Comparative Analysis of Metopograpsus quadridentatus (Crustacea: Decapoda: Grapsidae) Mitochondrial Genome Reveals Gene Rearrangement and Phylogeny
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and DNA Extraction
2.2. Sequencing and Analysis
2.3. Bioinformatics Analysis
2.4. Phylogenetic Analysis
3. Results and Discussion
3.1. Mitogenome Organization and Nucleotide Composition
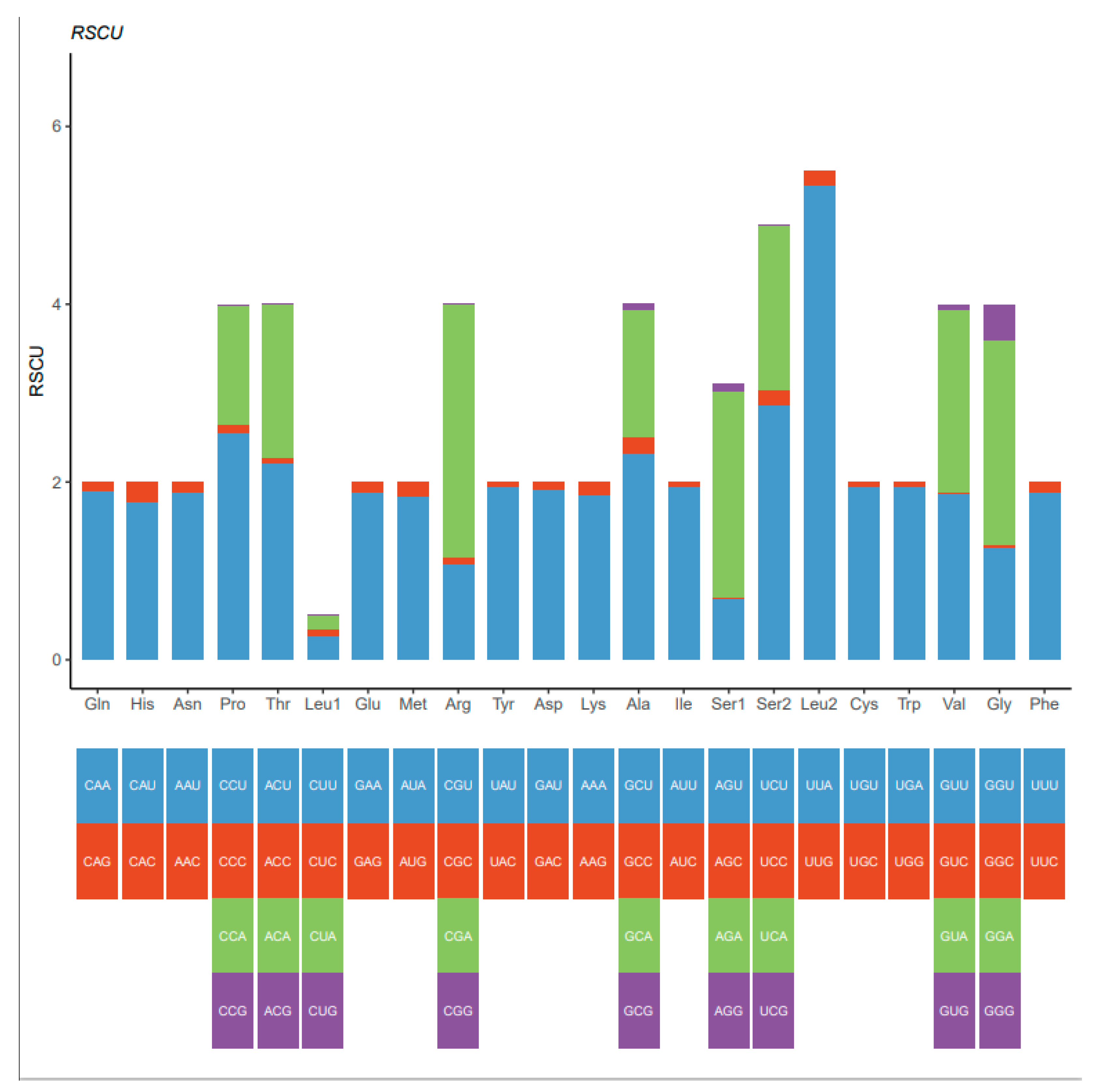
3.2. Protein-Coding Genes
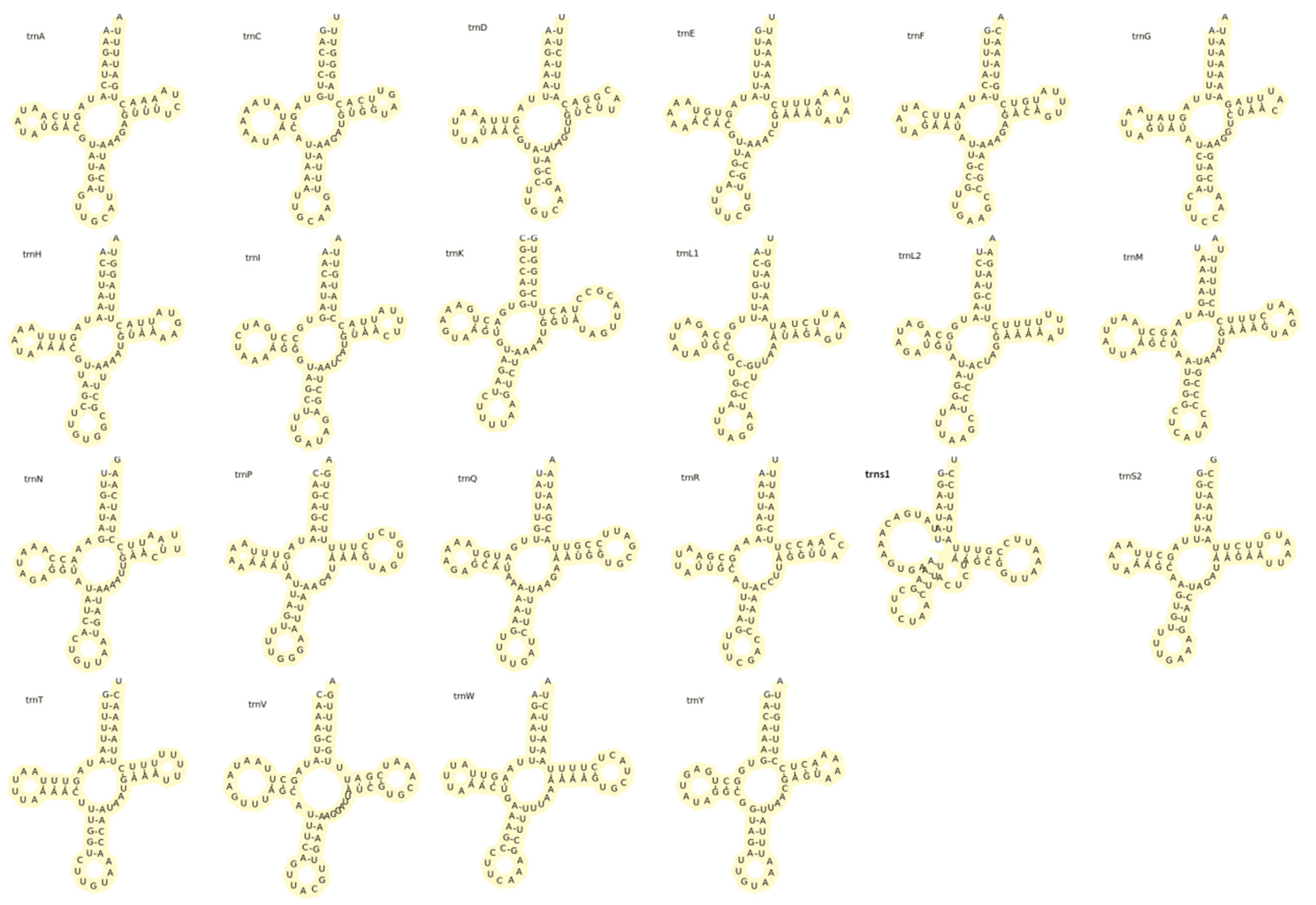
3.3. Transfer RNA and Ribosomal RNA Genes and Control Region
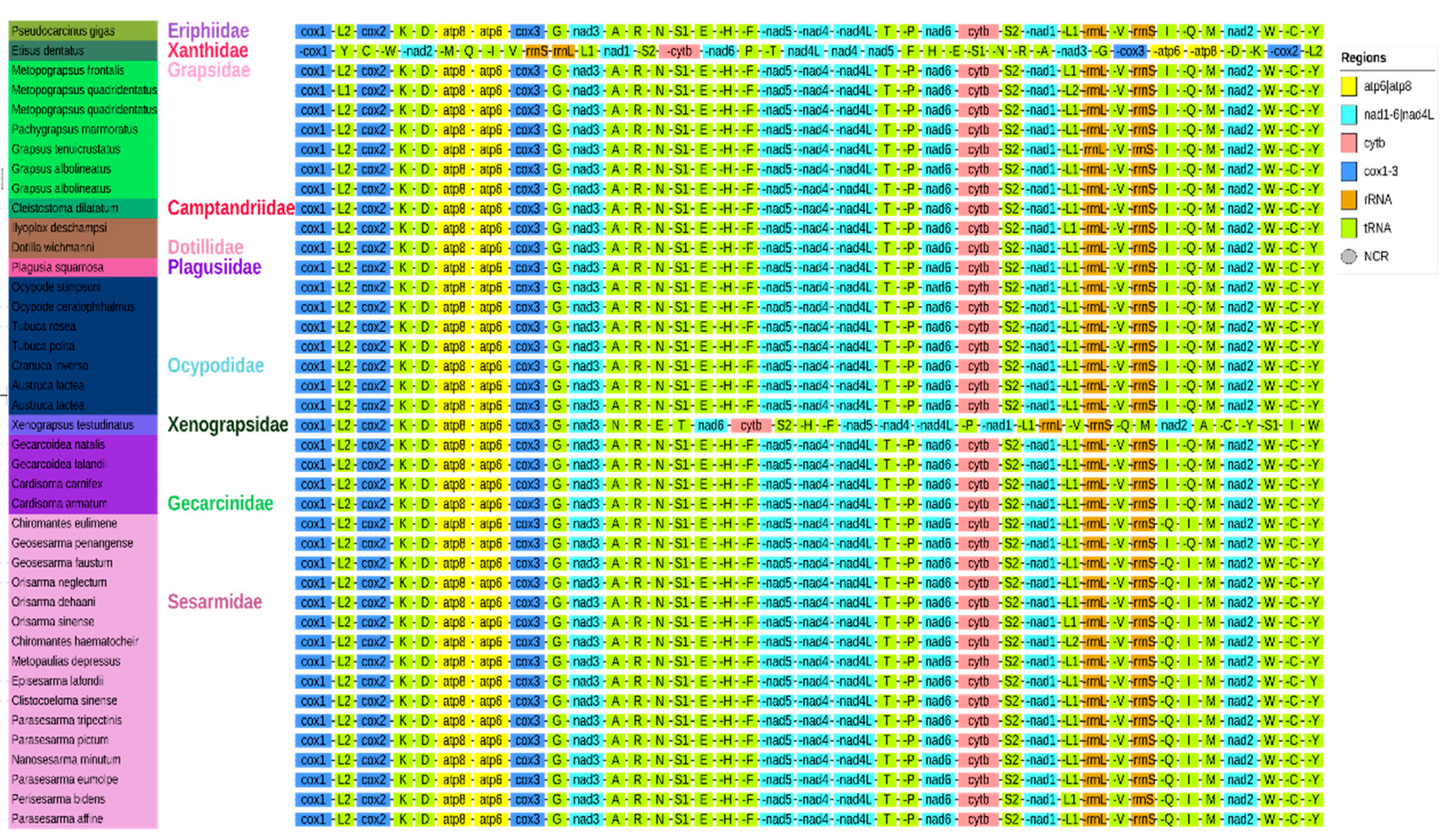
3.4. Gene Arrangement
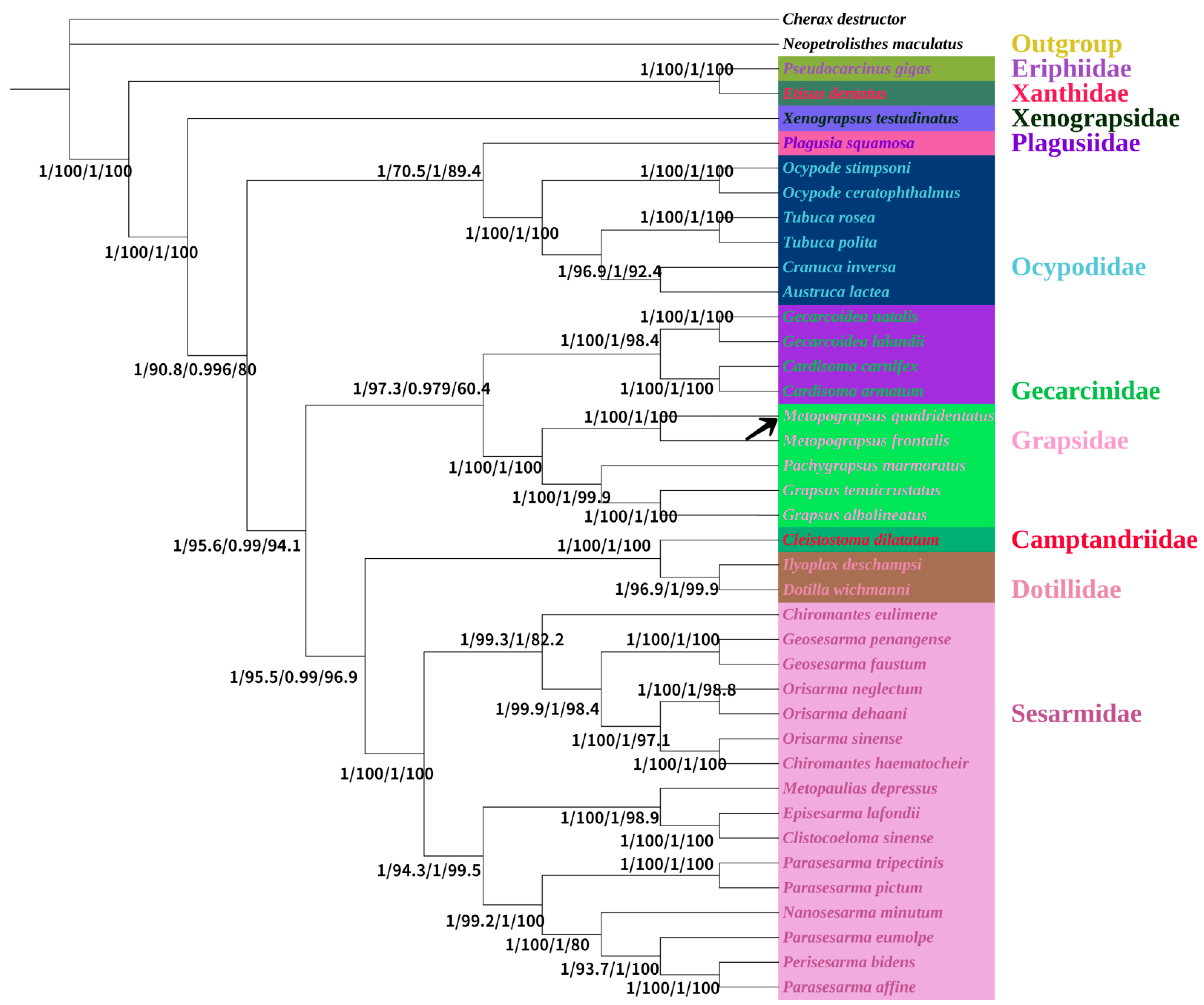
3.5. Phylogenetic Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tsang, L.M.; Schubart, C.D.; Ahyong, S.T.; Lai, J.C.Y.; Au, E.Y.C.; Chan, T.Y.; Ng, P.K.L.; Chu, K.H. Evolutionary History of True Crabs (Crustacea: Decapoda: Brachyura) and the Origin of Freshwater Crabs. Mol. Biol. Evol. 2014, 31, 1173–1187. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Ma, C.; Li, X.; Xu, Z.; Feng, N.; Ma, L. The complete mitochondrial genome sequence and gene organization of the mud crab (Scylla paramamosain) with phylogenetic consideration. Gene 2013, 519, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Rackham, O.; Filipovska, A. Organization and Expression of the Mammalian Mitochondrial Genome. Nat. Rev. Genet. 2022, 23, 606–623. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wei, L.; Liu, B.; Liu, L.; Lü, Z.; Gong, L. Two complete mitogenomes of Ocypodoidea (Decapoda: Brachyura), Cleistostoma dilatatum (Camptandriidae) and Euplax sp. (Macrophthalmidae) and its phylogenetic implications. Acta Oceanol. Sin. 2023, 42, 81–92. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, Y.; Gong, L.; Lu, X.; Jiang, L.; Liu, B.; Liu, L.; Lü, Z.; Li, P. Mitochondrial genome of Episesarma lafondii (Brachyura: Sesarmidae) and comparison with other sesarmid crabs. J. Ocean Univ. China 2021, 20, 1545–1556. [Google Scholar] [CrossRef]
- Guinot, D.; Jamieson, B.G.M.; Deforges, B.R. Relationship of Homolidae and Dromiidae: Evidence from Spermatozoal Ultrastructure (Crustacea, Decapoda). Acta Zool. 1994, 75, 255–267. [Google Scholar] [CrossRef]
- Shen, J.; Dai, A. Illustrated Fauna of China: Crustacea; Science Press: Beijing, China, 1964; Volume 2, pp. 1–142. [Google Scholar]
- Metzker, M.L. Sequencing Technologies—The Next Generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef]
- Fratini, S.; Cannicci, S.; Schubart, C.D. Molecular phylogeny of the crab genus Metopograpsus H. Milne Edwards, 1853 (Decapoda: Brachyura: Grapsidae) reveals high intraspecific genetic variation and distinct evolutionarily significant units. Invertebr. Syst. 2018, 32, 215–223. [Google Scholar] [CrossRef]
- Lei, R.H.; Frasier, C.L.; Hawkins, M.T.R.; Engberg, S.E.; Bailey, C.A.; Johnson, S.E.; McLain, A.T.; Groves, C.P.; Perry, G.H.; Nash, S.D.; et al. Phylogenomic Reconstruction of Sportive Lemurs (Genus Lepilemur) Recovered from Mitogenomes with Inferences for Madagascar Biogeography. J. Hered. 2017, 108, 107–119. [Google Scholar]
- Mueller, R.L.; Macey, J.R.; Jaekel, M.; Wake, D.B.; Boore, J.L. Morphological Homoplasy, Life History Evolution, and Historical Biogeography of Plethodontid Salamanders Inferred from Complete Mitochondrial Genomes. Proc. Natl. Acad. Sci. USA 2004, 101, 13820–13825. [Google Scholar] [CrossRef]
- Cerny, V.; Fernandes, V.; Costa, M.D.; Hajek, M.; Mulligan, C.J.; Pereira, L. Migration of Chadic Speaking Pastoralists Within Africa Based on Population Structure of Chad Basin and Phylogeography of Mitochondrial L3f Haplogroup. BMC Evol. Biol. 2009, 9, 63. [Google Scholar] [CrossRef]
- Inak, G.; Lorenz, C.; Lisowski, P.; Zink, A.; Mlody, B.; Prigione, A. Concise Review: Induced Pluripotent Stem Cell-Based Drug Discovery for Mitochondrial Disease. Stem Cells 2017, 35, 1655–1662. [Google Scholar] [CrossRef] [PubMed]
- Janzen, D.H.; Burns, J.M.; Cong, Q.; Hallwachs, W.; Dapkey, T.; Manjunath, R.; Hajibabaei, M.; Hebert, P.D.N.; Grishin, N.V. Nuclear Genomes Distinguish Cryptic Species Suggested by Their DNA Barcodes and Ecology. Proc. Natl. Acad. Sci. USA 2017, 114, 8313–8318. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.P.; Liu, Y.; Xin, Z.Z.; Zhang, D.Z.; Wang, Z.F.; Zhu, X.Y.; Wang, Y.; Zhang, H.B.; Zhou, C.L.; Chai, X.Y.; et al. Characterisation of the Complete Mitochondrial Genome of Helice wuana (Grapsoidea: Varunidae) and Comparison with Other Brachyuran Crabs. Genomics 2018, 110, 221–230. [Google Scholar] [CrossRef]
- Kobayashi, G.; Itoh, H.; Fukuda, H.; Kojima, S. The Complete Mitochondrial Genome of the Sand Bubbler Crab Scopimera globosa and Its Phylogenetic Position. Genomics 2021, 113, 831–839. [Google Scholar] [CrossRef]
- Ji, Y.T.; Zhou, X.J.; Yang, Q.; Lu, Y.B.; Wang, J.; Zou, J.X. Adaptive evolution characteristics of mitochondrial genomes in genus Aparapotamon (Brachyura, Potamidae) of freshwater crabs. BMC Genom. 2023, 24, 193. [Google Scholar] [CrossRef]
- Simon, C.; Buckley, T.R.; Frati, F.; Stewart, J.B.; Beckenbach, A.T. Incorporating Molecular Evolution into Phylogenetic Analysis, and a New Compilation of Conserved Polymerase Chain Reaction Primers for Animal Mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2016, 37, 545–579. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, J.; Wu, Q.; Xu, X.; Wang, P.; Wang, Z. Insights into the Evolution of Brachyura (Crustacea: Decapoda) from Mitochondrial Sequences and Gene Order Rearrangements. Int. J. Biol. Macromol. 2021, 170, 717–727. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) Version 1.3.1: Expanded Toolkit for the Graphical Visualization of Organellar Genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Kocher, T.D. Patterns of Nucleotide Composition at Fourfold Degenerate Sites of Animal Mitochondrial Genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Xiang, C.Y.; Gao, F.; Jakovlić, I.; Lei, H.P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An Integrated and Scalable Desktop Platform for Streamlined Molecular Sequence Data Management and Evolutionary Phylogenetics Studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P.; Teslenko, M. Draft MrBayes, Version 3.2 Manual; Tutorials and Model Summaries. Systematic Biology 2011. MrBayes 3.2 Manual. Available online: https://web-genobioinfo.toulouse.inrae.fr/~formation/11_Phylogeny/doc/mb3.2_manual.pdf (accessed on 19 January 2024).
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Liu, H.; Shen, M. The complete mitochondrial genome of spiny spooner Etisus dentatus (Herbst, 1785) using high-throughput sequencing. Mitochondrial DNA Part B 2021, 6, 985–987. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Xu, J.; Yang, M.; Wu, B.; Yan, B.; Xiong, Y. Population genetic diversity of sesarmid crab (Perisesarma bidens) in China based on mitochondrial DNA. Mitochondrial DNA A 2016, 27, 3255–3262. [Google Scholar] [CrossRef]
- Zhang, Y.; Gong, L.; Lu, X.; Jiang, L.; Liu, B.; Liu, L.; Lü, Z.; Li, P.; Zhang, X. Gene rearrangements in the mitochondrial genome of Chiromantes eulimene (Brachyura: Sesarmidae) and phylogenetic implications for Brachyura. Int. J. Biol. Macromol. 2020, 162, 704–714. [Google Scholar] [CrossRef]
- Kim, H.; Jung, J. Complete mitochondrial genome of the ghost crab Ocypode stimpsoni Ortmann, 1897 (Brachyura: Decapoda: Ocypodidae) and its phylogenetic relationship in Brachyura. Mitochondrial DNA B 2020, 5, 1699–1700. [Google Scholar] [CrossRef]
- Guan, M.; Liu, X.; Lin, F.; Xie, Z.; Fazhan, H.; Ikhwanuddin, M.; Tan, H.; Ma, H. The whole mitochondrial genome of the mangrove crab, Metopograpsus frontalis (Miers, 1880) (Decapoda, Grapsidae) and its phylogenetic relationship. Mitochondrial DNA B 2018, 3, 368–369. [Google Scholar] [CrossRef]
- Li, Q.; Xu, C.; Wang, C.; Liu, G. The complete mitochondrial genome of red-clawed crab Chiromantes haematochir (Sesarmidae: Grapsidae). Mitochondrial DNA B Resour. 2019, 4, 53–54. [Google Scholar] [CrossRef]
- Chen, J.Q.; Xing, Y.H.; Yao, W.J.; Xu, X.; Zhang, C.; Zhang, Z.; Liu, Q. Phylomitogenomics reconfirm the phylogenetic position of the genus Metaplax inferred from the two grapsid crabs (Decapoda: Brachyura: Grapsoidea). PLoS ONE 2019, 14, e0210763. [Google Scholar] [CrossRef]
- Wang, Z.F.; Wang, Z.Q.; Shi, X.J.; Wu, Q.; Tao, Y.; Guo, H.; Ji, C.; Bai, Y. Complete mitochondrial genome of Parasesarma affine (Brachyura: Sesarmidae): Gene rearrangements in Sesarmidae and phylogenetic analysis of the Brachyura. Int. J. Biol. Macromol. 2018, 118, 31–40. [Google Scholar] [CrossRef]
- Tan, M.H.; Gan, H.M.; Lee, Y.P.; Linton, S.; Grandjean, F.; Bartholomei-Santos, M.L.; Miller, A.D.; Austin, C.M. ORDER within the chaos: Insights into phylogenetic relationships within the Anomura (Crustacea: Decapoda) from mitochondrial sequences and gene order rearrangements. Mol. Phylogenet. Evol. 2018, 127, 320–331. [Google Scholar] [CrossRef]
- Chen, J.; Xing, Y.; Yao, W.; Zhang, C.; Zhang, Z.; Jiang, G.; Ding, Z. Characterization of four new mitogenomes from Ocypodoidea & Grapsoidea, and phylomitogenomic insights into thoracotreme evolution. Gene 2018, 675, 27–35. [Google Scholar]
- Wang, Z.; Shi, X.; Tao, Y.; Wu, Q.; Bai, Y.; Guo, H.; Tang, D. The complete mitochondrial genome of Parasesarma pictum (Brachyura: Grapsoidea: Sesarmidae) and comparison with other Brachyuran crabs. Genomics 2019, 111, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.S.; Sam, K.K.; Ahmad, A.B.; Siti, K.-A.; Zafir, A.W.A.; Shu-Chien, A.C. Gene Arrangement and Adaptive Evolution in the Mitochondrial Genomes of Terrestrial Sesarmid Crabs Geosesarma faustum and Geosesarma penangensis. Front. Ecol. Evol. 2021, 9, 778570. [Google Scholar] [CrossRef]
- Tan, M.H.; Gan, H.M.; Lee, Y.P.; Austin, C.M. The complete mitogenome of the ghost crab Ocypode ceratophthalmus (Pallas, 1772) (Crustacea: Decapoda: Ocypodidae). Mitochondrial DNA A 2016, 27, 2123–2124. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Shi, X.; Guo, H.; Tang, D.; Bai, Y.; Wang, Z. Characterization of the complete mitochondrial genome of Uca lacteus and comparison with other Brachyuran crabs. Genomics 2020, 112, 10–19. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, S.; Chen, Y.; Liu, Q.; Tang, B. Mitochondrial Genome of Grapsus albolineatus and Insights into the Phylogeny of Brachyura. Animals 2025, 15, 679. [Google Scholar] [CrossRef]
- Rodriguez-Pilco, M.A.; Leśny, P.; Podsiadłowski, L.; Schubart, C.D.; Baeza, J.A. Characterization of the Complete Mitochondrial Genome of the Bromeliad Crab Metopaulias depressus (Rathbun, 1896) (Crustacea: Decapoda: Brachyura: Sesarmidae). Genes 2022, 13, 299. [Google Scholar] [CrossRef]
- Sung, J.M.; Lee, J.; Kim, S.K.; Karagozlu, M.Z.; Kim, C.B. The complete mitochondrial genome of Grapsus tenuicrustatus (Herbst, 1783) (Decapoda, Grapsidae). Mitochondrial DNA B 2016, 1, 441–442. [Google Scholar] [CrossRef]
- Shen, H.; Braband, A.; Scholtz, G. Mitogenomic analysis of decapod crustacean phylogeny corroborates traditional views on their relationships. Mol. Phylogenet. Evol. 2013, 66, 776–789. [Google Scholar] [CrossRef]
- Ji, Y.-K.; Wang, A.; Lu, X.-L.; Song, D.-H.; Jin, Y.-H.; Lu, J.-J.; Sun, H.-Y. Mitochondrial genomes of two brachyuran crabs (Crustacea: Decapoda) and phylogenetic analysis. J. Crust. Biol. 2014, 34, 494–503. [Google Scholar] [CrossRef]
- Ki, J.S.; Dahms, H.U.; Hwang, J.S.; Lee, J.S. The complete mitogenome of the hydrothermal vent crab Xenograpsus testudinatus (Decapoda, Brachyura) and comparison with brachyuran crabs. Comp. Biochem. Physiol. D 2009, 4, 290–299. [Google Scholar] [CrossRef]
- Miller, A.D.; Murphy, N.P.; Burridge, C.P.; Austin, C.M. Complete mitochondrial DNA sequences of the decapod crustaceans Pseudocarcinus gigas (Menippidae) and Macrobrachium rosenbergii (Palaemonidae). Mar. Biotechnol. 2005, 7, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.D.; Nguyen, T.T.T.; Burridge, C.P.; Austin, C.M. Complete mitochondrial DNA sequence of the Australian freshwater crayfish, Cherax destructor (Crustacea: Decapoda: Parastacidae): A novel gene order revealed. Gene 2004, 331, 65–72. [Google Scholar] [CrossRef]
- Ingman, M.; Kaessmann, H.; Pääbo, S.; Gyllensten, U. Mitochondrial Genome Variation and the Origin of Modern Humans. Nature 2000, 408, 708–713. [Google Scholar] [CrossRef]
- Tang, B.P.; Xin, Z.Z.; Liu, Y.; Zhang, D.Z.; Wang, Z.F.; Zhang, H.B.; Chai, X.Y.; Zhou, C.L.; Liu, Q.N. The Complete Mitochondrial Genome of Sesarmops sinensis Reveals Gene Rearrangements and Phylogenetic Relationships in Brachyura. PLoS ONE 2017, 12, e0179800. [Google Scholar] [CrossRef]
- Zhang, J.; Kan, X.; Miao, G.; Hu, S.; Sun, Q.; Tian, W. qMGR: A New Approach for Quantifying Mitochondrial Genome Rearrangement. Mitochondrion 2020, 52, 20–23. [Google Scholar] [CrossRef]
- Jühling, F.; Pütz, J.; Bernt, M.; Donath, A.; Middendorf, M.; Florentz, C.; Stadler, P.F. Improved Systematic tRNA Gene Annotation Allows New Insights into the Evolution of Mitochondrial tRNA Structures and into the Mechanisms of Mitochondrial Genome Rearrangements. Nucleic Acids Res. 2012, 40, 2833–2845. [Google Scholar] [CrossRef]
- Pang, X.; Han, C.; Guo, B.; Liu, K.; Lin, X.; Lu, X. The First Complete Mitochondrial Genome of Eucrate crenata (Decapoda: Brachyura: Goneplacidae) and Phylogenetic Relationships within Infraorder Brachyura. Genes 2022, 13, 1127. [Google Scholar] [CrossRef]
- Rawlings, T.A.; Collins, T.M.; Bieler, R. Changing Identities: tRNA Duplication and Remolding within Animal Mitochondrial Genomes. Proc. Natl. Acad. Sci. USA 2003, 100, 15700–15705. [Google Scholar] [CrossRef]
- Ghanavi, H.R.; Rahimi, P.; Tavana, M.; Rezaei Tavabe, K.; Jouladeh-Roudbar, A.; Doadrio, I. The Evolutionary Journey of Freshwater Crabs of the Genus Potamon (Decapoda: Brachyura: Potamidae). Mol. Phylogenet. Evol. 2023, 180, 107690. [Google Scholar] [CrossRef]
- Reyes, A.; Gissi, C.; Pesole, G.; Saccone, C. Asymmetrical Directional Mutation Pressure in the Mitochondrial Genome of Mammals. Mol. Biol. Evol. 1998, 15, 957–966. [Google Scholar] [CrossRef]

| Accession No. | Species | Size (bp) | A + T% | Reference |
|---|---|---|---|---|
| MF198251.1 | Metopograpsus quadridentatus | 15,523 | 70.4 | this study |
| OL661264.1 | Cleistostoma dilatatum | 15,443 | 69.0 | [4] |
| NC_063602.1 | Plagusia squamosa | 15,460 | 69.9 | unpulished |
| NC_063149.1 | Episesarma lafondii | 15,640 | 75.9 | [5] |
| NC_061931.1 | Parasesarma eumolpe | 15,646 | 75.5 | [19] |
| NC_057477.1 | Cardisoma armatum | 15,586 | 69.0 | [19] |
| NC_057475.1 | Gecarcoidea lalandii | 15,575 | 75.0 | [19] |
| NC_054248.1 | Etisus dentatus | 15,884 | 71.9 | [33] |
| NC_051868.1 | Perisesarma bidens | 15,641 | 74.8 | [34] |
| NC_047209.1 | Chiromantes eulimene | 15,894 | 75.5 | [35] |
| NC_046797.1 | Ocypode stimpsoni | 15,557 | 67.8 | [36] |
| NC_042401.1 | Austruca lactea | 15,659 | 69.4 | unpublished |
| NC_042152.1 | Metopograpsus frontalis | 15,587 | 69.8 | [37] |
| NC_042142.1 | Chiromantes haematocheir | 15,899 | 75.6 | [38] |
| NC_041212.1 | Orisarma dehaani | 15,917 | 75.7 | unpublished |
| NC_040977.1 | Nanosesarma minutum | 15,637 | 77.7 | [39] |
| NC_039990.1 | Parasesarma affine | 15,638 | 74.8 | [40] |
| NC_039111.1 | Cranuca inversa | 15,677 | 71.0 | [41] |
| NC_039106.1 | Tubuca polita | 15,672 | 71.6 | [41] |
| NC_039105.1 | Cardisoma carnifex | 15,597 | 68.8 | [41] |
| NC_038180.1 | Dotilla wichmanni | 15,600 | 68.5 | [42] |
| NC_038066.1 | Parasesarma pictum | 15,611 | 75.6 | [43] |
| MZ725941.1 | Geosesarma penangense | 15,955 | 78.4 | unpublished |
| MZ725940.1 | Geosesarma faustum | 15,880 | 78.4 | [44] |
| MW255974.1 | Ocypode ceratophthalmus | 15,555 | 69.5 | [45] |
| MN072632.2 | Tubuca rosea | 15,643 | 71.1 | unpublished |
| MH816962.2 | Gecarcoidea natalis | 15,545 | 72.2 | unpublished |
| MH796169.1 | Austruca lactea | 15,661 | 69.6 | [46] |
| MF198247.1 | Grapsus albolineatus | 15,580 | 67.4 | [47] |
| KX156954.1 | Orisarma neglectum | 15,920 | 75.6 | unpublished |
| KX118277.1 | Metopaulias depressus | 15,765 | 77.3 | [48] |
| KU589292.1 | Clistocoeloma sinense | 15,706 | 75.7 | unpublished |
| KU343209.2 | Parasesarma tripectinis | 15,612 | 74.2 | unpublished |
| KT878721.1 | Grapsus tenuicrustatus | 15,858 | 65.0 | [49] |
| KR336554.1 | Orisarma sinense | 15,905 | 75.7 | unpublished |
| KC107816.1 | Neopetrolisthes maculatus | 15,324 | 71.2 | [50] |
| JF909979.1 | Ilyoplax deschampsi | 15,460 | 69.6 | [51] |
| EU727203.1 | Xenograpsus testudinatus | 15,798 | 73.9 | [52] |
| AY562127.1 | Pseudocarcinus gigas | 15,515 | 70.5 | [53] |
| AY383557.2 | Cherax destructor | 15,894 | 62.4 | [54] |
| Gene | From | To | Size | Intergenic Nucleotides | Start | Stop | Strand |
|---|---|---|---|---|---|---|---|
| cox1 | 1 | 1534 | 1534 | ATG | T | H | |
| trnL1 | 1535 | 1601 | 67 | H | |||
| cox2 | 1610 | 2297 | 688 | 8 | ATG | T | H |
| trnK | 2298 | 2366 | 69 | H | |||
| trnD | 2367 | 2429 | 63 | H | |||
| atp8 | 2430 | 2588 | 159 | ATG | TAA | H | |
| atp6 | 2582 | 3255 | 674 | −7 | ATT | TA | H |
| cox3 | 3256 | 4046 | 791 | ATG | TA | H | |
| trnG | 4047 | 4112 | 66 | H | |||
| nad3 | 4113 | 4463 | 351 | ATT | TAA | H | |
| trnA | 4462 | 4525 | 64 | −2 | H | ||
| trnR | 4528 | 4591 | 64 | 2 | H | ||
| trnN | 4600 | 4668 | 69 | 8 | H | ||
| trnS1 | 4673 | 4740 | 68 | 4 | H | ||
| trnE | 4742 | 4810 | 69 | 1 | H | ||
| trnH | 4813 | 4877 | 65 | 2 | L | ||
| trnF | 4882 | 4944 | 63 | 4 | L | ||
| nad5 | 4970 | 6703 | 1734 | 25 | ATG | TAG | L |
| nad4 | 6736 | 8085 | 1350 | 32 | GTG | TAA | L |
| nad4L | 8079 | 8381 | 303 | −7 | ATG | TAA | L |
| trnT | 8399 | 8464 | 66 | 17 | H | ||
| trnP | 8465 | 8531 | 67 | L | |||
| nad6 | 8534 | 9039 | 506 | 2 | ATT | TA | H |
| cytb | 9040 | 10,174 | 1135 | ATG | T | H | |
| trnS2 | 10,175 | 10,241 | 67 | H | |||
| nad1 | 10,260 | 11,207 | 948 | 18 | ATG | TAA | L |
| trnL2 | 11,230 | 11,296 | 67 | 22 | L | ||
| rrnL | 11,297 | 12,619 | 1323 | L | |||
| trnV | 12,623 | 12,696 | 74 | 3 | L | ||
| rrnS | 12,698 | 13,529 | 832 | 1 | L | ||
| trnI | 14,098 | 14,166 | 69 | 568 | H | ||
| trnQ | 14,164 | 14,232 | 69 | −3 | L | ||
| trnM | 14,240 | 14,309 | 70 | 7 | H | ||
| nad2 | 14,313 | 15,320 | 1008 | 3 | ATT | TAG | H |
| trnW | 15,319 | 15,386 | 68 | −2 | H | ||
| trnC | 15,390 | 15,452 | 63 | 3 | L | ||
| trnY | 15,456 | 15,523 | 68 | 3 | L |
| Codon | Count | RSCU | Codon | Count | RSCU | Codon | Count | RSCU | Codon | Count | RSCU |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UUU(F) | 264 | 1.6 | UCU(S) | 118 | 2.59 | UAU(Y) | 125 | 1.64 | UGU(C) | 33 | 1.69 |
| UUC(F) | 65 | 0.4 | UCC(S) | 24 | 0.53 | UAC(Y) | 27 | 0.36 | UGC(C) | 6 | 0.31 |
| UUA(L) | 311 | 3.15 | UCA(S) | 67 | 1.47 | UAA(*) | 5 | 1.43 | UGA(W) | 82 | 1.69 |
| UUG(L) | 64 | 0.65 | UCG(S) | 8 | 0.18 | UAG(*) | 2 | 0.57 | UGG(W) | 15 | 0.31 |
| CUU(L) | 110 | 1.11 | CCU(P) | 62 | 1.65 | CAU(H) | 46 | 1.08 | CGU(R) | 19 | 1.38 |
| CUC(L) | 25 | 0.25 | CCC(P) | 24 | 0.64 | CAC(H) | 39 | 0.92 | CGC(R) | 4 | 0.29 |
| CUA(L) | 73 | 0.74 | CCA(P) | 56 | 1.49 | CAA(Q) | 63 | 1.73 | CGA(R) | 27 | 1.96 |
| CUG(L) | 9 | 0.09 | CCG(P) | 8 | 0.21 | CAG(Q) | 10 | 0.27 | CGG(R) | 5 | 0.36 |
| AUU(I) | 258 | 1.73 | ACU(T) | 95 | 2.01 | AAU(N) | 97 | 1.4 | AGU(S) | 30 | 0.66 |
| AUC(I) | 41 | 0.27 | ACC(T) | 31 | 0.66 | AAC(N) | 42 | 0.6 | AGC(S) | 9 | 0.2 |
| AUA(M) | 173 | 1.61 | ACA(T) | 56 | 1.19 | AAA(K) | 76 | 1.69 | AGA(S) | 72 | 1.58 |
| AUG(M) | 42 | 0.39 | ACG(T) | 7 | 0.15 | AAG(K) | 14 | 0.31 | AGG(S) | 37 | 0.81 |
| GUU(V) | 115 | 1.71 | GCU(A) | 116 | 2.25 | GAU(D) | 43 | 1.25 | GGU(G) | 60 | 1.04 |
| GUC(V) | 8 | 0.12 | GCC(A) | 33 | 0.64 | GAC(D) | 26 | 0.75 | GGC(G) | 18 | 0.31 |
| GUA(V) | 121 | 1.8 | GCA(A) | 49 | 0.95 | GAA(E) | 55 | 1.49 | GGA(G) | 108 | 1.88 |
| GUG(V) | 25 | 0.37 | GCG(A) | 8 | 0.16 | GAG(E) | 19 | 0.51 | GGG(G) | 44 | 0.77 |
| Species | Family | Size (bp) | A (%) | T (%) | C (%) | G (%) | AT Skew |
|---|---|---|---|---|---|---|---|
| Metopograpsus frontalis | Grapsidae | 15,587 | 32.8 | 37 | 19.3 | 11 | −0.06 |
| Orisarma dehaani | Sesarmidae | 15,917 | 37.5 | 38.2 | 14.8 | 9.5 | −0.01 |
| Orisarma neglectum | Sesarmidae | 15,920 | 37.4 | 38.2 | 14.9 | 9.5 | −0.01 |
| Orisarma sinense | Sesarmidae | 15,905 | 37.4 | 38.3 | 14.9 | 9.4 | −0.012 |
| Chiromantes eulimene | Sesarmidae | 15,894 | 37.1 | 38.4 | 14.8 | 9.7 | −0.017 |
| Episesarma lafondii | Sesarmidae | 15,640 | 37 | 38.9 | 14.7 | 9.4 | −0.025 |
| Chiromantes haematocheir | Sesarmidae | 15,899 | 37.3 | 38.3 | 15 | 9.4 | −0.013 |
| Clistocoeloma sinense | Sesarmidae | 15,706 | 37.1 | 38.6 | 14.9 | 9.4 | −0.02 |
| Geosesarma penangense | Sesarmidae | 15,955 | 38.3 | 40.1 | 13.1 | 8.5 | −0.023 |
| Cardisoma armatum | Gecarcinidae | 15,586 | 35.3 | 33.7 | 20.7 | 10.3 | 0.023 |
| Perisesarma bidens | Sesarmidae | 15,641 | 36.6 | 38.2 | 15.1 | 10.1 | −0.021 |
| Cardisoma carnifex | Gecarcinidae | 15,597 | 35.4 | 33.4 | 21 | 10.1 | 0.028 |
| Parasesarma eumolpe | Sesarmidae | 15,646 | 36.7 | 38.8 | 14.7 | 9.8 | −0.027 |
| Parasesarma tripectinis | Sesarmidae | 15,612 | 36.2 | 38 | 15.7 | 10.1 | −0.024 |
| Nanosesarma minutum | Sesarmidae | 15,637 | 38 | 39.7 | 13.4 | 8.9 | −0.022 |
| Geosesarma faustum | Sesarmidae | 15,880 | 38.7 | 39.7 | 13.2 | 8.3 | −0.013 |
| Metopaulias depressus | Sesarmidae | 15,765 | 37.9 | 39.4 | 14 | 8.7 | −0.019 |
| Plagusia squamosa | Plagusiidae | 15,460 | 34.3 | 35.6 | 18.9 | 11.2 | −0.018 |
| Cleistostoma dilatatum | Camptandriidae | 15,443 | 34.3 | 34.7 | 19.5 | 11.4 | −0.006 |
| Grapsus albolineatus | Grapsidae | 15,580 | 33.4 | 34 | 20.5 | 12.1 | −0.01 |
| Cranuca inversa | Ocypodidae | 15,677 | 35.8 | 35.2 | 18.1 | 10.9 | 0.008 |
| Dotilla wichmanni | Dotillidae | 15,600 | 33.8 | 34.7 | 20.7 | 10.8 | −0.014 |
| Ilyoplax deschampsi | Dotillidae | 15,460 | 34.1 | 35.5 | 19.7 | 10.7 | −0.019 |
| Austruca lactea | Ocypodidae | 15,659 | 34.8 | 34.6 | 18.5 | 12 | 0.003 |
| Ocypode stimpsoni | Ocypodidae | 15,557 | 33.7 | 34.1 | 20.8 | 11.4 | −0.006 |
| Ocypode ceratophthalmus | Ocypodidae | 15,555 | 33.8 | 35.7 | 19.4 | 11.1 | −0.028 |
| Pseudocarcinus gigas | Eriphiidae | 15,515 | 35 | 35.5 | 18.7 | 10.8 | −0.006 |
| Gecarcoidea lalandii | Gecarcinidae | 15,575 | 37.7 | 37.3 | 15.8 | 9.2 | 0.005 |
| Gecarcoidea natalis | Gecarcinidae | 15,545 | 36 | 36.2 | 15.1 | 9.1 | −0.003 |
| Parasesarma pictum | Sesarmidae | 15,611 | 36.6 | 39 | 14.6 | 9.8 | −0.032 |
| Tubuca rosea | Ocypodidae | 15,643 | 36.6 | 34.5 | 18.6 | 10.3 | 0.029 |
| Parasesarma affine | Sesarmidae | 15,638 | 36.6 | 38.2 | 15.1 | 10.1 | −0.022 |
| Tubuca polita | Ocypodidae | 15,672 | 36.6 | 35 | 17.5 | 10.9 | 0.022 |
| Austruca lactea | Ocypodidae | 15,661 | 34.9 | 34.7 | 18.5 | 12 | 0.003 |
| Etisus dentatus | Xanthidae | 15,884 | 37.9 | 34 | 10.4 | 17.8 | 0.054 |
| Grapsus tenuicrustatus | Grapsidae | 15,858 | 31.9 | 33.1 | 22.8 | 12.1 | −0.018 |
| Xenograpsus testudinatus | Xenograpsidae | 15,798 | 36.7 | 37.2 | 16.8 | 9.3 | −0.007 |
| Neopetrolisthes maculatus | Porcellanidae | 15,324 | 34.9 | 36.3 | 17.4 | 11.4 | −0.02 |
| Cherax destructor | Parastacidae | 15,894 | 32.1 | 30.3 | 24.1 | 13.5 | 0.029 |
| Metopograpsus quadridentatus | Grapsidae | 15,523 | 34.3 | 36.1 | 19.5 | 10.2 | −0.025 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bian, D.-D.; Tang, S.; Wang, S.-N.; Liu, Q.-N.; Tang, B.-P. Comparative Analysis of Metopograpsus quadridentatus (Crustacea: Decapoda: Grapsidae) Mitochondrial Genome Reveals Gene Rearrangement and Phylogeny. Animals 2025, 15, 1162. https://doi.org/10.3390/ani15081162
Bian D-D, Tang S, Wang S-N, Liu Q-N, Tang B-P. Comparative Analysis of Metopograpsus quadridentatus (Crustacea: Decapoda: Grapsidae) Mitochondrial Genome Reveals Gene Rearrangement and Phylogeny. Animals. 2025; 15(8):1162. https://doi.org/10.3390/ani15081162
Chicago/Turabian StyleBian, Dan-Dan, Sheng Tang, Song-Nan Wang, Qiu-Ning Liu, and Bo-Ping Tang. 2025. "Comparative Analysis of Metopograpsus quadridentatus (Crustacea: Decapoda: Grapsidae) Mitochondrial Genome Reveals Gene Rearrangement and Phylogeny" Animals 15, no. 8: 1162. https://doi.org/10.3390/ani15081162
APA StyleBian, D.-D., Tang, S., Wang, S.-N., Liu, Q.-N., & Tang, B.-P. (2025). Comparative Analysis of Metopograpsus quadridentatus (Crustacea: Decapoda: Grapsidae) Mitochondrial Genome Reveals Gene Rearrangement and Phylogeny. Animals, 15(8), 1162. https://doi.org/10.3390/ani15081162







