Effect of High Energy Low Protein Diet on Lipid Metabolism and Inflammation in the Liver and Abdominal Adipose Tissue of Laying Hens
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Treatments
2.2. Sample Collection and Measurement
2.3. Morphological and Histological Observation of the Liver and AATs
2.4. Ultrastructural Observation of the Liver and AATs
2.5. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
2.6. Determination of Biochemical Parameters
2.7. Statistical Analysis
3. Results
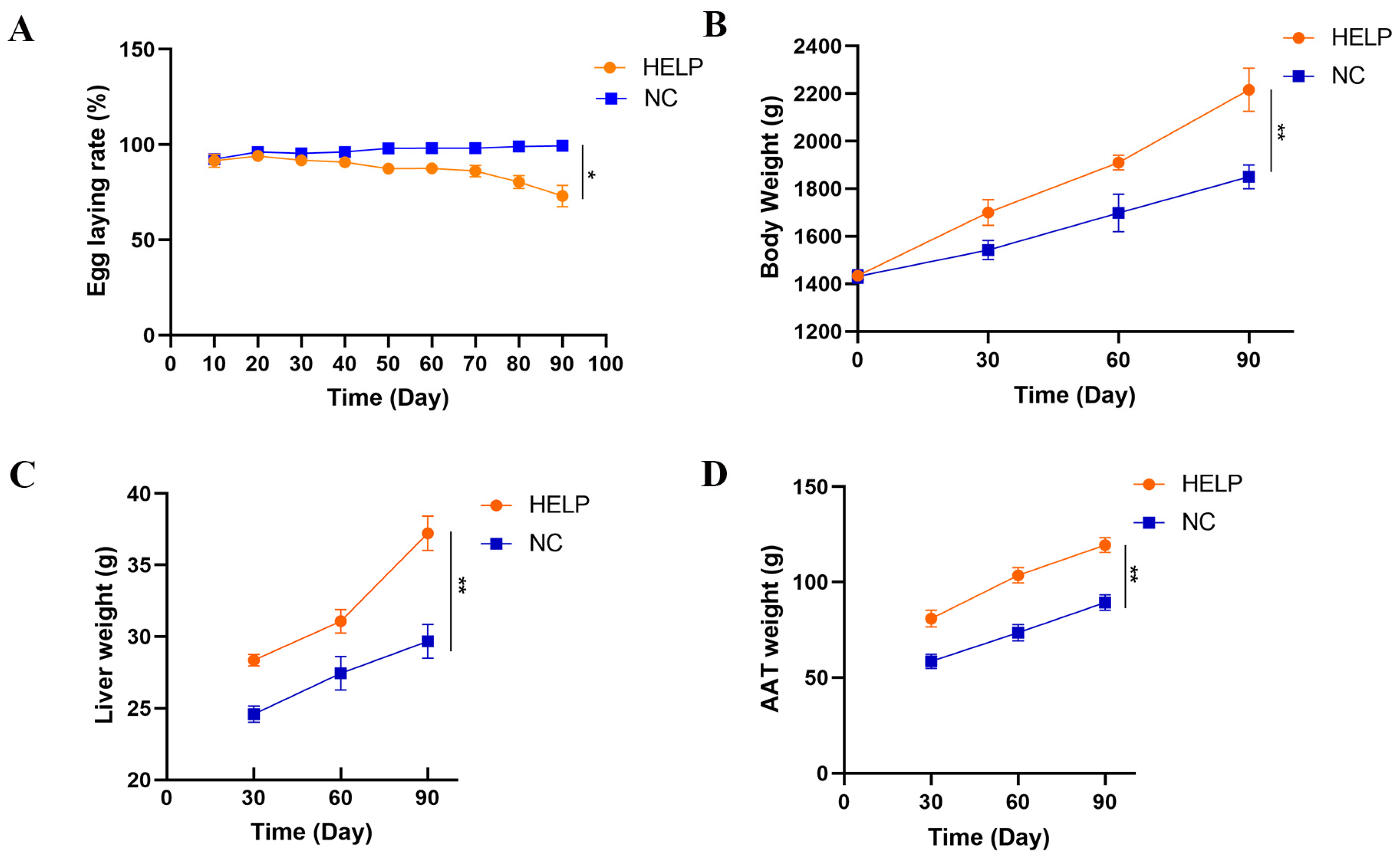
3.1. Production Performance and Organ Weights
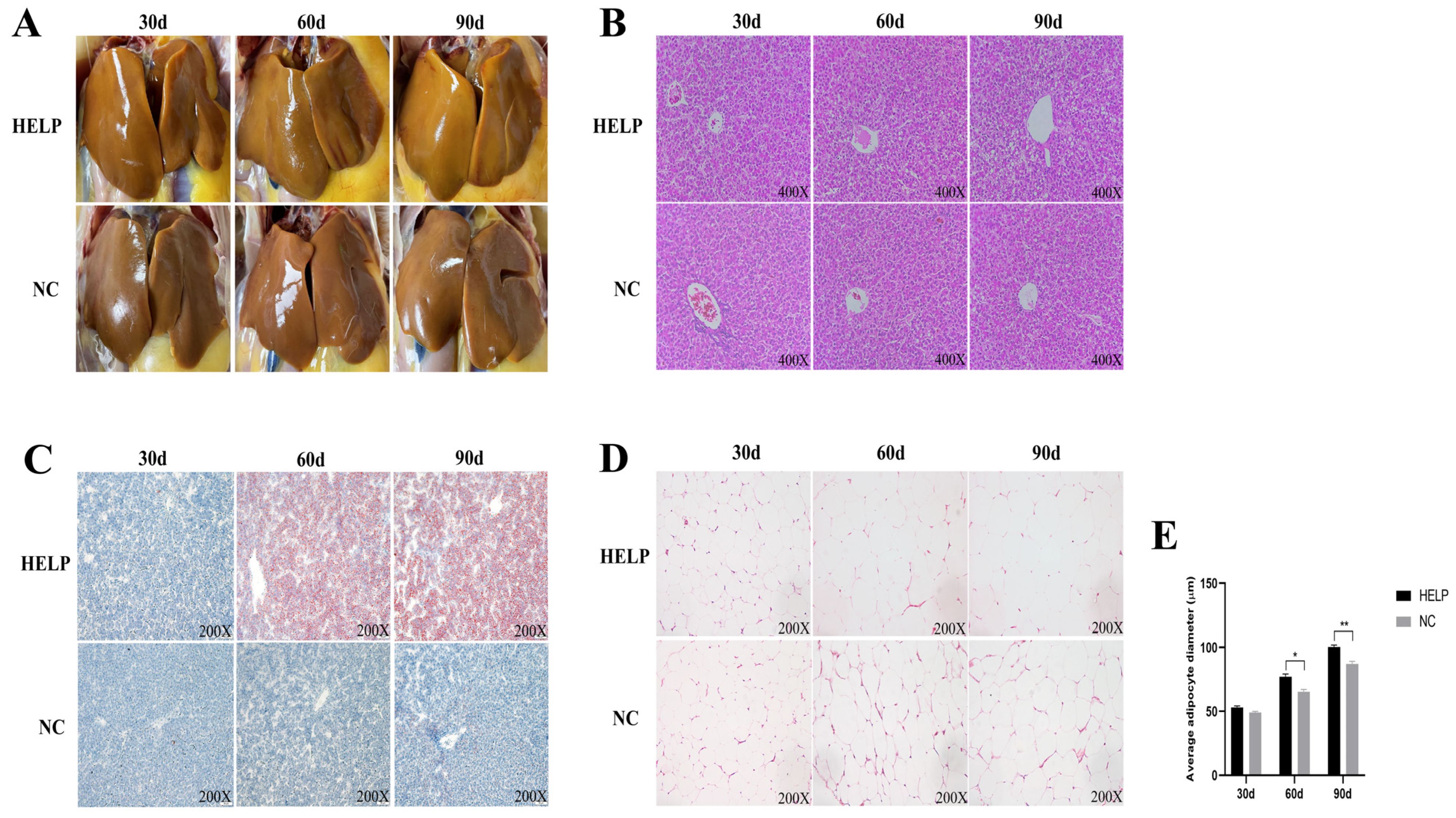
3.2. Morphological and Histological Observations of the Chicken Liver and AAT
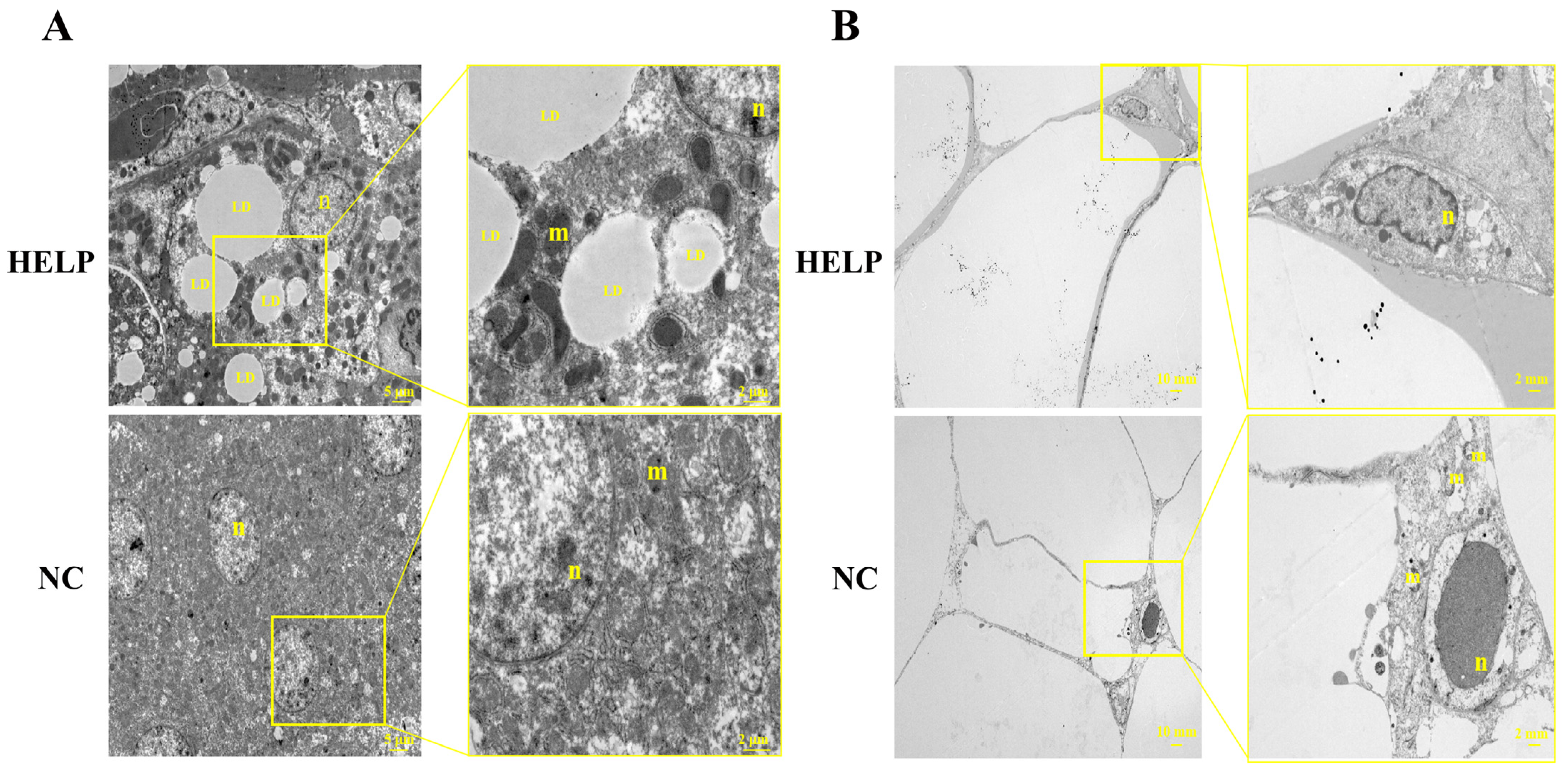
3.3. Ultrastructural Observation of the Chicken Liver and AAT
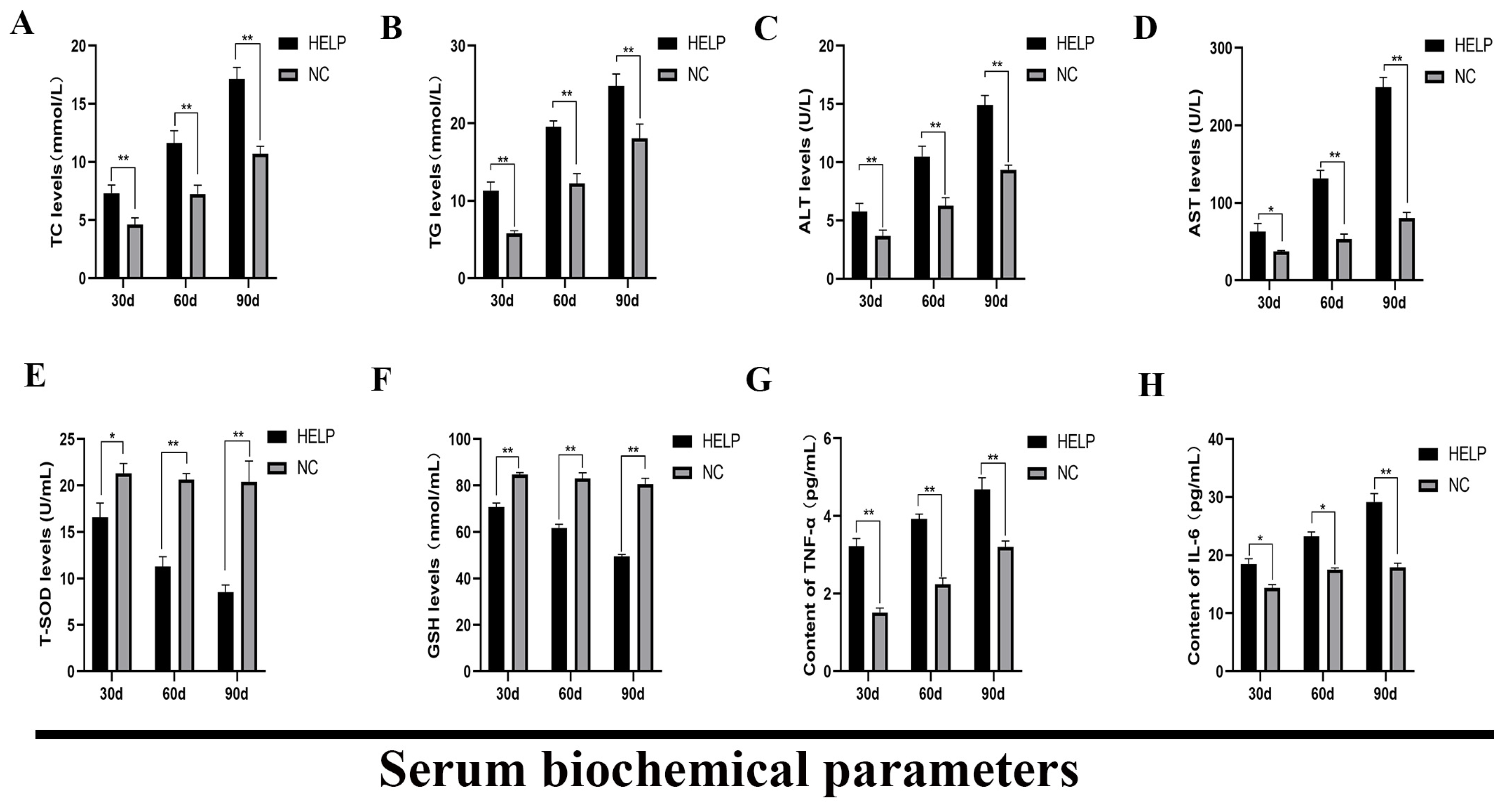
3.4. Determination of the Serum Biochemical Parameters
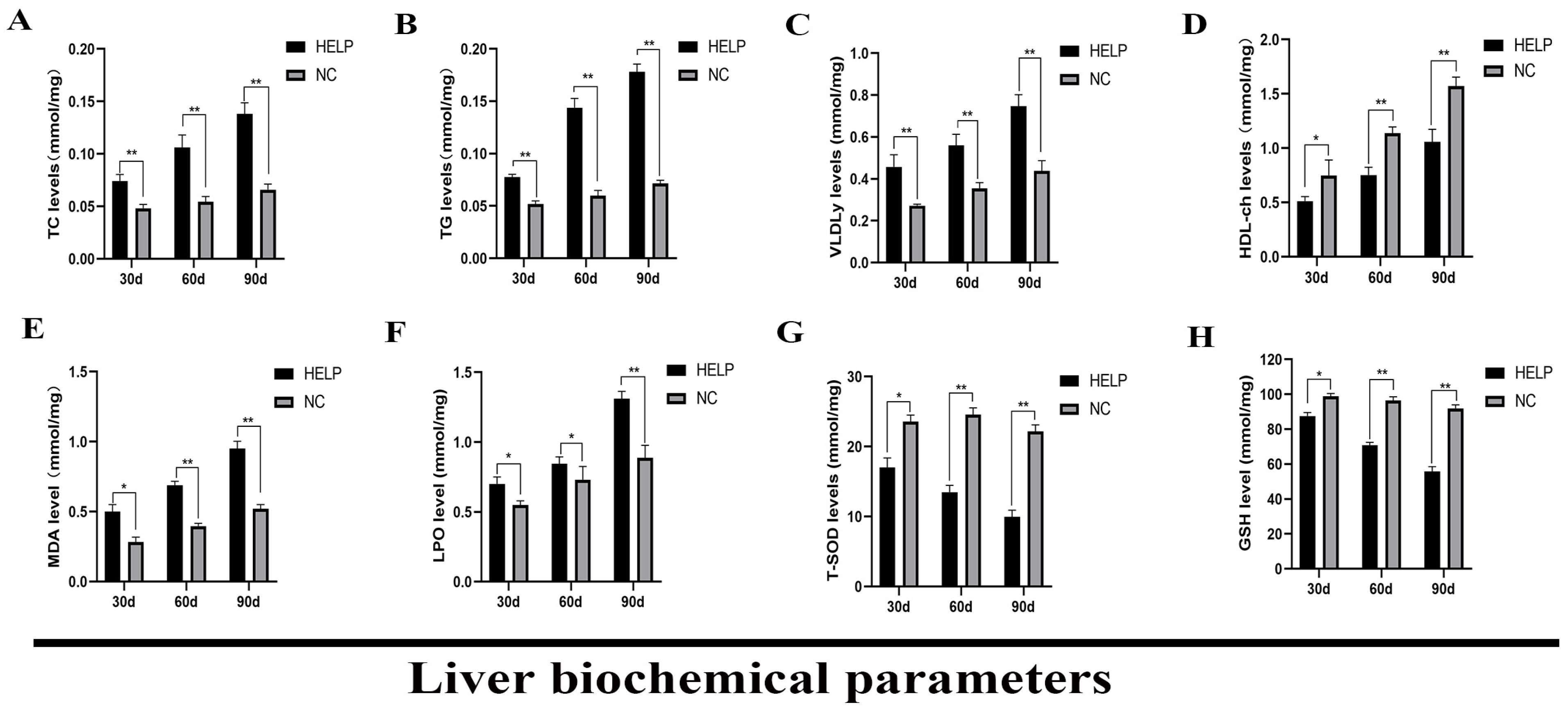
3.5. Determination of Liver Biochemical Parameters
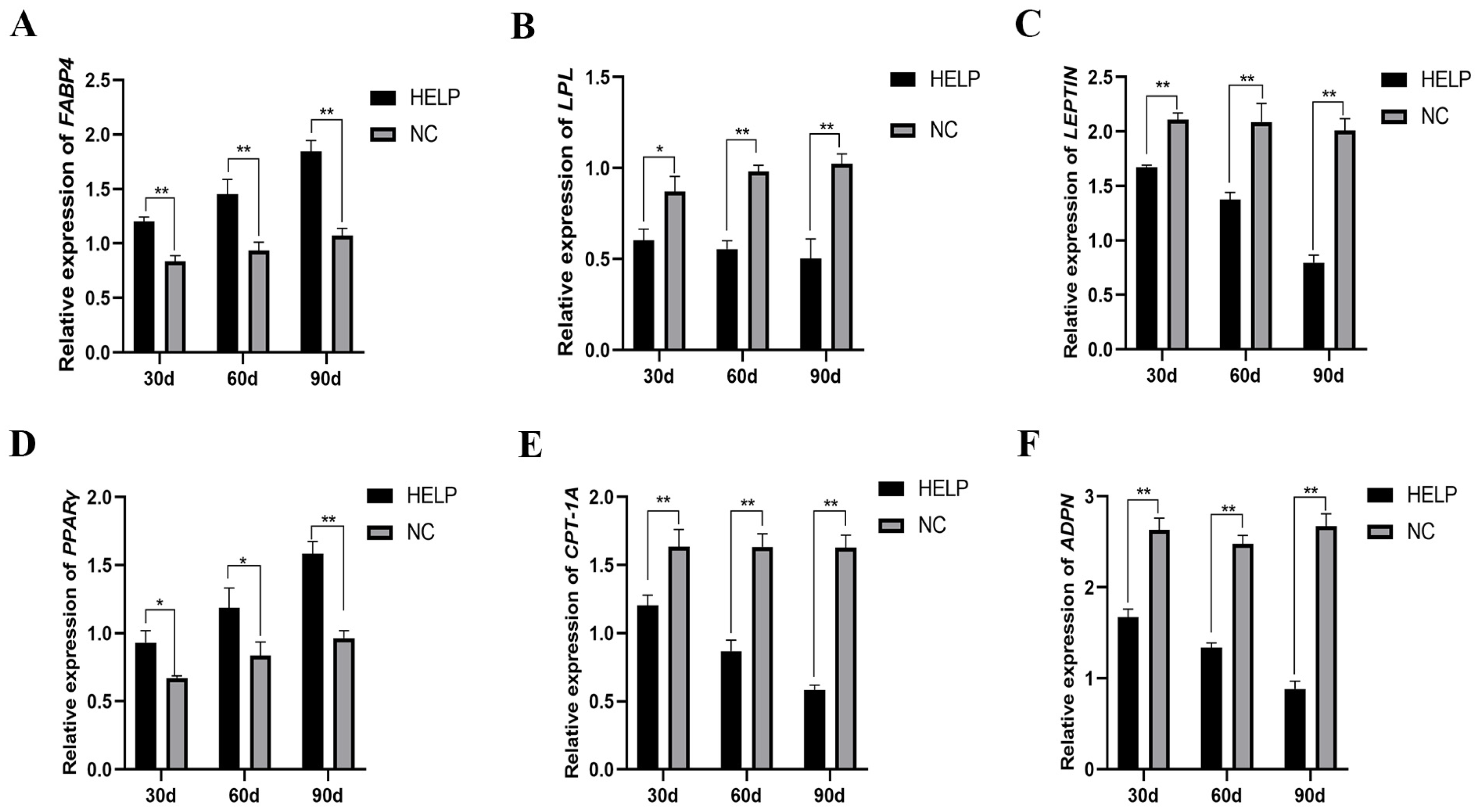
3.6. Expression Abundance of Genes Related to Lipid Metabolism in the Liver
3.7. Expression Abundance of Genes Related to Lipid Metabolism and Adipokine Synthesis in the AAT
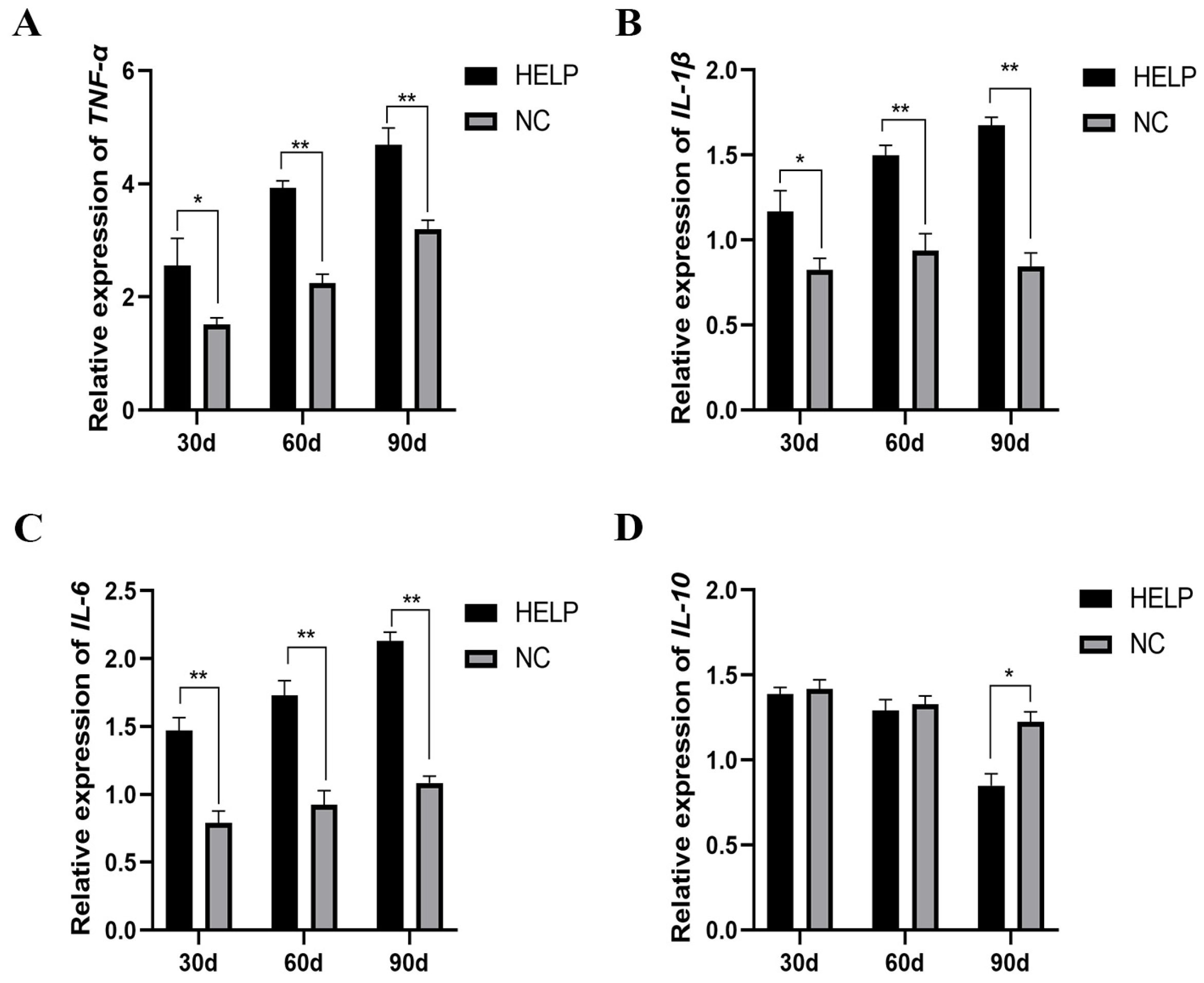
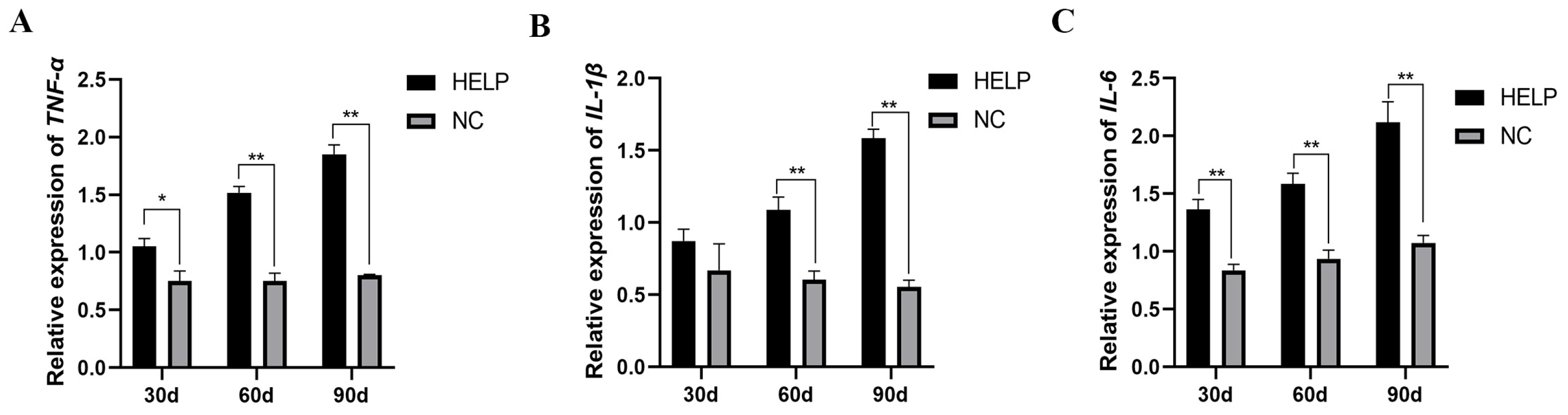
3.8. Expression Abundance of Inflammation-Related Genes in the Liver and AAT
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harms, R.H.; Arafa, A.S.; Simpson, C.F.; Miles, R.D. Influence of Protamone on fatty liver syndrome in commercial laying hens. Poult. Sci. 1982, 61, 2370–2374. [Google Scholar] [CrossRef]
- O’Hea, E.K.; Leveille, G.A. Lipid biosynthesis and transport in the domestic chick (Gallus domesticus). Comp. Biochem. Physiol. 1969, 30, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Kartsoli, S.; Kostara, C.E.; Tsimihodimos, V.; Bairaktari, E.T.; Christodoulou, D.K. Lipidomics in non-alcoholic fatty liver disease. World J. Hepatol. 2020, 12, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, Z.; Zhang, S.; Zhang, H.; Teng, X. miR-187-5p/apaf-1 axis was involved in oxidative stress-mediated apoptosis caused by ammonia via mitochondrial pathway in chicken livers. Toxicol. Appl. Pharmacol. 2020, 388, 114869. [Google Scholar] [CrossRef]
- Simon, J.; Leroith, D. Insulin receptors of chicken liver and brain. Characterization of alpha and beta subunit properties. Eur. J. Biochem. 1986, 158, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Huang, E.; Ruan, J.; Huang, L.; Liang, H.; Wei, Q.; Xie, X.; Zeng, Q.; Huang, J. Effects of a high energy and low protein diet on hepatic and plasma characteristics and Cidea and Cidec mRNA expression in liver and adipose tissue of laying hens with fatty liver hemorrhagic syndrome. Anim. Sci. J. 2019, 90, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.W.; Huang, T.W.; Peng, Y.J.; Lin, Y.Y.; Mersmann, H.J.; Ding, S.T. A novel chicken model of fatty liver disease induced by high cholesterol and low choline diets. Poult. Sci. 2021, 100, 100869. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wang, H.; Yang, Y.; Jiang, Z.; Ma, H. Dehydroepiandrosterone activates the GPER-mediated AMPK signaling pathway to alleviate the oxidative stress and inflammatory response in laying hens fed with high-energy and low-protein diets. Life Sci. 2022, 308, 120926. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, Y.; Chen, J.; Dai, X.; Xing, C.; Zhang, C.; Cao, H.; Guo, X.; Hu, G.; Zhuang, Y. Berberine Protects against High-Energy and Low-Protein Diet-Induced Hepatic Steatosis: Modulation of Gut Microbiota and Bile Acid Metabolism in Laying Hens. Int. J. Mol. Sci. 2023, 24, 17304. [Google Scholar] [CrossRef]
- Tseng, Y.H. Adipose tissue in communication: Within and without. Nat. Rev. Endocrinol. 2023, 19, 70–71. [Google Scholar] [CrossRef]
- Tsuji, T.; Tseng, Y.H. Adipose tissue-derived lipokines in metabolism. Curr. Opin. Genet. Dev. 2023, 81, 102089. [Google Scholar] [CrossRef]
- Weschenfelder, C.; Schaan, D.Q.A.; Lorenzon, D.S.J.; Bueno, G.S.; Marcadenti, A. Adipokines and Adipose Tissue-Related Metabolites, Nuts and Cardiovascular Disease. Metabolites 2020, 10, 32. [Google Scholar] [CrossRef]
- Galley, J.C.; Singh, S.; Awata, W.; Alves, J.V.; Bruder-Nascimento, T. Adipokines: Deciphering the cardiovascular signature of adipose tissue. Biochem. Pharmacol. 2022, 206, 115324. [Google Scholar] [CrossRef]
- Engin, A. The Pathogenesis of Obesity-Associated Adipose Tissue Inflammation. Adv. Exp. Med. Biol. 2017, 960, 221–245. [Google Scholar]
- Chavakis, T.; Alexaki, V.I.; Ferrante, A.J. Macrophage function in adipose tissue homeostasis and metabolic inflammation. Nat. Immunol. 2023, 24, 757–766. [Google Scholar] [CrossRef]
- Francisco, V.; Sanz, M.J.; Real, J.T.; Marques, P.; Capuozzo, M.; Ait, E.D.; Gualillo, O. Adipokines in Non-Alcoholic Fatty Liver Disease: Are We on the Road toward New Biomarkers and Therapeutic Targets? Biology 2022, 11, 1237. [Google Scholar] [CrossRef]
- Marra, F.; Bertolani, C. Adipokines in liver diseases. Hepatology 2009, 50, 957–969. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Scupakova, K.; Soons, Z.; Ertaylan, G.; Pierzchalski, K.A.; Eijkel, G.B.; Ellis, S.R.; Greve, J.W.; Driessen, A.; Verheij, J.; De Kok, T.M.; et al. Spatial Systems Lipidomics Reveals Nonalcoholic Fatty Liver Disease Heterogeneity. Anal. Chem. 2018, 90, 5130–5138. [Google Scholar] [CrossRef]
- Trott, K.A.; Giannitti, F.; Rimoldi, G.; Hill, A.; Woods, L.; Barr, B.; Anderson, M.; Mete, A. Fatty liver hemorrhagic syndrome in the backyard chicken: A retrospective histopathologic case series. Vet. Pathol. 2014, 51, 787–795. [Google Scholar] [CrossRef]
- Wan, Z.; Perry, C.G.; Macdonald, T.; Chan, C.B.; Holloway, G.P.; Wright, D.C. IL-6 is not necessary for the regulation of adipose tissue mitochondrial content. PLoS ONE 2012, 7, e51233. [Google Scholar] [CrossRef]
- Sadashiv; Tiwari, S.; Gupta, V.; Paul, B.N.; Kumar, S.; Chandra, A.; Dhananjai, S.; Negi, M.P.; Ghatak, A. IL-6 gene expression in adipose tissue of postmenopausal women and its association with metabolic risk factors. Mol. Cell. Endocrinol. 2015, 399, 87–94. [Google Scholar] [CrossRef]
- Macpherson, R.E.; Huber, J.S.; Frendo-Cumbo, S.; Simpson, J.A.; Wright, D.C. Adipose Tissue Insulin Action and IL-6 Signaling after Exercise in Obese Mice. Med. Sci. Sports. Exerc. 2015, 47, 2034–2042. [Google Scholar] [CrossRef]
- Polyzos, S.A.; Kountouras, J.; Mantzoros, C.S. Adipose tissue, obesity and non-alcoholic fatty liver disease. Minerva Endocrinol. 2017, 42, 92–108. [Google Scholar] [CrossRef]
- Mellouk, N.; Rame, C.; Delaveau, J.; Rat, C.; Maurer, E.; Froment, P.; Dupont, J. Adipokines expression profile in liver, adipose tissue and muscle during chicken embryo development. Gen. Comp. Endocr. 2018, 267, 146–156. [Google Scholar] [CrossRef]
- Na, W.; Wang, Y.; Gong, P.; Zhang, X.; Zhang, K.; Zhang, H.; Wang, N.; Li, H. Screening of Reference Genes for RT-qPCR in Chicken Adipose Tissue and Adipocytes. Front. Physiol. 2021, 12, 676864. [Google Scholar] [CrossRef]
- Simoes, I.; Fontes, A.; Pinton, P.; Zischka, H.; Wieckowski, M.R. Mitochondria in non-alcoholic fatty liver disease. Int. J. Biochem. Cell Biol. 2018, 95, 93–99. [Google Scholar] [CrossRef]
- Ajith, T.A. Role of mitochondria and mitochondria-targeted agents in non-alcoholic fatty liver disease. Clin. Exp. Pharmacol. Physiol. 2018, 45, 413–421. [Google Scholar] [CrossRef]
- Shami, G.J.; Cheng, D.; Verhaegh, P.; Koek, G.; Wisse, E.; Braet, F. Three-dimensional ultrastructure of giant mitochondria in human non-alcoholic fatty liver disease. Sci. Rep. 2021, 11, 3319. [Google Scholar] [CrossRef]

| Items % | NC Group | HELP Group |
|---|---|---|
| Corn (maize) | 64.00 | 70.00 |
| Wheat bran | 2.00 | 1.20 |
| Soybean meal | 24.00 | 14.58 |
| Soybean oil | 0 | 5 |
| Calcium | 8.00 | 8.00 |
| * Premix | 2.00 | 2.00 |
| Total | 100.00 | 100.00 |
| Nutrient level | ||
| Crude protein (CP) | 15.58 | 12.00 |
| Available phosphate (AP) | 0.51 | 0.46 |
| Arginine (Arg) | 1.03 | 0.74 |
| Methionine (Met) | 0.37 | 0.32 |
| Valine (Val) | 0.77 | 0.58 |
| Metabolic energy (kcal/kg) | 2678.99 | 3100.00 |
| Methionine + cysteine (Met + Cys) | 0.67 | 0.56 |
| Gene | Sequence (5′–3′) | Product Length (bp) | Annealing Temperature (°C) | Accession Number |
|---|---|---|---|---|
| PPARα | F: AGGCCAAGTTGAAAGCAGAA R: TTTCCCTGCAAGGATGACTC | 155 | 60 | NM_001001464.1 |
| PPARγ | F: TGACAGGAAAGACGACAGACA R: CTCCACAGAGCGAAACTGAC | 164 | 59 | NM_001001460.1 |
| MTTP | F: GTTCTGAAGGACATGCGTGC R: GATGTCTAGGCCGTACGTGG | 120 | 58 | NM_001109784.2 |
| SREBP1 | F: CTACCGCTCATCCATCAACG R: CTGCTTCAGCTTCTGGTTGC | 136 | 60 | NM_204126.3 |
| FASN | F: TGCTATGCTTGCCAACAGGA R: ACTGTCCGTGACGAATTGCT | 128 | 59 | NM_205155.3 |
| SCD | F: CTATGCGGGGCTACTT R: GGATGGCTGGAATGAA | 167 | 58 | NM_204890.2 |
| VTGII | F: AACTACTCGATGCCCGCAAA R: ACCAGCAGTTTCACCTGTCC | 179 | 58 | NM_001031276.1 |
| ApoB | F: GGTTACTCCCACGATGGCAA R: TCGCAGAAATGCCCTTCCTT | 120 | 60 | NM_001044633.2 |
| LDLR | F: GTGGACGAGTGCTCTCAGG R: ATAGAGGTTCCCTTCGGCCA | 167 | 58 | NM_204452.1 |
| FABP4 | F: GCCTGACAAAATGTGCGACC R: ATTAGGCTTGGCCACACCAG | 130 | 59 | NM_204290.2 |
| LPL | F: GCATTCACCATTCAGAGAGTCAG R: AACTGCTAAAGAGGAACTGATGG | 130 | 57 | NM_205282.2 |
| CPT-1A | F: TGAGCACTCTTGGGCAGATG R: TCTCCTTTGCAGTGTCCGTC | 108 | 56 | NM_001012898.1 |
| LEPTIN | F: GCAGTGCCGTGCCA GACT C R: GA ATGTCCTGCAGAGAGCCC | 284 | 58 | LN794246.1 |
| ADPN | F: CCCAGAGCAGTGGCTGTTTA R: TGGGTATTTCCAAGGGACGC | 116 | 59 | NM_206991.2 |
| TNFα | F: TGTGCTGTGTGCAACGACTA R: CAGGCCTGGCAACTCTTTCT | 144 | 60 | NM_205183.2 |
| IL-1β | F: TGCCTGCAGAAGAAGCCTCG R: GACGGGCTCAAAAACCTCCT | 205 | 56 | NM_204524.1 |
| IL-6 | F: CTGCAGGACGAGATGTGCAA R: AGGTCTGAAAGGCGAACAGG | 166 | 57 | NM_204628.1 |
| IL-10 | F: AGTTTAAGGGGACCTTTGGCT R: AACTCCCCCATGGCTTTGTAG | 265 | 59 | XM_025143715.1 |
| β-actin | F: TATTGCTGCGCTCGTTGTTG R: TGGCCCATACCAACCATCAC | 144 | 60 | NM_205518.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, X.; Wang, Y.; Amevor, F.K.; Ning, Z.; Deng, X.; Wu, Y.; Wei, S.; Cao, X.; Xu, D.; Tian, Y.; et al. Effect of High Energy Low Protein Diet on Lipid Metabolism and Inflammation in the Liver and Abdominal Adipose Tissue of Laying Hens. Animals 2024, 14, 1199. https://doi.org/10.3390/ani14081199
Du X, Wang Y, Amevor FK, Ning Z, Deng X, Wu Y, Wei S, Cao X, Xu D, Tian Y, et al. Effect of High Energy Low Protein Diet on Lipid Metabolism and Inflammation in the Liver and Abdominal Adipose Tissue of Laying Hens. Animals. 2024; 14(8):1199. https://doi.org/10.3390/ani14081199
Chicago/Turabian StyleDu, Xiaxia, Yinuo Wang, Felix Kwame Amevor, Zifan Ning, Xun Deng, Youhao Wu, Shuo Wei, Xueqing Cao, Dan Xu, Yaofu Tian, and et al. 2024. "Effect of High Energy Low Protein Diet on Lipid Metabolism and Inflammation in the Liver and Abdominal Adipose Tissue of Laying Hens" Animals 14, no. 8: 1199. https://doi.org/10.3390/ani14081199
APA StyleDu, X., Wang, Y., Amevor, F. K., Ning, Z., Deng, X., Wu, Y., Wei, S., Cao, X., Xu, D., Tian, Y., Ye, L., Shu, G., & Zhao, X. (2024). Effect of High Energy Low Protein Diet on Lipid Metabolism and Inflammation in the Liver and Abdominal Adipose Tissue of Laying Hens. Animals, 14(8), 1199. https://doi.org/10.3390/ani14081199







