Molecular Characterization and Expression Analysis of the C-Type Lectin Domain Family 4 Member F in Litopenaeus vannamei against White Spot Syndrome Virus
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. RNA Extraction and cDNA Synthesis
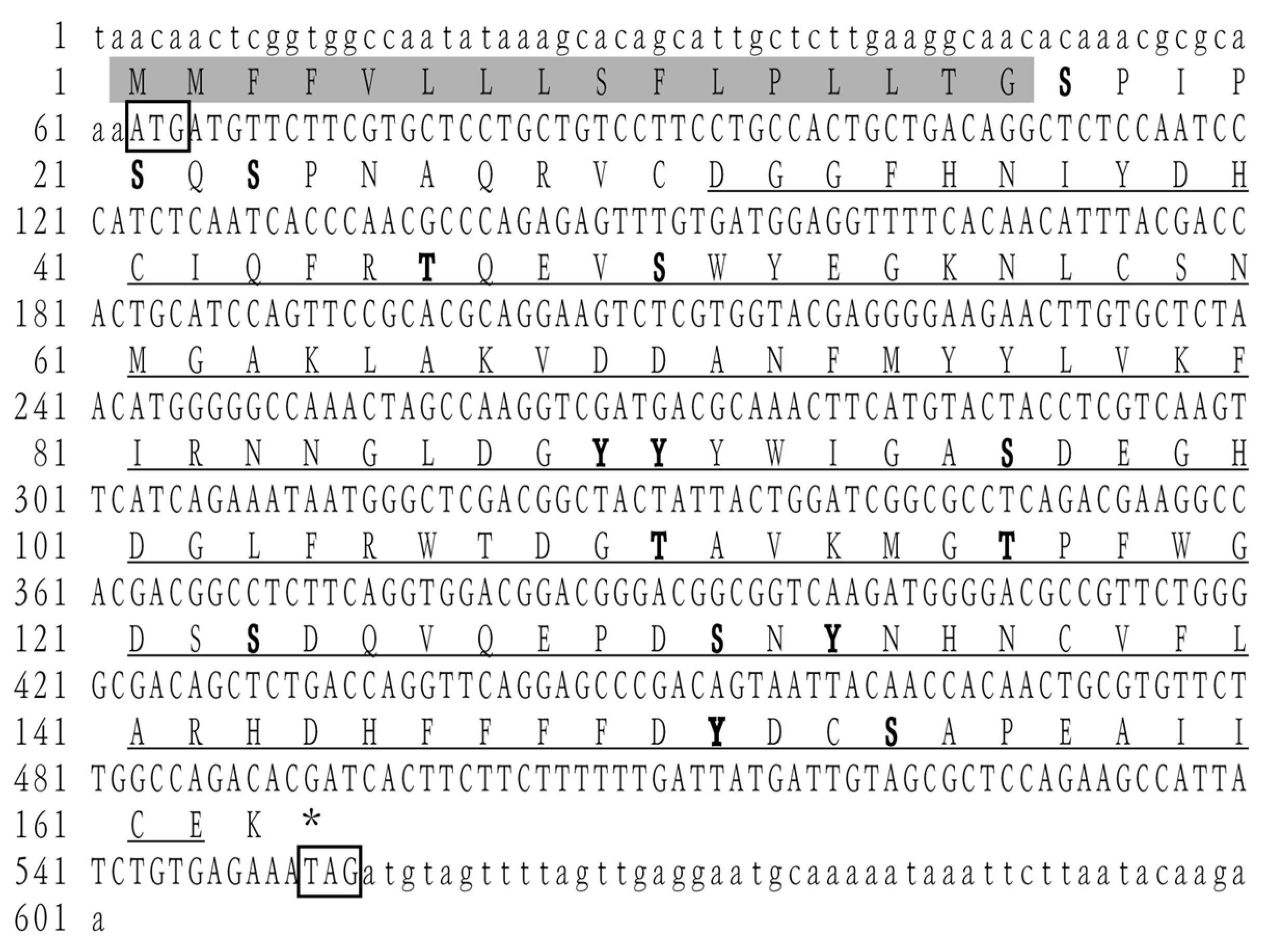
2.3. Molecular Cloning and Sequencing of LvCLEC4F
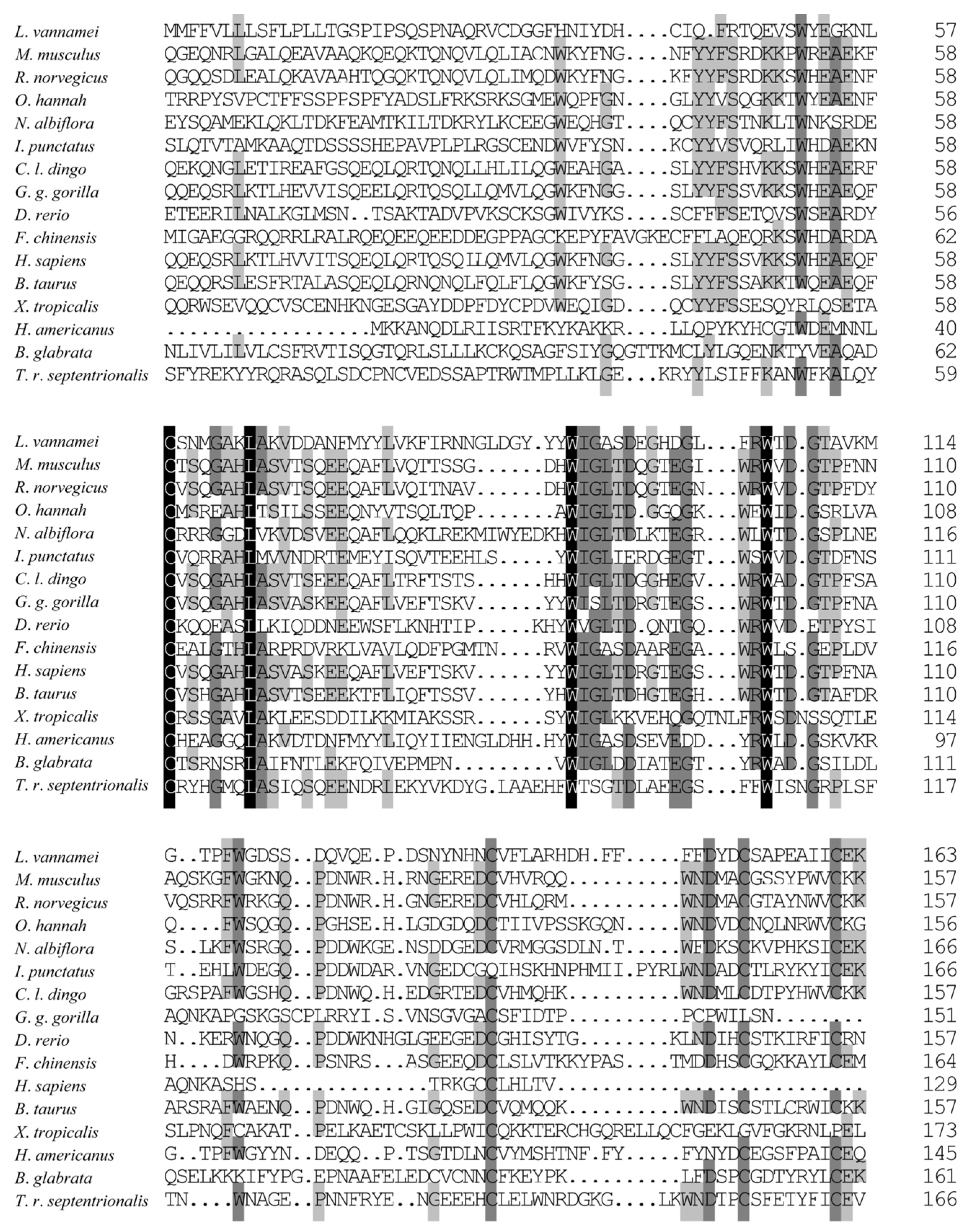
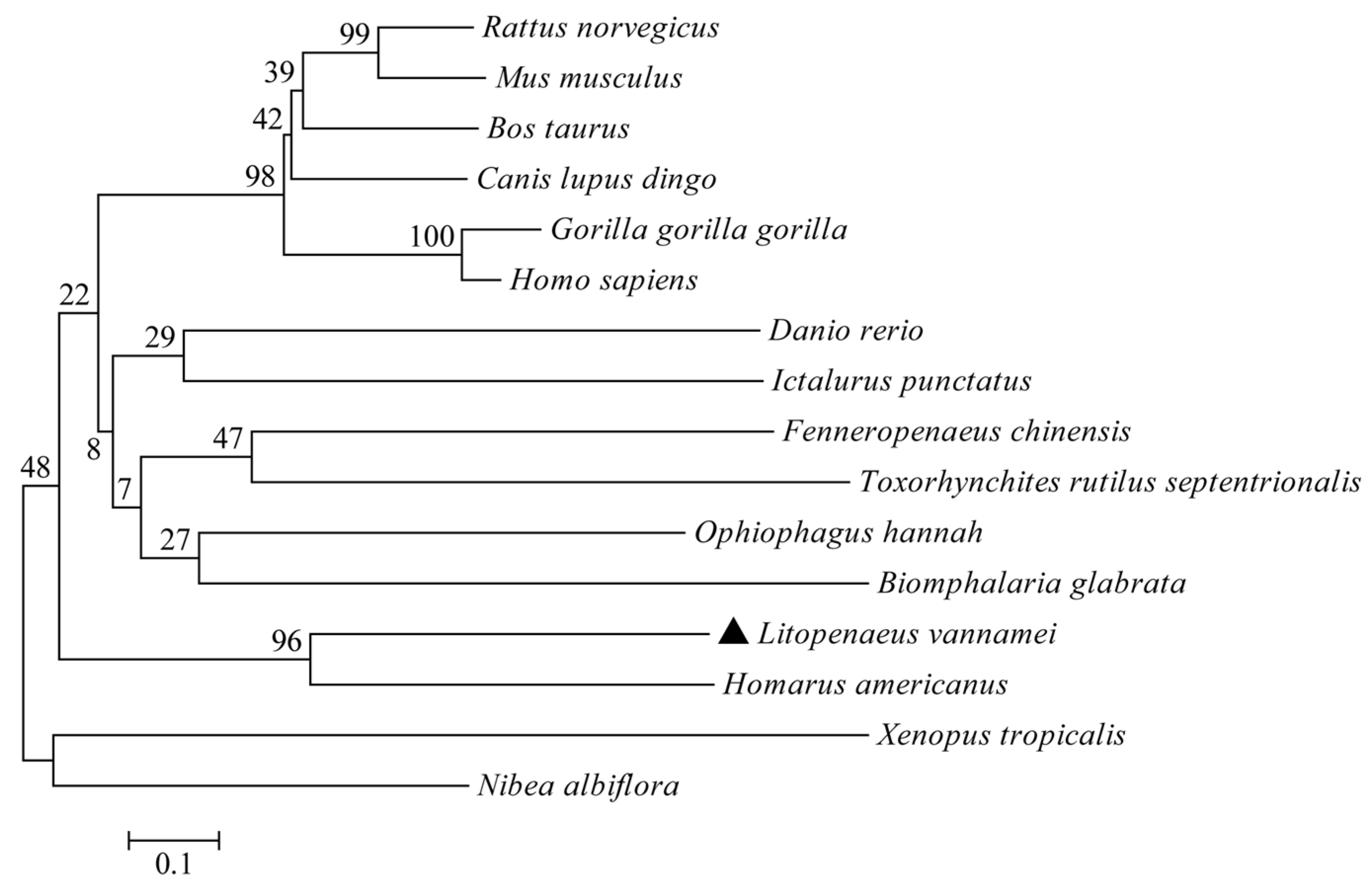
2.4. Bioinformatics Analysis
2.5. WSSV Challenge Experiment
2.6. RT-PCR
2.7. Synthesis of Double-Stranded RNA
2.8. Knockdown of LvCLEC4F by dsRNA
2.9. Survival Rate Analysis after LvCLEC4F Knockdown
2.10. Investigating Immune Response and Gene Expression after LvCLEC4F Knockdown
2.10.1. Experimental Setting
2.10.2. Detection of Hemolymph Agglutination
2.10.3. The WSSV Viral Load Analysis with TaqMan RT-PCR
2.10.4. Gene Expression Analysis
3. Results
3.1. Characterization of LvCLEC4F
3.2. Multiple Alignments and Phylogenetic Analysis
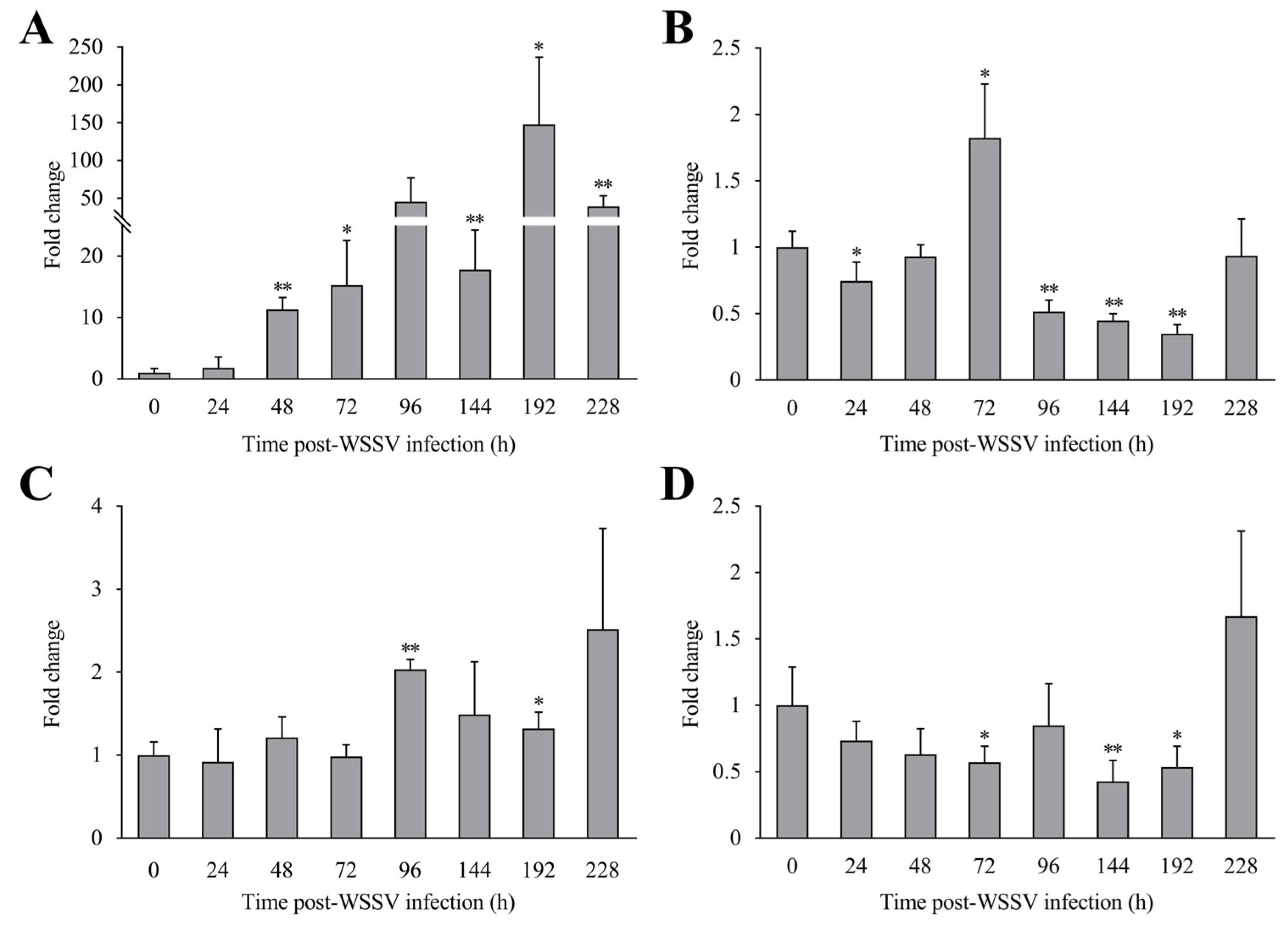
3.3. The Expression Profiles of LvCLEC4F Post-WSSV Infection
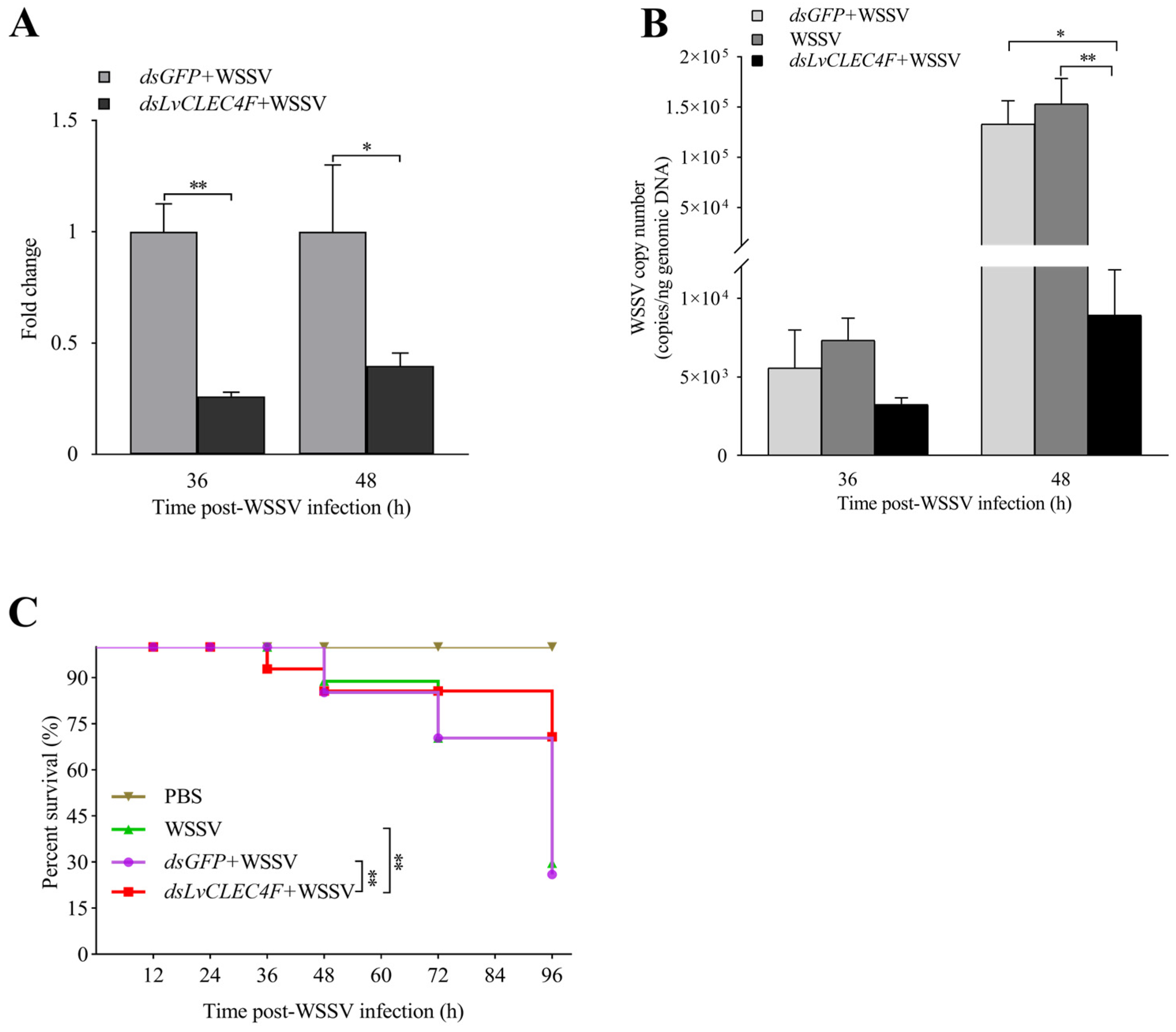
3.4. WSSV Infection Was Suppressed after Knocking down LvCLEC4F
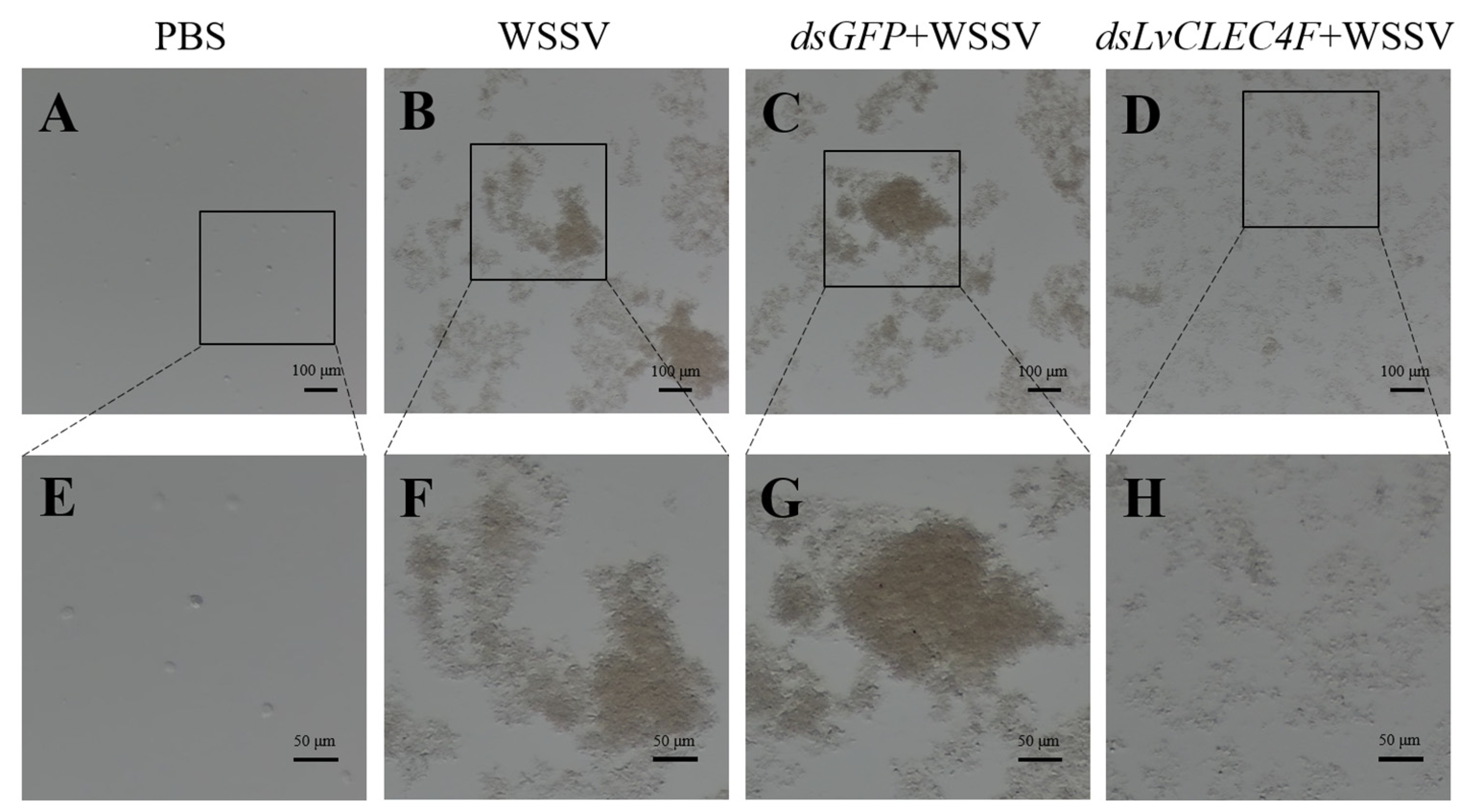
3.5. Hemolymph Agglutination In Vivo Assay
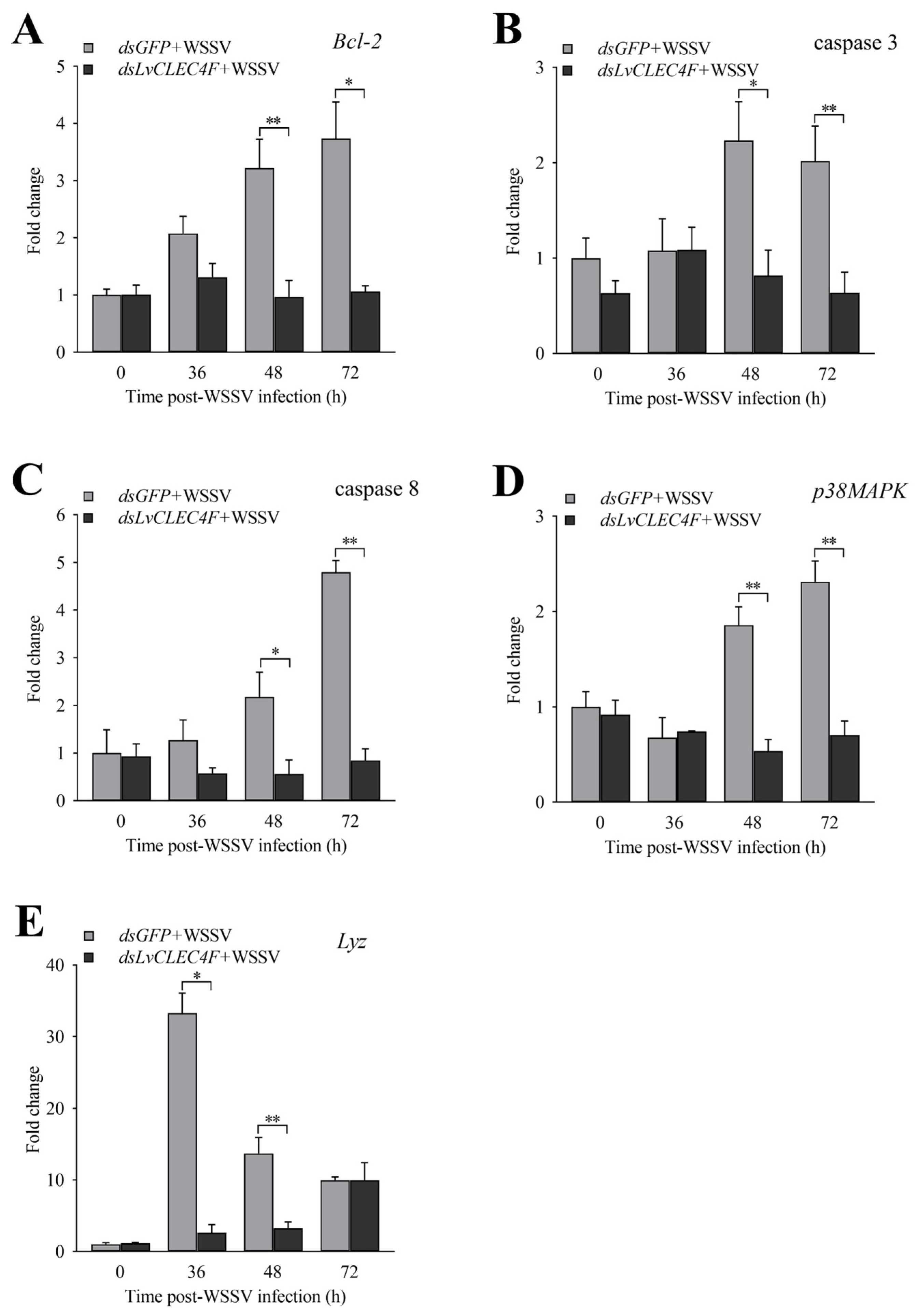
3.6. The Expression Profiles of Genes Related to Innate Immunity after Knocking down LvCLEC4F
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hou, C.; Song, W.; Yuan, H.; Hu, N.; Tan, B.; Zhang, S. Comparative transcriptome analysis revealed that dietary zymosan-A improved the immunity of Penaeus vannamei by regulating the TLR signaling pathway. Aquaculture 2022, 561, 738603. [Google Scholar] [CrossRef]
- Lightner, D.V. Global transboundary disease politics: The OIE perspective. J. Invertebr. Pathol. 2012, 110, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Lightner, D.V. Virus diseases of farmed shrimp in the Western Hemisphere (the Americas): A review. J. Invertebr. Pathol. 2011, 106, 110–130. [Google Scholar] [CrossRef]
- Chen, J.; Li, Z.; Hew, C. Characterization of a novel envelope protein WSV010 of shrimp white spot syndrome virus and its interaction with a major viral structural protein VP24. Virology 2007, 364, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Verbruggen, B.; Bickley, L.; van Aerle, R.; Bateman, K.S.; Stentiford, G.D.; Santos, E.M.; Tyler, C.R. Molecular Mechanisms of White Spot Syndrome Virus Infection and Perspectives on Treatments. Viruses 2016, 8, 23. [Google Scholar] [CrossRef]
- Luo, J.; Chen, Y.; Huang, Y.; Feng, J.; Yuan, Y.; Jian, J.; Cai, S.; Yang, S. A novel C-type lectin for Litopenaeus vannamei involved in the innate immune response against Vibrio infection. Fish Shellfish Immunol. 2023, 135, 108621. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, J.A.; Kafatos, F.C.; Janeway, C.A., Jr.; Ezekowitz, R.A.B. Phylogenetic Perspectives in Innate Immunity. Science 1999, 284, 1313–1318. [Google Scholar] [CrossRef] [PubMed]
- De Gregorio, E.; Spellman, P.T.; Rubin, G.M.; Lemaitr, B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. USA 2001, 98, 12590–12595. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, J. Pattern recognition receptors acting in innate immune system of shrimp against pathogen infections. Fish Shellfish Immunol. 2013, 34, 981–989. [Google Scholar] [CrossRef]
- Sancho, D.; Reis e Sousa, C. Signaling by Myeloid C-type Lectin Receptors in Immunity and Homeostasis. Annu. Rev. Immunol. 2012, 30, 491–529. [Google Scholar] [CrossRef]
- Drickamer, K. Evolution of Ca2+-dependent Animal Lectins. Prog. Nucl. Res. Molec. Biol. 1993, 45, 207–232. [Google Scholar]
- Drickamer, K. Two Distinct Classes of Carbohydrate-recognition Domains in Animal Lectins. J. Biol. Chem. 1988, 263, 9557–9560. [Google Scholar] [CrossRef] [PubMed]
- Rogers, N.C.; Slack, E.C.; Edwards, A.D.; Nolte, M.A.; Schulz, O.; Schweighoffer, E.; Williams, D.L.; Gordon, S.; Tybulewicz, V.L.; Brown, G.D.; et al. Syk-Dependent Cytokine Induction by Dectin-1 Reveals a Novel Pattern Recognition Pathway for C Type Lectins. Immunity 2005, 22, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.C.; Azumi, K.; Nonaka, M. Complement systems in invertebrates. The ancient alternative and lectin pathways. Immunopharmacology 1999, 42, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, S.J.; García-Vallejo, J.J.; van Kooy, Y. Dendritic cells and C-type lectin receptors: Coupling innate to adaptive immune responses. Immunol. Cell Biol. 2008, 86, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, Y.; Xu, J.; Zhao, X.; Wang, J. Collaboration between a soluble C-type lectin and calreticulin facilitates white spot syndrome virus infection in shrimp. J. Immunol. 2014, 193, 2106–2117. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pan, L.; Yu, J. The injection of one recombinant C-type lectin (LvLec) induced the immune response of hemocytes in Litopenaeus vannamei. Fish Shellfish Immunol. 2022, 124, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Yang, L.; Wang, Z.; Zuo, H.; Weng, S.; He, J.; Xu, X. A novel C-type lectin with microbiostatic and immune regulatory functions from Litopenaeus vannamei. Fish Shellfish Immunol. 2019, 93, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Lan, J.; Zhao, X.; Vasta, G.R.; Wang, J. Binding of a C-type lectin’s coiled-coil domain to the Domeless receptor directly activates the JAK/STAT pathway in the shrimp immune response to bacterial infection. PLoS Pathog. 2017, 13, e1006626. [Google Scholar] [CrossRef]
- Huang, Y.H.; Kumar, R.; Liu, C.H.; Lin, S.S.; Wang, H.C. A novel C-type lectin LvCTL 4.2 has antibacterial activity but facilitates WSSV infection in shrimp (L. vannamei). Dev. Comp. Immunol. 2022, 126, 104239. [Google Scholar] [CrossRef]
- Durand, S.V.; Lightner, D.V. Quantitative real time PCR for the measurement of white spot syndrome virus in shrimp. J. Fish Dis. 2002, 25, 381–389. [Google Scholar] [CrossRef]
- Chen, D.; Meng, X.; Xu, J.; Yu, J.; Meng, M.; Wang, J. PcLT, a novel C-type lectin from Procambarus clarkii, is involved in the innate defense against Vibrio alginolyticus and WSSV. Dev. Comp. Immunol. 2013, 39, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Li, D.; Zhang, M.; Yang, H.; Ruan, L.; Xu, X. Cloning and characterization of three novel WSSV recognizing lectins from shrimp Marsupenaeus japonicus. Fish Shellfish Immunol. 2010, 28, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, H.; Li, X.; Zhao, X.; Wang, J. Characterization of a C-type lectin (PcLec2) as an upstream detector in the prophenoloxidase activating system of red swamp crayfish. Fish Shellfish Immunol. 2011, 30, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, X.; Sun, C.; Zhao, X.; Wang, J. C-type lectin from red swamp crayfish Procambarus clarkii participates in cellular immune response. Arch. Insect Biochem. Physiol. 2011, 76, 168–184. [Google Scholar] [CrossRef]
- Alenton, R.R.R.; Koiwai, K.; Nakamura, R.; Thawonsuwan, J.; Kondo, H.; Hirono, I. A Hint of Primitive Mucosal Immunity in Shrimp through Marsupenaeus japonicus Gill C-Type Lectin. J. Immunol. 2019, 203, 2310–2318. [Google Scholar] [CrossRef]
- Mahla, R.S.; Reddy, M.C.; Prasad, D.V.; Kumar, H. Sweeten PAMPs: Role of Sugar Complexed PAMPs in Innate Immunity and Vaccine Biology. Front. Immunol. 2013, 4, 248. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, W.; Wang, X.; Mu, Y.; Zhao, X.; Yu, X.; Wang, J. A novel C-type lectin with two CRD domains from Chinese shrimp Fenneropenaeus chinensis functions as a pattern recognition protein. Mol. Immunol. 2009, 46, 1626–1637. [Google Scholar] [CrossRef]
- Jaree, P.; Somboonwiwat, K. DnaJC16, the molecular chaperone, is implicated in hemocyte apoptosis and facilitates of WSSV infection in shrimp. Fish Shellfish Immunol. 2023, 137, 108770. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, H.; Zhu, J.; Liu, H.; Yang, Y.; Sun, B.; Wu, T.; Zhang, Y.; Yao, D. The transcription factor CEBP homolog of Penaeus vannamei contributes to WSSV replication. Fish Shellfish Immunol. 2023, 134, 108571. [Google Scholar] [CrossRef]
- Liang, Z.; Yang, L.; Zheng, J.; Zuo, H.; Weng, S.; He, J.; Xu, X. A low-density lipoprotein receptor (LDLR) class A domain-containing C-type lectin from Litopenaeus vannamei plays opposite roles in antibacterial and antiviral responses. Dev. Comp. Immunol. 2019, 92, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Dostert, C.; Jouanguy, E.; Irving, P.; Troxler, L.; Galiana-Arnoux, D.; Hetru, C.; Hoffmann, J.A.; Imler, J.L. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat. Immunol. 2005, 6, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Huang, Y.; Wei, S.; Huang, X.; Ye, F.; Fu, J.; Qin, Q. Characterization of p38 MAPKs from orange-spotted grouper, Epinephelus coioides involved in SGIV infection. Fish Shellfish Immunol. 2011, 31, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yao, W.; Liu, P.; Li, J.; Wang, Q. Expression profiles of the p38 MAPK signaling pathway from Chinese shrimp Fenneropenaeus chinensis in response to viral and bacterial infections. Gene 2018, 642, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Leu, J.H.; Lin, S.J.; Huang, J.Y.; Chen, T.C.; Lo, C.F. A model for apoptotic interaction between white spot syndrome virus and shrimp. Fish Shellfish Immunol. 2013, 34, 1011–1017. [Google Scholar] [CrossRef] [PubMed]
- Janewanthanakul, S.; Supungul, P.; Tang, S.; Tassanakajon, A. Heat shock protein 70 from Litopenaeus vannamei (LvHSP70) is involved in the innate immune response against white spot syndrome virus (WSSV) infection. Dev. Comp. Immunol. 2020, 102, 103476. [Google Scholar] [CrossRef]
- Huang, X.; Li, W.; Jin, M.; Ma, F.T.; Huang, Y.; Shi, Y.R.; Zhao, L.L.; Feng, J.L.; Ren, Q.; Wang, W. Single CRD containing lectin from Macrobrachium rosenbergii (MrLec) participates in innate immunity against pathogen infections. Fish Shellfish Immunol. 2016, 51, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Li, W.; Meng, L.; Sha, Z.X.; Wang, Z.J.; Ren, G.C. Molecular cloning and expression analysis of a hepcidin antimicrobial peptide gene from turbot (Scophthalmus maximus). Fish Shellfish Immunol. 2007, 22, 172–181. [Google Scholar] [CrossRef]

| Primer or Probe Name | Sequences (5′–3′) |
|---|---|
| LvCLEC4F-F | CTTGAAGGCAACACAAACGC |
| LvCLEC4F-R | TTTGCATTCCTCAACTAAAACTACA |
| qLvCLEC4F-F | TTCGTGCTCCTGCTGTCCTT |
| qLvCLEC4F-R | CAAACTCTCTGGGCGTTGGGT |
| 18S-F | TATACGCTAGTGGAGCTGGAA |
| 18S-R | GGGGAGGTAGTGACGAAAAAT |
| WSSV-F | TGGTCCCGTCCTCATCTCAG |
| WSSV-R | GCTGCCTTGCCGGAAATTA |
| WSSV-probe | AGCCATGAAGAATGCCGTCTATCACACA |
| Bcl-2-F | GCTATGTGTCCTTTGTGGCT |
| Bcl-2-R | TGAACTTGGCAATGGTAACTG |
| caspase3-F | AGTTAGTACAAACAGATTGGAGCG |
| caspase3-R | CGGTCCTTGTGGACAGACAG |
| caspase 8-F | GGCGACAAGATGAGGCAA |
| caspase 8-R | CAGGGTGAGGGAGAGAAAACT |
| p38MAPK-F | GTCGGCTCGCAACTACATAC |
| p38MAPK-R | CCGTTACACGCCTTTCACT |
| Lyz-F | ACTGGTGCGGAAGCGACTA |
| Lyz-R | GGCGGATAGTCTCGGCG |
| dsGFP-Fi | GCGTAATACGACTCACTATAGGCATCTTCTTCAAGGACGACGG |
| dsGFP-Ri | GCGTAATACGACTCACTATAGGAGTTCACCTTGATGCCGTTCT |
| LvCLEC4F-Fi | GCGTAATACGACTCACTATAGGCGCGCAAAATGATGTTCTTCGT |
| LvCLEC4F-Ri | GCGTAATACGACTCACTATAGGTGGAGCGCTACAATCATAATCAAA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, Q.; Yang, B.; Luo, K.; Luan, S.; Kong, J.; Li, X.; Meng, X. Molecular Characterization and Expression Analysis of the C-Type Lectin Domain Family 4 Member F in Litopenaeus vannamei against White Spot Syndrome Virus. Animals 2024, 14, 1137. https://doi.org/10.3390/ani14081137
Xue Q, Yang B, Luo K, Luan S, Kong J, Li X, Meng X. Molecular Characterization and Expression Analysis of the C-Type Lectin Domain Family 4 Member F in Litopenaeus vannamei against White Spot Syndrome Virus. Animals. 2024; 14(8):1137. https://doi.org/10.3390/ani14081137
Chicago/Turabian StyleXue, Qian, Bingbing Yang, Kun Luo, Sheng Luan, Jie Kong, Xupeng Li, and Xianhong Meng. 2024. "Molecular Characterization and Expression Analysis of the C-Type Lectin Domain Family 4 Member F in Litopenaeus vannamei against White Spot Syndrome Virus" Animals 14, no. 8: 1137. https://doi.org/10.3390/ani14081137





