gould, T.; Hurst, N.; MenTaLgu, Y. Inkscape (version 0.92.2 5c3e80d, 6 August 2017). 2004. Available online: http://inkscape.org/ (accessed on 30 March 2024).
- Bonhomme, V.; Picq, S.; Gaucherel, C.; Claude, J. Momocs: Outline Analysis Using R. J. Stat. Soft. 2014, 56, 1–24. [Google Scholar] [CrossRef]
- Ibañez, A.L.; Cowx, I.G.; O’Higgins, P. Geometric Morphometric Analysis of Fish Scales for Identifying Genera, Species, and Local Populations within the Mugilidae. Can. J. Fish. Aquat. Sci. 2007, 64, 1091–1100. [Google Scholar] [CrossRef]
- Ibañez, A.L.; Cowx, I.G.; O’Higgins, P. Variation in Elasmoid Fish Scale Patterns Is Informative with Regard to Taxon and Swimming Mode. Zool. J. Linn. Soc. 2009, 155, 834–844. [Google Scholar] [CrossRef]
- Requieron, E.A.; Torres, M.A.J.; Demayo, C.G. Applications of Relative Warp Analysis in Describing of Scale Shape Morphology Between Sexes of the Snakehead Fish Channa Striata. Int. J. Biol. Ecol. Environ. Sci. 2012, 1, 205–209. [Google Scholar]
- Grady, J.T.; Bower, L.M.; Gienger, C.M.; Blanton, R.E. Fish Scale Shape Follows Predictable Patterns of Variation Based on Water Column Position, Body Size, and Phylogeny. Evol. Ecol. 2022, 36, 93–116. [Google Scholar] [CrossRef]
- Ibáñez, A.L.; Jawad, L.A.; David, B.; Rowe, D.; Ünlü, E. The Morphometry of Fish Scales Collected from New Zealand and Turkey. N. Z. J. Zool. 2023, 50, 318–328. [Google Scholar] [CrossRef]
- Zelditch, M.L.; Swiderski, D.L.; Sheets, H.D. Geometric Morphometrics for Biologists: A Primer, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Arbour, J.H.; Brown, C.M. Incomplete Specimens in Geometric Morphometric Analyses. Methods Ecol. Evol. 2014, 5, 16–26. [Google Scholar] [CrossRef]
- Gunz, P.; Mitteroecker, P. Semilandmarks: A method for quantifying curves and surfaces. Hystrix Ital. J. Mammal. 2013, 24, 103–109. [Google Scholar] [CrossRef]
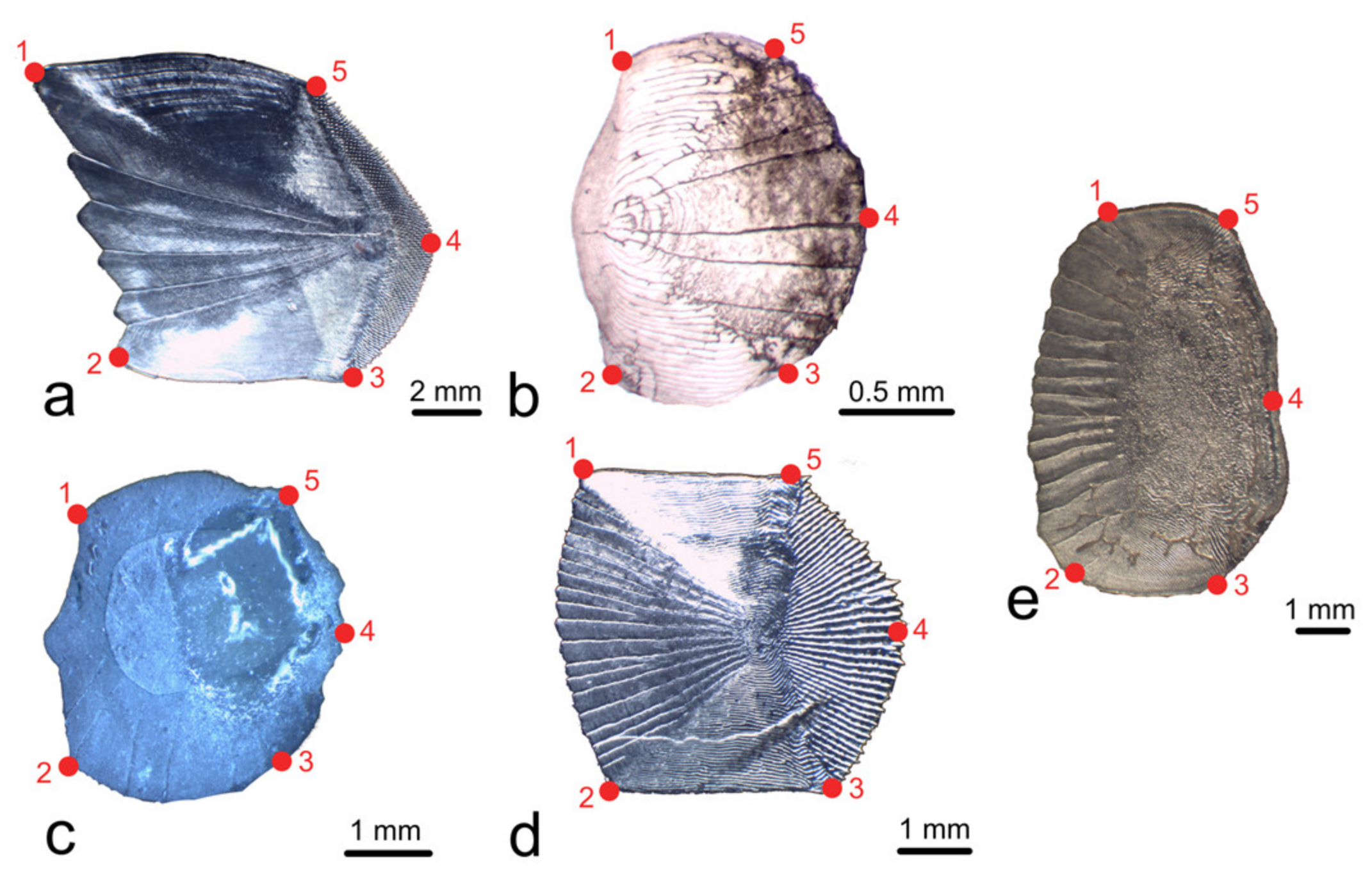

| Species | Specimens | Total Length (cm) | Standard Length (cm) | Source |
|---|---|---|---|---|
| Mullus surmuletus (Linnaeus, 1758) | Mulsur1 | 21.2 | 19.2 | Professional fisheries |
| Mulsur2 | 25.0 | 21.0 | Professional fisheries | |
| Mulsur3 | 22.4 | 18.7 | Professional fisheries | |
| Dicentrarchus labrax (Linnaeus, 1758) | Diclab1 | 34.6 | 29.0 | Professional fisheries |
| Diclab2 | 32.9 | 28.8 | Professional fisheries | |
| Sardina pilchardus (Walbaum, 1792) | Sarpil1 | 12.1 | 10.4 | Professional fisheries |
| Sarpil2 | 11.9 | 10.3 | Professional fisheries | |
| Sarpil3 | 11.5 | 9.9 | Professional fisheries | |
| Sarpil4 | 10.6 | 9.1 | Professional fisheries | |
| Sparus aurata (Linnaeus, 1758) | Spaaur21.1 | 26.5 | 23.9 | Lavagna |
| Spaaur21.2 | 27.4 | 23.5 | Turkey | |
| Spaaur21.3 | 18.6 | 15.6 | Nutritech 1 | |
| Spaaur21.4 | 17.5 | 14.5 | Nutritech 1 | |
| Spaaur21.5 | 19.1 | 16.2 | Nutritech 1 | |
| Spaaur21.6 | 17.8 | 15.4 | Nutritech 1 | |
| Spaaur21.7 | 23.8 | 20.7 | Nutritech 2 | |
| Spaaur21.8 | 23.1 | 19.6 | Nutritech 2 | |
| Spaaur21.9 | 22.4 | 18.6 | Nutritech 2 | |
| Spaaur21.10 | 22.5 | 18.7 | Nutritech 2 | |
| Spaaur21.11 | 23.8 | 19.9 | Nutritech 1 | |
| Spaaur21.12 | 25.2 | 22.1 | Nutritech 1 | |
| Spaaur21.13 | 22.7 | 20.2 | Nutritech 1 | |
| Spaaur21.14 | 27.5 | 24.2 | Nutritech 1 | |
| Spaaur21.15 | 28.5 | 24.7 | Nutritech 1 | |
| Spaaur21.16 | 27.6 | 24.3 | Nutritech 1 | |
| Spaaur21.17 | 30.2 | 26.7 | Nutritech 1 | |
| Spaaur21.18 | 34.2 | 29.1 | Nutritech 1 | |
| Danio rerio (Hamilton, 1822) | 10 specimens | 2.5–3.5 | Not recorded | University of Genoa |
| Species |  | Sampling Area | |||
|---|---|---|---|---|---|
| C | D | E | H | ||
| Mullus surmuletus (Linnaeus, 1758) | 3 | 35 | 3 | 12 | |
| Dicentrarchus labrax (Linnaeus, 1758) | 3 | 29 | 13 | 14 | |
| Sardina pilchardus (Walbaum, 1792) | 11 | ||||
| Sparus aurata (Linnaeus, 1758) | 81 | ||||
| Danio rerio (Hamilton, 1822) | 28 | ||||
| Pairs | LM-Based GM | OL-Based GM | ||
|---|---|---|---|---|
| R2 | Adj. p-Value | R2 | Adj. p-Value | |
| T vs. N1 | 0.03 | 0.534 | 0.04 | 0.642 |
| T vs. N2 | 0.10 | 0.852 | 0.08 | 1.000 |
| T vs. L | 0.25 | 0.816 | 0.62 | 0.234 |
| N1 vs. N2 | 0.02 | 0.726 | 0.05 | 0.234 |
| N1 vs. L | 0.05 | 0.120 | 0.17 | 0.006 ** |
| N2 vs. L | 0.20 | 0.012 * | 0.50 | 0.006 ** |
| Pairs | LM-Based GM | OL-Based GM | LM-Based GM | OL-Based GM | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R2 | Adj. p-Value | R2 | Adj. p-Value | R2 | Adj. p-Value | R2 | Adj. p-Value | |||
| D vs. C | D. labrax | 0.23 | 0.006 ** | 0.08 | 0.222 | M. surmuletus | 0.21 | 0.006 ** | 0.23 | 0.006 ** |
| D vs. E | 0.26 | 0.006 ** | 0.16 | 0.006 ** | 0.33 | 0.006 ** | 0.28 | 0.006 ** | ||
| D vs. H | 0.34 | 0.006 ** | 0.28 | 0.006 ** | 0.25 | 0.006 ** | 0.26 | 0.006 ** | ||
| C vs. E | 0.23 | 0.018 * | 0.05 | 1.000 | 0.54 | 0.600 | 0.40 | 0.600 | ||
| C vs. H | 0.38 | 0.030 * | 0.24 | 0.030 * | 0.69 | 0.012 * | 0.51 | 0.012 * | ||
| E vs. H | 0.45 | 0.006 ** | 0.22 | 0.006 ** | 0.79 | 0.018 * | 0.53 | 0.024 * | ||
| Pairs | LM-Based GM | OL-Based GM | LM-Based GM | OL-Based GM | LM-Based GM | OL-Based GM | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 | p-Value | R2 | p-Value | R2 | Adj. p-Value | R2 | Adj. p-Value | R2 | Adj. p-Value | R2 | Adj. p-Value | ||||
| Dl/Sa | Pairwise | 0.70 | 0.001 *** | 0.48 | 0.001 *** | Altogether | 0.70 | 0.01 * | 0.48 | 0.01 * | Altogether without D. rerio | 0.70 | 0.006 ** | 0.48 | 0.006 ** |
| Dl/Sp | 0.36 | 0.001 *** | 0.30 | 0.001 *** | 0.35 | 0.01 * | 0.30 | 0.01 * | 0.35 | 0.006 ** | 0.30 | 0.006 ** | |||
| Dl/Ms | 0.20 | 0.001 *** | 0.35 | 0.001 *** | 0.19 | 0.01 * | 0.36 | 0.01 * | 0.19 | 0.006 ** | 0.36 | 0.006** | |||
| Dl/Dr | 0.62 | 0.001 *** | 0.37 | 0.001 *** | 0.61 | 0.01 * | 0.37 | 0.01 * | |||||||
| Sa/Sp | 0.38 | 0.001 *** | 0.32 | 0.001 *** | 0.39 | 0.01 * | 0.32 | 0.01 * | 0.39 | 0.006 ** | 0.32 | 0.006 ** | |||
| Sa/Dr | 0.34 | 0.001 *** | 0.36 | 0.001 *** | 0.34 | 0.01 * | 0.36 | 0.01 * | |||||||
| Sa/Ms | 0.70 | 0.001 *** | 0.34 | 0.001 *** | 0.71 | 0.01 * | 0.34 | 0.01 * | 0.70 | 0.006 ** | 0.34 | 0.006 ** | |||
| Sp/Ms | 0.29 | 0.001 *** | 0.34 | 0.001 *** | 0.31 | 0.01 * | 0.34 | 0.01 * | 0.31 | 0.006 ** | 0.34 | 0.006 ** | |||
| Dr/Ms | 0.57 | 0.001 *** | 0.41 | 0.001 *** | 0.59 | 0.01 * | 0.41 | 0.01 * | |||||||
| Dr/Sp | 0.24 | 0.001 *** | 0.12 | 0.001 *** | 0.26 | 0.01 * | 0.12 | 0.01 * | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Traverso, F.; Aicardi, S.; Bozzo, M.; Zinni, M.; Amaroli, A.; Galli, L.; Candiani, S.; Vanin, S.; Ferrando, S. New Insights into Geometric Morphometry Applied to Fish Scales for Species Identification. Animals 2024, 14, 1090. https://doi.org/10.3390/ani14071090
Traverso F, Aicardi S, Bozzo M, Zinni M, Amaroli A, Galli L, Candiani S, Vanin S, Ferrando S. New Insights into Geometric Morphometry Applied to Fish Scales for Species Identification. Animals. 2024; 14(7):1090. https://doi.org/10.3390/ani14071090
Chicago/Turabian StyleTraverso, Francesca, Stefano Aicardi, Matteo Bozzo, Matteo Zinni, Andrea Amaroli, Loris Galli, Simona Candiani, Stefano Vanin, and Sara Ferrando. 2024. "New Insights into Geometric Morphometry Applied to Fish Scales for Species Identification" Animals 14, no. 7: 1090. https://doi.org/10.3390/ani14071090
APA StyleTraverso, F., Aicardi, S., Bozzo, M., Zinni, M., Amaroli, A., Galli, L., Candiani, S., Vanin, S., & Ferrando, S. (2024). New Insights into Geometric Morphometry Applied to Fish Scales for Species Identification. Animals, 14(7), 1090. https://doi.org/10.3390/ani14071090











