Integrated Whole-Transcriptome Analysis to Elucidate the Core Regulatory Network of circRNA Involved in Ovarian Development and Reproductive Capacity Differences in Sheep: circRNA2058-miR-9226-5p-MET Axis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Total RNA Extraction and Library Construction
2.3. Sequencing Data Quality Control and Analysis
2.4. Differential Expression Analysis and Target Gene Prediction
2.5. Construction and Bioinformatics Analysis of the ceRNA Network
2.6. qRT-PCR Validation
2.7. Binding Site Prediction
2.8. Recombinant Plasmids Preparation
2.9. Cell Transfection
2.10. Reporter Signal Measurement
2.11. Normalization Control Measurement
2.12. Data Normalization
3. Results
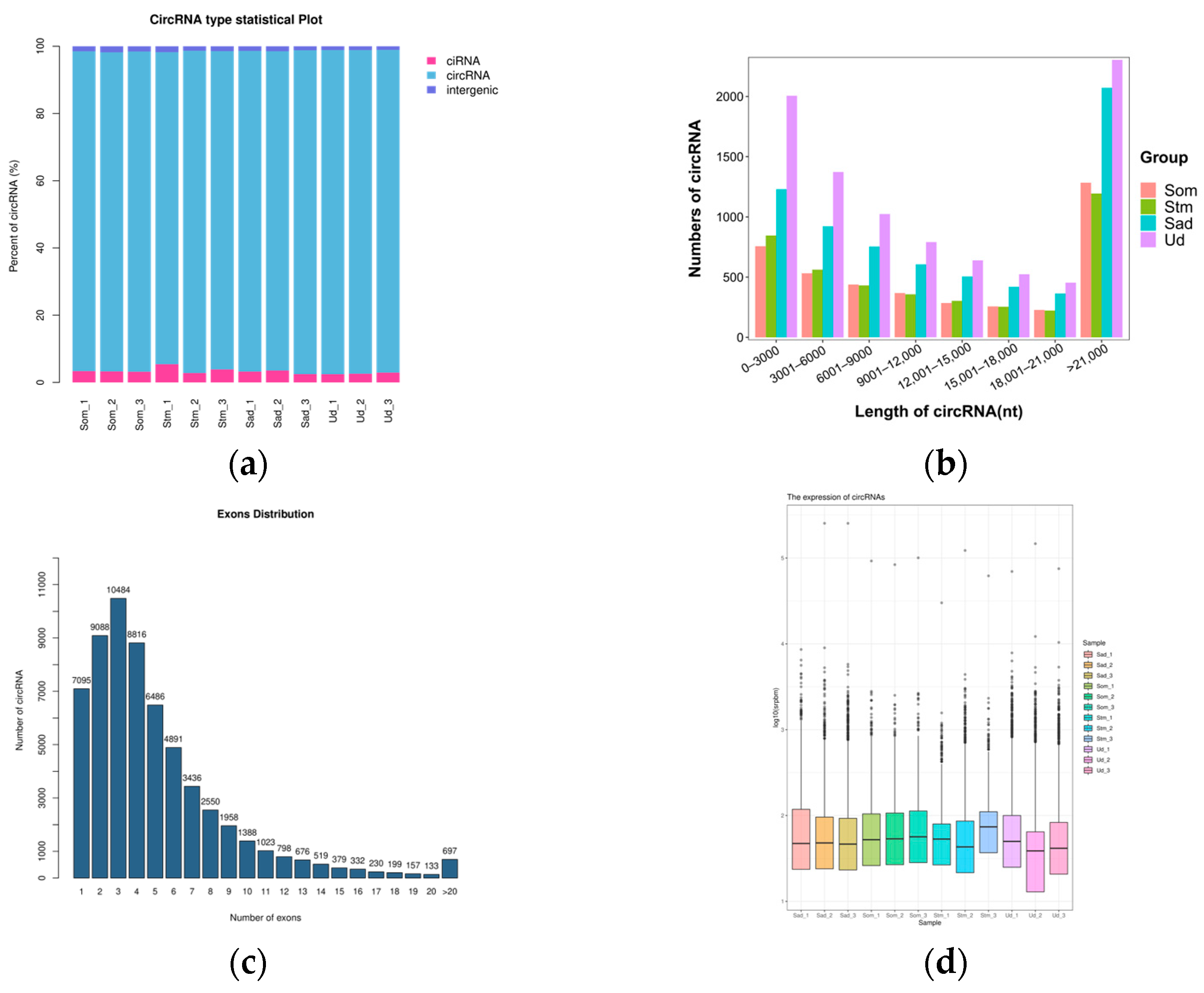
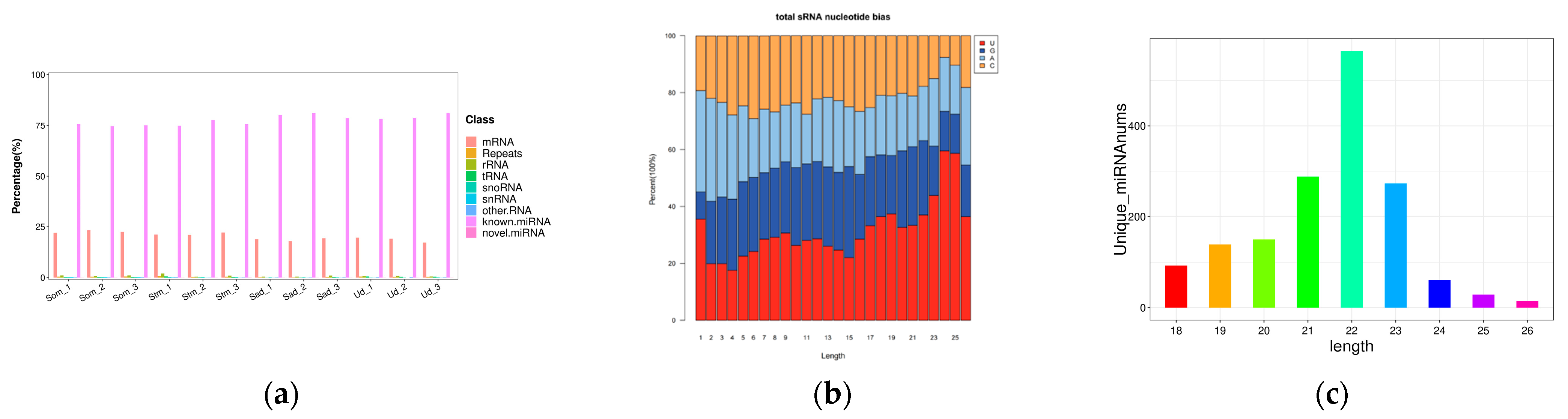
3.1. Sequencing Data Analysis
3.2. Screening and Identification of circRNAs and miRNAs
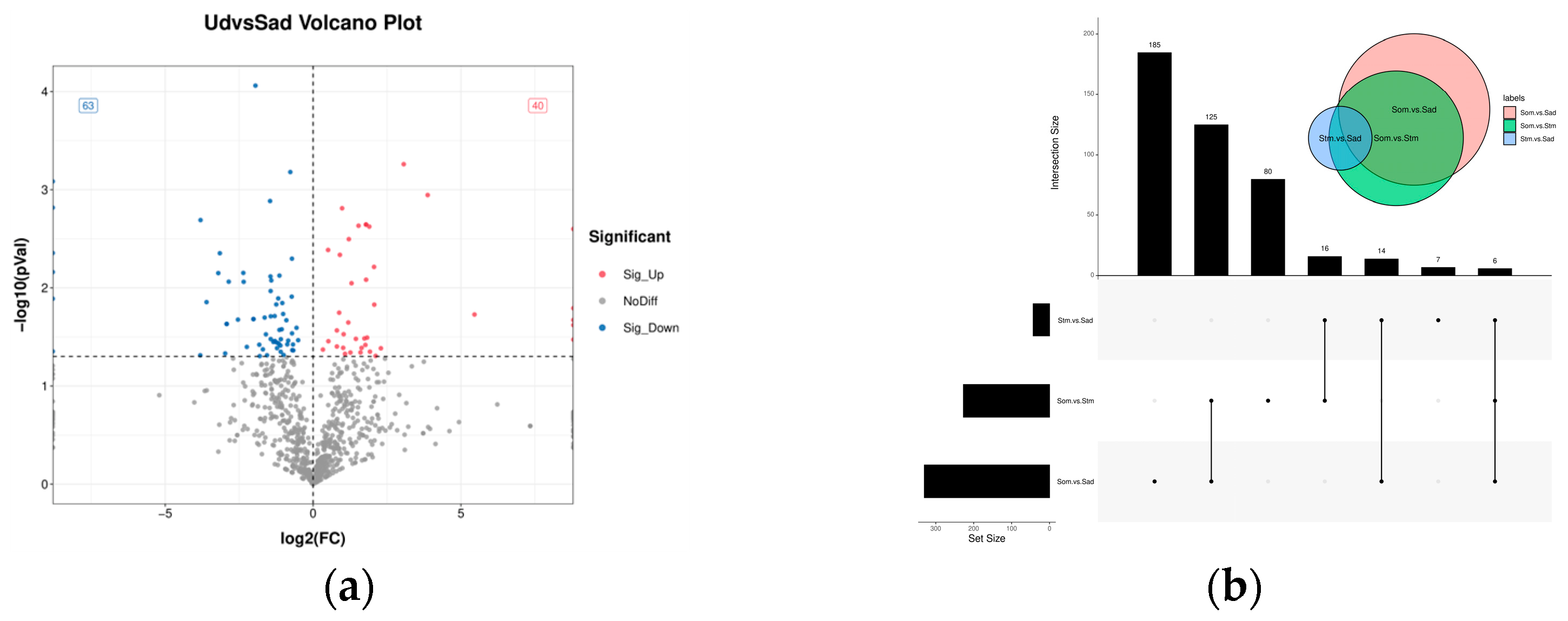
3.3. Differential Expression Analysis
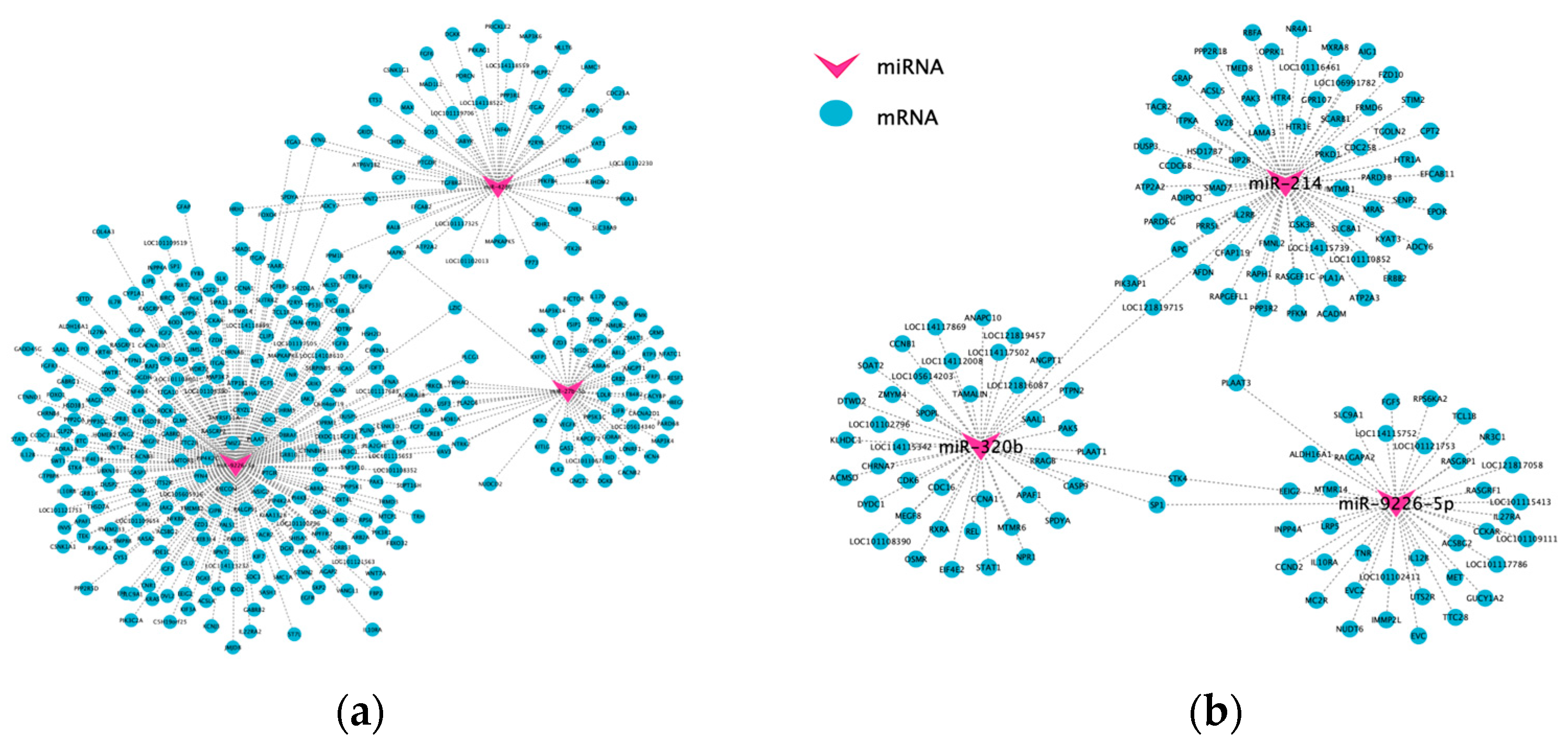
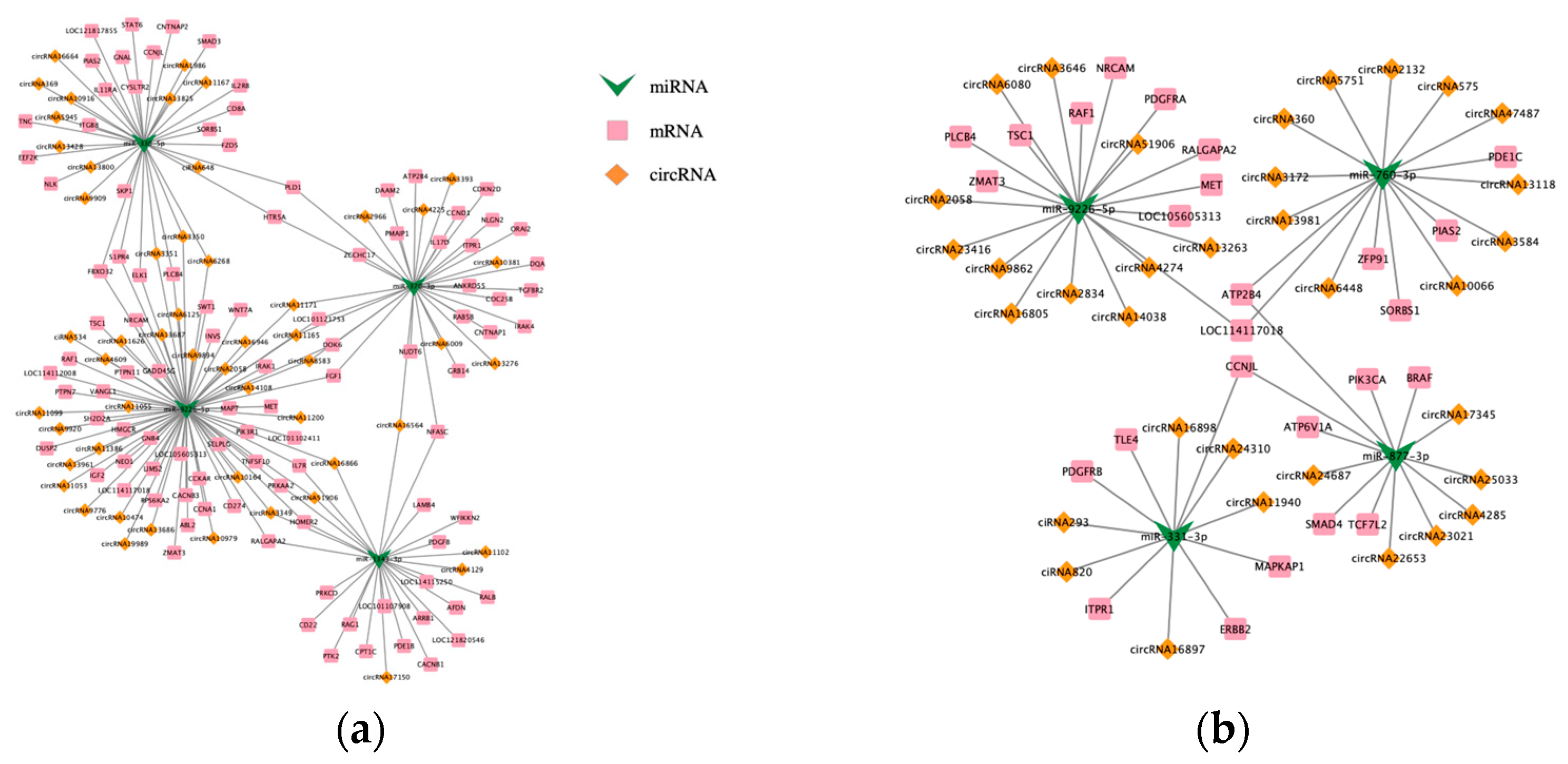
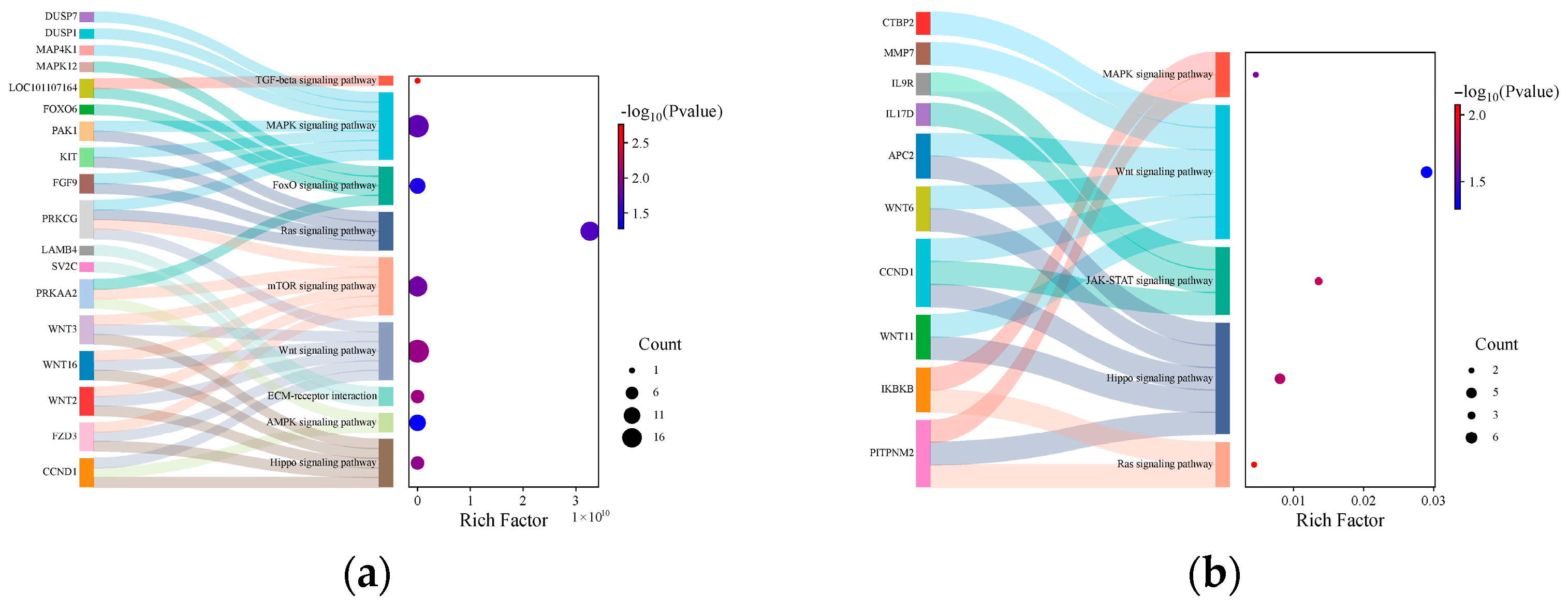
3.4. ceRNA Network Analysis
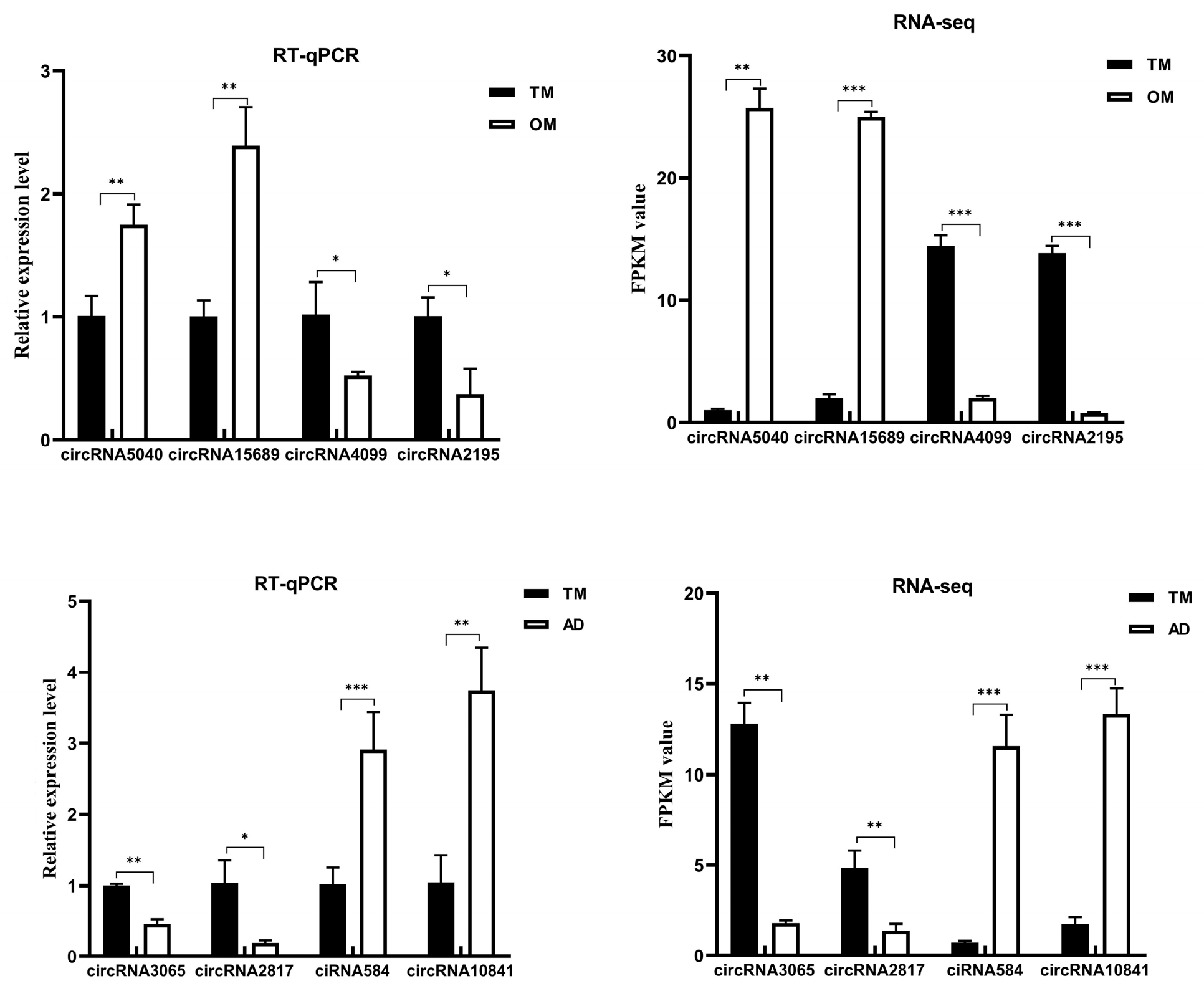
3.5. qRT-PCR and Sanger Verification
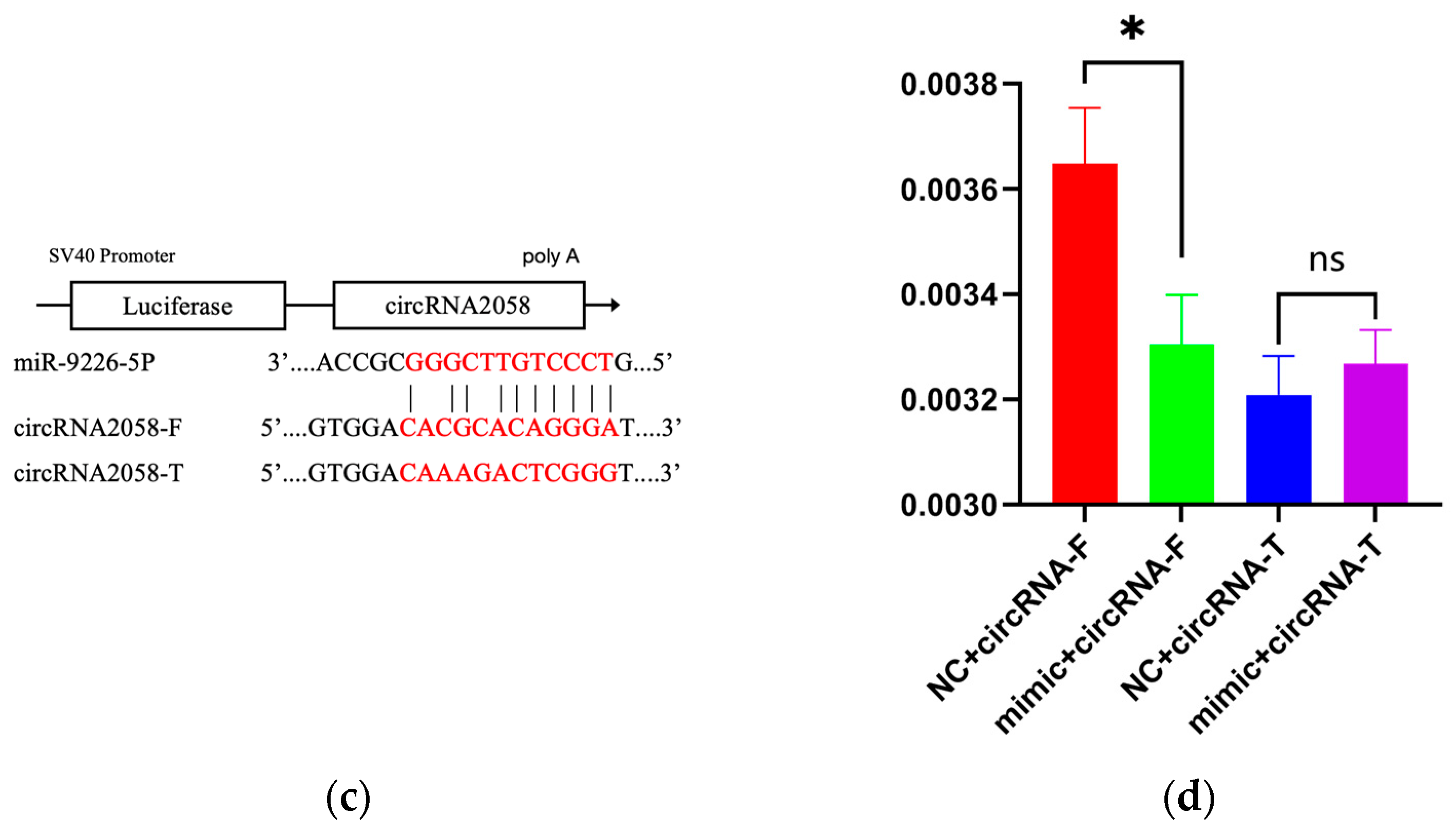
3.6. Dual-Luciferase Reporter Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DECs | differentially expressed circRNAs |
| DEMs | differentially expressed miRNAs |
| DEGs | differentially expressed mRNAs |
| ncRNA | non-coding RNA |
| ceRNA | competing endogenous RNA |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| NGS | Next-generation sequencing |
References
- Yang, Y.; Ma, Y.; Lv, C.; Zhou, L. An Introduction to High-Fertility Sheep Breeds. China Anim. Husb. Vet. Med. 2003, 30, 30–33. [Google Scholar]
- Zhang, Y.; Luo, H.; Fan, X.; Wang, Y.; Zheng, Y.; Zhang, H. Research Progress on Genetic Diversity of Xilingol Sheep Breeds. Chin. Livest. Poult. Breed. 2020, 16, 3–6. [Google Scholar]
- Tao, H.; Xiong, Q.; Zhang, F.; Zhang, N.; Liu, Y.; Suo, X.; Li, X.; Yang, Q.; Chen, M. Circular RNA profiling reveals chi_circ_0008219 function as microRNA sponges in pre-ovulatory ovarian follicles of goats (Capra hircus). Genomics 2017, 110, 257–266. [Google Scholar] [CrossRef]
- Niu, X.; Huang, Y.; Lu, H.; Li, S.; Huang, S.; Ran, X.; Wang, J. CircRNAs in Xiang pig ovaries among diestrus and estrus stages. Porc. Health Manag. 2022, 8, 29. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, J.; Han, D.; Wang, H.Q.; Liu, J.B.; Xiao, Y.; Jiang, H.; Gao, Y.; Yuan, B. CiRS-187 regulates BMPR2 expression by targeting miR-187 in bovine cumulus cells treated with BMP15 and GDF9. Theriogenology 2023, 197, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Y.; Zhang, J.; Du, X.; Li, Q.; Pan, Z. circSLC41A1 Resists Porcine Granulosa Cell Apoptosis and Follicular Atresia by Promoting SRSF1 through miR-9820-5p Sponging. Int. J. Mol. Sci. 2022, 23, 1509. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, P.; Zhou, Z.; He, X.; Tao, L.; Jiang, Y.; Lan, R.; Hong, Q.; Chu, M. Expression Profile Analysis to Identify Circular RNA Expression Signatures in the Prolificacy Trait of Yunshang Black Goat Pituitary in the Estrus Cycle. Front. Genet. 2022, 12, 801357. [Google Scholar] [CrossRef]
- Wang, J.; Chen, H.; Zhang, Y.; Jiang, S.; Zeng, X.; Shen, H. Comprehensive Analysis of Differentially Expressed CircRNAs in the Ovaries of Low- and High-Fertility Sheep. Animals 2023, 13, 236. [Google Scholar] [CrossRef]
- Lv, W.; An, R.; Li, X.; Zhang, Z.; Geri, W.; Xiong, X.; Yin, S.; Fu, W.; Liu, W.; Lin, Y.; et al. Multi-Omics Approaches Uncovered Critical mRNA-miRNA-lncRNA Networks Regulating Multiple Birth Traits in Goat Ovaries. Int. J. Mol. Sci. 2024, 25, 12466. [Google Scholar] [CrossRef] [PubMed]
- Kechin, A.; Boyarskikh, U.; Kel, A.; Filipenko, M. cutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J. Comput. Biol. 2017, 24, 1138–1143. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Dong, R.; Zhang, Y.; Zhang, J.L.; Luo, Z.; Zhang, J.; Chen, L.L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef]
- Ye, J.; Yao, Z.; Si, W.; Gao, X.; Yang, C.; Liu, Y.; Ding, J.; Huang, W.; Fang, F.; Zhou, J. Identification and characterization of microRNAs in the pituitary of pubescent goats. Reprod. Biol. Endocrinol. 2018, 16, 51. [Google Scholar] [CrossRef]
- Cao, Z.; Gao, D.; Xu, T.; Zhang, L.; Tong, X.; Zhang, D.; Wang, Y.; Ning, W.; Qi, X.; Ma, Y.; et al. Circular RNA profiling in the oocyte and cumulus cells reveals that circARMC4 is essential for porcine oocyte maturation. Aging 2019, 11, 8015–8034. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Feng, G.; Guo, C.; Li, Z.; Liu, D.; Liu, G.; Zou, X.; Sun, B.; Guo, Y.; Deng, M.; et al. Identification of functional circRNAs regulating ovarian follicle development in goats. BMC Genom. 2024, 25, 893. [Google Scholar] [CrossRef]
- Huang, C.; Feng, F.; Dai, R.; Ren, W.; Li, X.; Zhaxi, T.; Ma, X.; Wu, X.; Chu, M.; La, Y.; et al. Whole-transcriptome analysis of longissimus dorsi muscle in cattle-yaks reveals the regulatory functions of ADAMTS6 gene in myoblasts. Int. J. Biol. Macromol. 2024, 262, 129985. [Google Scholar] [CrossRef]
- Li, P.; Wang, J.; Zhi, L.; Cai, F. Linc00887 suppresses tumorigenesis of cervical cancer through regulating the miR-454-3p/FRMD6-Hippo axis. Cancer Cell Int. 2021, 21, 33. [Google Scholar] [CrossRef]
- Visser-Grieve, S.; Hao, Y.; Yang, X. Human homolog of Drosophila expanded, hEx, functions as a putative tumor suppressor in human cancer cell lines independently of the Hippo pathway. Oncogene 2012, 31, 1189–1195. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wei, L.; Liang, Q.; Liu, X.; Li, N. Research Progress on Cyclins and Cyclin-dependent Kinases Related with Primordial Follicle Activation. Genom. Appl. Biol. 2024, 43, 1321–1331. [Google Scholar]
- Lalonde-Larue, A.; Boyer, A.; Dos Santos, E.C.; Boerboom, D.; Bernard, D.J.; Zamberlam, G. The Hippo Pathway Effectors YAP and TAZ Regulate LH Release by Pituitary Gonadotrope Cells in Mice. Endocrinology 2022, 163, bqab238. [Google Scholar] [CrossRef]
- Plewes, M.R.; Hou, X.; Zhang, P.; Liang, A.; Hua, G.; Wood, J.R.; Cupp, A.S.; Lv, X.; Wang, C.; Davis, J.S. Yes-associated protein 1 is required for proliferation and function of bovine granulosa cells in vitro. Biol. Reprod. 2019, 101, 1001–1017. [Google Scholar] [CrossRef] [PubMed]
- Ndiaye, K.; Castonguay, A.; Benoit, G.; Silversides, D.W.; Lussier, J.G. Differential regulation of Janus kinase 3 (JAK3) in bovine preovulatory follicles and identification of JAK3 interacting proteins in granulosa cells. J. Ovarian Res. 2016, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; López, J.M. Understanding MAPK Signaling Pathways in Apoptosis. Int. J. Mol. Sci. 2020, 21, 2346. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Cai, Y.; Yang, H.; Zhang, C.; Liu, W.; Zhao, B.; Wang, F.; Zhang, Y. AMH regulates granulosa cell function via ESR2/p38-MAPK signaling pathway in sheep. Commun. Biol. 2025, 8, 824. [Google Scholar] [CrossRef]
- Zhang, X.; Li, M.; Huang, M.; Peng, H.; Song, X.; Chen, L.; Hu, W.; Xu, W.; Luo, R.; Han, D.; et al. Effect of RFRP-3, the mammalian ortholog of GnIH, on apoptosis and autophagy in porcine ovarian granulosa cells via the p38MAPK pathway. Theriogenology 2022, 180, 137–145. [Google Scholar] [CrossRef]
- Wang, J.; Bai, J.; Liu, Y.; Zhang, Z.; Lv, Z.; Hai, G.; Li, H.; Liu, W.; Tang, L.; Hua, Y.; et al. Transcriptomic characterization of lamb ovaries and oocytes reveals key biomarkers after superovulation. J. Anim. Sci. 2025, skaf193. [Google Scholar] [CrossRef]
- Li, G.; Yan, L.; Wang, L.; Ma, W.; Wu, H.; Guan, S.; Yao, Y.; Deng, S.; Yang, H.; Zhang, J.; et al. Ovarian overexpression of ASMT gene increases follicle numbers in transgenic sheep: Association with lipid metabolism. Int. J. Biol. Macromol. 2024, 269, 131803. [Google Scholar] [CrossRef]
- Song, Z. Association Analysis of SNPs in CTNNB1 and pri-miR-458b with Laying Traits in Layers. Master’s Thesis, Shandong Agricultural University, Taian, China, 2020. [Google Scholar]
- Liu, T.; Wen, Y.; Cui, Z.; Chen, H.; Lin, J.; Xu, J.; Chen, D.; Zhu, Y.; Yu, Z.; Wang, C.; et al. MicroRNA-3061 downregulates the expression of PAX7/Wnt/Ca2+ signalling axis genes to induce premature ovarian failure in mice. Cell Prolif. 2024, 57, e13686. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Li, Q.; Yang, L.; Liu, L.; Cao, Q.; Li, Q. SMAD4 activates Wnt signaling pathway to inhibit granulosa cell apoptosis. Cell Death Dis. 2020, 11, 373. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Chen, X.; Dai, M.; Li, M.; Huang, Y.; Bai, L.; Wu, Y.; He, G. The research progress of HGF/c-MET signaling pathway regulating reproductive cytogenesis. J. Pract. Med. 2019, 35, 3413–3417. [Google Scholar]
- Xu, J.; Xu, H.; Hu, L.; Zhang, F.; Gao, Q.; Luo, H.; Zhang, H.; Shi, R.; Li, X.; Liu, L.; et al. Association Analysis of MER Gene Single Nucleotide Polymorphism with Reproduction and Milk Production on Traits in Chinese Holstein Cattle. Acta Vet. Et Zootech. Sin. 2022, 53, 3769–3785. [Google Scholar]
- Ling, L.; Feng, X.; Wei, T.; Wang, Y.; Wang, Y.; Wang, Z.; Tang, D.; Luo, Y.; Xiong, Z. Human amnion-derived mesenchymal stem cell (hAD-MSC) transplantation improves ovarian function in rats with premature ovarian insufficiency (POI) at least partly through a paracrine mechanism. Stem Cell Res. Ther. 2019, 10, 46. [Google Scholar] [CrossRef]
- Liu, H.; Li, S.R.; Si, Q. Regulation of miRNAs on c-met protein expression in ovarian cancer and its implication. Eur. Rev. Med. Pharmacol. Sci. 2024, 28, 3891. [Google Scholar]
- Parrott, J.A.; Skinner, M.K. Developmental and hormonal regulation of hepatocyte growth factor expression and action in the bovine ovarian follicle. Biol. Reprod. 1998, 59, 553–560. [Google Scholar] [CrossRef]





| Sample | Raw Data | Valid Data | Mapped Reads/% | Q20% | Q30% | GC Content/% |
|---|---|---|---|---|---|---|
| Som_1 | 95,584,162 | 92,544,232 | 87,671,478 (94.73%) | 99.97 | 97.81 | 45.00 |
| Som_2 | 93,194,002 | 90,150,580 | 85,397,972 (94.73%) | 99.97 | 97.84 | 44.00 |
| Som_3 | 93,566,726 | 90,707,218 | 84,656,216 (93.33%) | 99.96 | 97.58 | 45.00 |
| Stm_1 | 93,780,650 | 90,919,404 | 86,062,585 (94.66%) | 99.97 | 97.78 | 45.00 |
| Stm_2 | 91,917,068 | 88,670,432 | 82,933,703 (93.53%) | 99.98 | 98.30 | 49.00 |
| Stm_3 | 71,302,642 | 67,362,198 | 60,153,911 (89.30%) | 98.50 | 94.59 | 45.00 |
| Sad_1 | 94,092,386 | 91,107,390 | 86,158,306 (94.57%) | 99.96 | 97.78 | 45.00 |
| Sad_2 | 97,790,686 | 94,803,474 | 89,533,252 (94.44%) | 99.96 | 97.71 | 45.00 |
| Sad_3 | 89,825,602 | 86,701,034 | 80,875,819 (93.28%) | 99.97 | 98.16 | 48.50 |
| Ud_1 | 73,320,580 | 70,898,902 | 65,359,324 (92.19%) | 99.85 | 97.83 | 47.00 |
| Ud_2 | 92,249,184 | 87,955,300 | 82,065,791 (93.30%) | 99.77 | 97.86 | 50.50 |
| Ud_3 | 71,516,612 | 68,959,698 | 63,802,517 (92.52%) | 99.81 | 97.60 | 48.00 |
| Sample | Raw Data | Valid Data | Mapped Reads/% | Q20% | Q30% | GC Content/% |
|---|---|---|---|---|---|---|
| Som_1 | 13,916,096 | 9,366,661 | 9,257,071 (98.83%) | 97.91 | 96.89 | 51.78 |
| Som_2 | 12,153,463 | 7,973,465 | 7,861,836 (98.60%) | 97.67 | 96.68 | 51.60 |
| Som_3 | 15,083,990 | 9,398,231 | 9,259,137 (98.52%) | 97.02 | 95.67 | 51.78 |
| Stm_1 | 9,076,908 | 5,229,613 | 5,161,628 (98.70%) | 97.74 | 96.52 | 51.98 |
| Stm_2 | 8,729,542 | 7,235,145 | 7,180,158 (99.24%) | 97.63 | 96.44 | 51.03 |
| Stm_3 | 10,569,235 | 5,463,213 | 5,393,284 (98.72%) | 97.35 | 96.04 | 50.62 |
| Sad_1 | 10,567,624 | 6,971,109 | 6,898,609 (98.96%) | 97.92 | 96.91 | 51.40 |
| Sad_2 | 9,796,916 | 7,951,578 | 7,863,315 (98.89%) | 97.92 | 96.96 | 51.22 |
| Sad_3 | 10,036,758 | 6,868,427 | 6,794,248 (98.92%) | 97.29 | 96.07 | 51.11 |
| Ud_1 | 11,079,361 | 6,789,313 | 6,743,146 (99.32%) | 99.24 | 97.13 | 51.14 |
| Ud_2 | 11,405,958 | 7,210,142 | 7,160,392 (99.31%) | 99.26 | 97.10 | 50.87 |
| Ud_3 | 11,156,801 | 7,387,622 | 7,321,133 (99.10%) | 99.28 | 97.18 | 50.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, B.; Wang, A.; Yu, X.; Li, Y.; Cong, Y.; Jiang, H. Integrated Whole-Transcriptome Analysis to Elucidate the Core Regulatory Network of circRNA Involved in Ovarian Development and Reproductive Capacity Differences in Sheep: circRNA2058-miR-9226-5p-MET Axis. Animals 2025, 15, 3077. https://doi.org/10.3390/ani15213077
Gu B, Wang A, Yu X, Li Y, Cong Y, Jiang H. Integrated Whole-Transcriptome Analysis to Elucidate the Core Regulatory Network of circRNA Involved in Ovarian Development and Reproductive Capacity Differences in Sheep: circRNA2058-miR-9226-5p-MET Axis. Animals. 2025; 15(21):3077. https://doi.org/10.3390/ani15213077
Chicago/Turabian StyleGu, Bo, Anqi Wang, Xinmiao Yu, Ying Li, Yao Cong, and Huaizhi Jiang. 2025. "Integrated Whole-Transcriptome Analysis to Elucidate the Core Regulatory Network of circRNA Involved in Ovarian Development and Reproductive Capacity Differences in Sheep: circRNA2058-miR-9226-5p-MET Axis" Animals 15, no. 21: 3077. https://doi.org/10.3390/ani15213077
APA StyleGu, B., Wang, A., Yu, X., Li, Y., Cong, Y., & Jiang, H. (2025). Integrated Whole-Transcriptome Analysis to Elucidate the Core Regulatory Network of circRNA Involved in Ovarian Development and Reproductive Capacity Differences in Sheep: circRNA2058-miR-9226-5p-MET Axis. Animals, 15(21), 3077. https://doi.org/10.3390/ani15213077






