Effect of Dietary Carbohydrate-to-Protein Ratio on Gut Microbiota in Atlantic Salmon (Salmo salar)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Method
2.1. Fish and Rearing Conditions
2.2. Feeding Trial
2.3. Sampling Procedure
2.4. DNA Extraction and Sequencing
2.5. Bioinformatics Analysis
2.6. Ethical Notes
2.7. Statistical Analysis
3. Results
3.1. Fish Growth Performance
3.2. High-Throughput Sequence Data
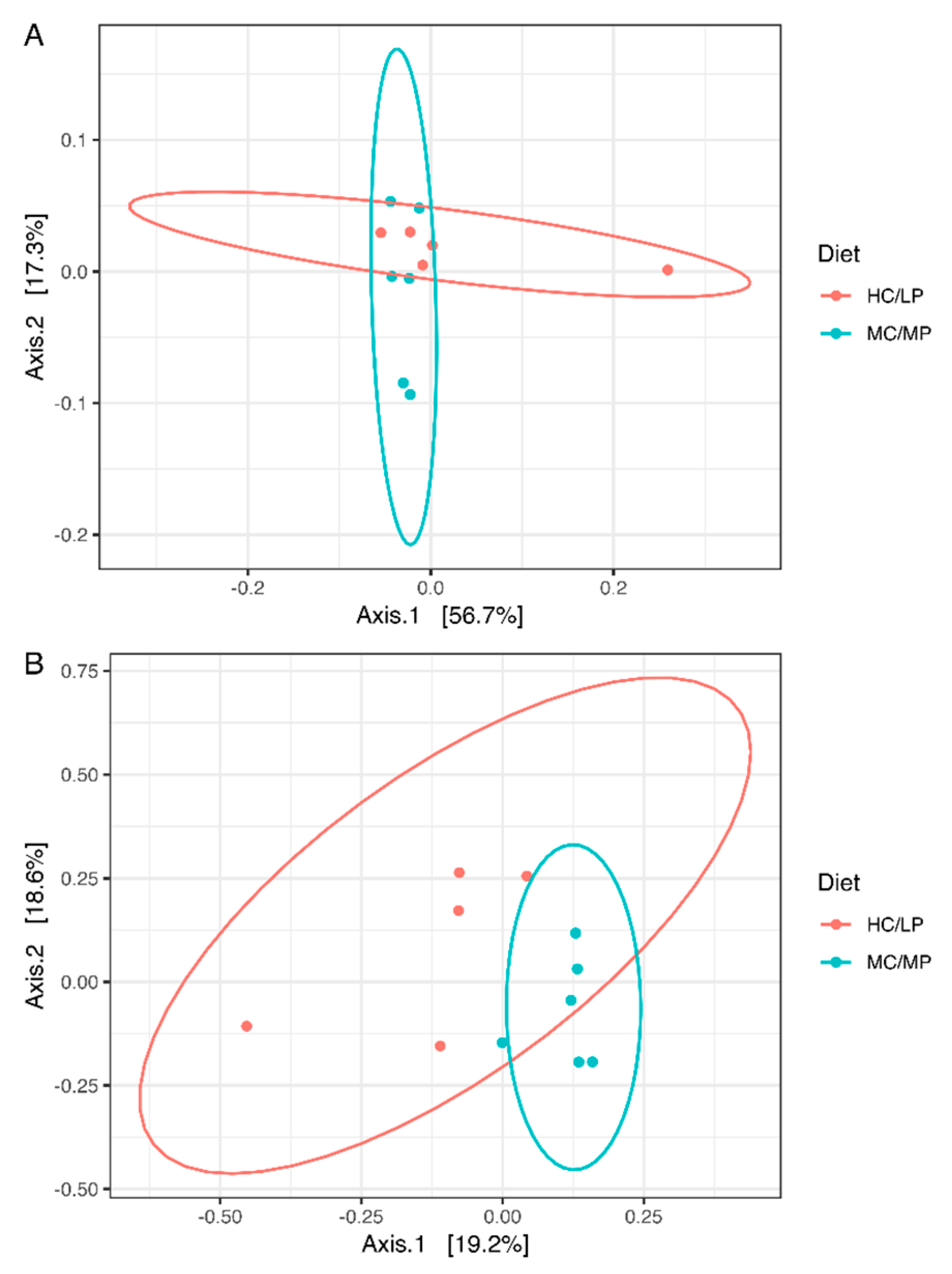
3.3. Diversity Analysis of Microbiota
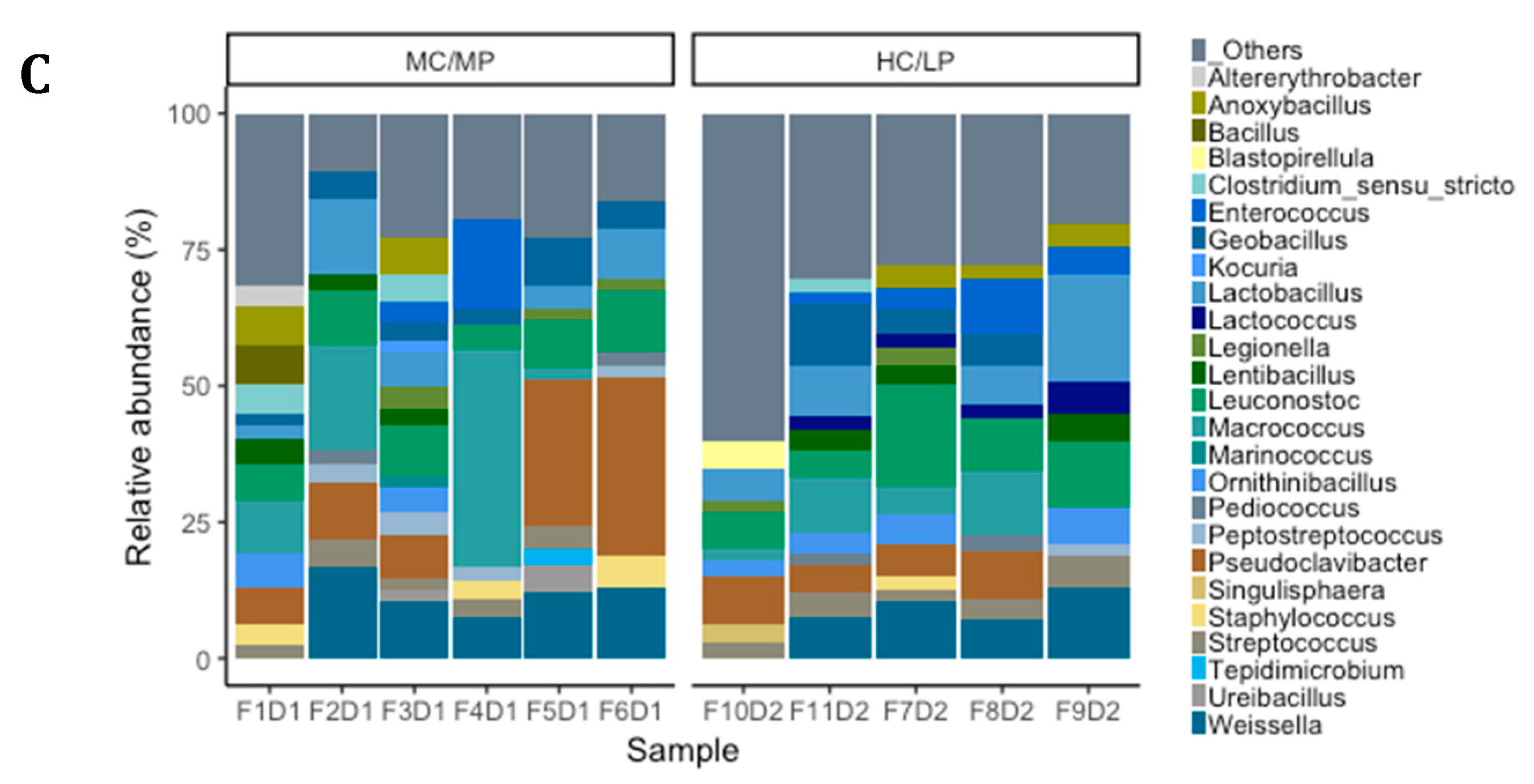
3.4. Taxonomic Composition and Differential Abundance of Bacterial Communities of Distal Intestine Digesta of Fish Fed Experimental Diets p
3.5. Core Microbiome for Distal Intestine Digesta of Fish Fed Experimental Diets
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Naylor, R.L.; Hardy, R.W.; Bureau, D.P.; Chiu, A.; Elliott, M.; Farrell, A.P.; Forster, I.; Gatlin, D.M.; Goldburg, R.J.; Hua, K.; et al. Feeding aquaculture in an era of finite resources. Proc. Natl. Acad. Sci. USA 2009, 106, 15103–15110. [Google Scholar] [CrossRef] [PubMed]
- Small, B.C.; Hardy, R.W.; Tucker, C.S. Enhancing fish performance in aquaculture. Anim. Front. 2016, 6, 42–49. [Google Scholar] [CrossRef]
- National Research Council. Nutrient Requirements of Fish and Shrimp, 1st ed.; The National Academic Press: Washington, DC, USA, 2011; pp. 299–304. [Google Scholar]
- Stone, D.A.J. Dietary Carbohydrate Utilization by Fish. Rev. Fish. Sci. 2003, 11, 337–369. [Google Scholar] [CrossRef]
- Kamalam, B.S.; Medale, F.; Panserat, S. Utilisation of dietary carbohydrates in farmed fishes: New insights on influencing factors, biological limitations and future strategies. Aquaculture 2017, 467, 3–27. [Google Scholar] [CrossRef]
- Wilson, R.P. Utilization of dietary carbohydrate by fish. Aquaculture 1994, 124, 67–80. [Google Scholar] [CrossRef]
- Polakof, S.; Panserat, S.; Soengas, J.L.; Moon, T. Glucose metabolism in fish: A review. J. Comp. Physiol. B 2012, 182, 1015–1045. [Google Scholar] [CrossRef] [PubMed]
- Dworkin, M.; Falkow, S.; Rosenberg, E.; Schleifer, K.-H.; Stackebrandt, E. The Prokaryotes a Handbook on the Biology of Bacteria, Volume 1: Symbiotic Associations, Biotechnology, Applied Microbiology, 3rd ed.; Springer Science + Business Media, Inc.: New York, NY, USA, 2006; pp. 137–185. [Google Scholar]
- Gatesoupe, F.J.; Huelvan, C.; Le Bayon, N.; Le Delliou, H.; Madec, L.; Mouchel, O.; Quazuguel, P.; Mazurais, D.; Zambonino-Infante, J.L. The highly variable microbiota associated to intestinal mucosa correlates with growth and hypoxia resistance of sea bass, Dicentrarchus labrax, submitted to different nutritional histories. BMC Microbiol. 2016, 16, 266. [Google Scholar] [CrossRef] [PubMed]
- Geurden, I.; Mennigen, J.; Plagnes-Juan, E.; Veron, V.; Cerezo, T.; Mazurais, D.; Zambonino-Infante, J.; Gatesoupe, J.S.; Skiba-Cassy, S. High or low dietary carbohydrate: protein ratios during first-feeding affect glucose metabolism and intestinal microbiota in juvenile rainbow trout. J. Exp. Biol. 2014, 217, 3396–3406. [Google Scholar] [CrossRef] [PubMed]
- Rimoldi, S.; Terova, G.; Ascione, C.; Giannico, R.; Brambilla, F. Next generation sequencing for gut microbiome characterization in rainbow trout (Oncorhynchus mykiss) fed animal by-product meals as an alternative to fishmeal protein sources. PLoS ONE 2018, 13. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuczynski, J.; Lauber, C.L.; Walters, W.A.; Parfrey, L.W.; Clemente, J.C.; Gevers, D.; Knight, R. Experimental and analytical tools for studying the human microbiome. Nat. Rev. Genet. 2012, 13, 47–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lokesh, J.; Kiron, V. Transition from freshwater to seawater reshapes the skin-associated microbiota of Atlantic salmon. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. 2011. R Package Version 1.17–10. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 10 March 2019).
- Segata, N.; Bornigen, D.; Morgan, X.C.; Huttenhower, C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Marandel, L.; Veron, V.; Surget, A.; Plagnes-Juan, E.; Panserat, S. Glucose metabolism ontogenesis in rainbow trout (Oncorhynchus mykiss) in the light of the recently sequenced genome: New tools for intermediary metabolism programming. J. Exp. Biol. 2016, 219, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Dimitroglou, A.; Merrifield, D.L.; Moate, R.; Davies, S.J.; Spring, P.; Sweetman, J.; Bradley, G. Dietary mannan oligosaccharide supplementation modulates intestinal microbial ecology and improves gut morphology of rainbow trout, Oncorhynchus mykiss (Walbaum). J. Anim. Sci. 2009, 87, 3226–3234. [Google Scholar] [CrossRef] [PubMed]
- Torrecillas, S.; Montero, D.; Izquierdo, M. Improved health and growth of fish fed mannan oligosaccharides: Potential mode of action. Fish Shellfish Immunol. 2014, 36, 525–544. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.R.; Links, M.G.; Collins, S.A.; Mansfield, G.S.; Drew, M.D.; Van Kessel, A.G.; Hill, J.E. Effects of plant-based diets on the distal gut microbiome of rainbow trout (Oncorhynchus mykiss). Aquaculture 2012, 350, 134–142. [Google Scholar] [CrossRef]
- Bruce, T.J.; Neiger, R.D.; Brown, M.L. Gut histology, immunology and the intestinal microbiota of rainbow trout, Oncorhynchus mykiss (Walbaum), fed process variants of soybean meal. Aquac. Res. 2018, 49, 492–504. [Google Scholar] [CrossRef]
- Gajardo, K.; Jaramillo-Torres, A.; Kortner, T.M.; Merrifield, D.L.; Tinsley, J.; Bakke, A.M.; Krogdahl, Å. Alternative protein sources in the diet modulate microbiota and functionality in the distal intestine of Atlantic salmon (Salmo salar). Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Gatesoupe, F.-J.; Fauconneau, B.; Deborde, C.; Madji Hounoum, B.; Jacob, D.; Moing, A.; Médale, F. Intestinal microbiota in rainbow trout, Oncorhynchus mykiss fed diets with different levels of fish-based and plant ingredients: A correlative approach with some plasma metabolites. Aquac. Nutr. 2018, 6. [Google Scholar] [CrossRef]
- Michl, S.C.; Ratten, J.M.; Beyer, M.; Hasler, M.; LaRoche, J.; Schulz, C. The malleable gut microbiome of juvenile rainbow trout (Oncorhynchus mykiss): Diet-dependent shifts of bacterial community structures. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Panserat, S.; Kaushik, S.; Médale, F. Rainbow trout as a model for nutrition and nutrient metabolism studies. In Trout: From Physiology to Conservation; Polakof, S., Moon, T.W., Eds.; Nova Science Publishers: Hauppauge, NY, USA, 2013; pp. 131–153. [Google Scholar]
- Limborg, M.T.; Heeb, P. Special Issue: Coevolution of Hosts and Their Microbiome. Genes 2018, 9, 549. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Guo, X.; Gooneratne, R.; Lai, R.; Zeng, C.; Zhan, F.; Wang, W. The gut microbiome and degradation enzyme activity of wild freshwater fishes influenced by their trophic levels. Sci Rep. 2016, 6, 24340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reveco, F.E.; Øverland, M.; Romarheim, O.H.; Mydland, L.T. Intestinal bacterial community structure differs between healthy and inflamed intestines in Atlantic salmon (Salmo salar L.). Aquaculture 2014, 420, 262–269. [Google Scholar] [CrossRef]
- Abid, A.; Davies, S.J.; Waines, P.; Emery, M.; Castex, M.; Gioacchini, G.; Carnevali, O.; Bickerdike, R.; Romero, J.; Merrifield, D.L. Dietary synbiotic application modulates Atlantic salmon (Salmo salar) intestinal microbial communities and intestinal immunity. Fish Shellfish Immunol. 2013, 35, 1948–1956. [Google Scholar] [CrossRef] [PubMed]
- Green, T.J.; Smullen, R.; Barnes, A.C. Dietary soybean protein concentrate-induced intestinal disorder in marine farmed Atlantic salmon, Salmo salar is associated with alterations in gut microbiota. Vet. Microbiol. 2013, 166, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Ingerslev, H.-C.; von Gersdorff Jørgensen, L.; Lenz Strube, M.; Larsen, N.; Dalsgaard, I.; Boye, M.; Madsen, L. The development of the gut microbiota in rainbow trout (Oncorhynchus mykiss) is affected by first feeding and diet type. Aquaculture 2014, 424, 24–34. [Google Scholar] [CrossRef]
- Dehler, C.E.; Secombes, C.J.; Martin, S.A.M. Environmental and physiological factors shape the gut microbiota of Atlantic salmon parr (Salmo salar L.). Aquaculture 2017, 467, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, C.; Courcelle, M.; Redivo, B.; Derome, N. Structural and compositional mismatch between captive and wild Atlantic salmon (Salmo salar) parrs gut microbiota highlights the relevance of integrating molecular ecology for management and conservation methods. Evol. Appl. 2018, 11, 1671–1685. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, C.; Romero, J. The microbiome of Seriola lalandi of wild and aquaculture origin reveals differences in composition and potential function. Front. Microbiol. 2017, 8, 1844. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, C.; Romero, J. Fine flounder (Paralichthys adspersus) microbiome showed important differences between wild and reared specimens. Front. Microbiol. 2017, 8, 271. [Google Scholar] [CrossRef] [PubMed]
- Boedeker, C.; Schuler, M.; Reintjes, G.; Jeske, O.; van Teeseling, M.C.; Jogler, M.; Rast, P.; Borchert, D.; Devos, D.P.; Kucklick, M.; et al. Determining the bacterial cell biology of Planctomycetes. Nat. Commun. 2017, 8, 14853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elshahed, M.S.; Youssef, N.H.; Luo, Q.; Najar, F.Z.; Roe, B.A.; Sisk, T.M.; Buhring, S.I.; Hinrichs, K.U.; Krumholz, L.R. Phylogenetic and metabolic diversity of Planctomycetes from anaerobic, sulfide-and sulfurrich Zodletone Spring, Oklahoma. Appl. Environ. Microbiol. 2007, 73, 4707–4716. [Google Scholar] [CrossRef] [PubMed]
- Merrifield, D.L.; Balcázar, J.L.; Daniels, C.; Zhou, Z.; Carnevali, O.; Sun, Y.-Z.; Hoseinifar, S.H.; Ringo, E. Indigenous Lactic Acid Bacteria in Fish and Crustaceans, Aquaculture Nutrition, Gut Health, Probiotics and Prebiotics, 1st ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2014; pp. 128–168. [Google Scholar]
- Kim, J.D.; Kaushik, S. Contribution of digestible energy from carbohydrates and estimation of protein/energy requirements for growth of rainbow trout (Oncorhynchus mykiss). Aquaculture 1992, 106, 161–169. [Google Scholar] [CrossRef]
- Hemre, G.I.; Sandnes, K.; Lie, Ø.; Torrissen, O.; Waagbø, A.R. Carbohydrate nutrition in Atlantic salmon, Salmo salar L., growth and feed utilisation. Aquac. Nutr. 1995, 26, 149–154. [Google Scholar] [CrossRef]
- Hemre, G.-I.; Mommsen, T.P.; Krogdahl, A. Carbohydrates in fish nutrition: Effects on growth, glucose metabolism and hepatic enzymes. Aquac. Nutr. 2002, 8, 175–194. [Google Scholar] [CrossRef]
- Krogdahl, A.; Sundby, A.; Olli, J.J. Atlantic salmon (Salmo salar, L) and rainbow trout (Oncorhynchus mykiss) digest and metabolize nutrients differently depending on water salinity and dietary starch level. Aquaculture 2004, 229, 335–360. [Google Scholar] [CrossRef]
- Hehemann, J.H.; Correc, G.; Barbeyron, T.; Helbert, W.; Czjzek, M.; Michel, G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 2010, 464, 908–912. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, E.S.; DeLoache, W.C.; Pruss, K.M.; Whitaker, W.R.; Sonnenburg, J.L. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 2018, 557, 434–438. [Google Scholar] [CrossRef] [PubMed]




| Ingredients (%, Diet) | Diet | |
|---|---|---|
| MC/MP | HC/LP | |
| Fish meal 1 | 30 | 21 |
| Soy protein concentrate 1 | 30 | 23 |
| Wheat gluten 1 | 7.5 | 7.5 |
| Wheat starch, gelatinized 1 | 15 | 30 |
| Fish oil 1 | 14 | 15 |
| Vitamin C (35%) 2 | 0.2 | 0.2 |
| Vitamin Premix 2,3 | 0.8 | 0.8 |
| Mineral Premix 2,4 | 0.2 | 0.2 |
| Choline chloride 2 | 0.3 | 0.3 |
| Dicalcium phosphate 2 | 2 | 1.7 |
| L-Methionine 5 | 0 | 0.2 |
| Lysine 6 | 0 | 0.1 |
| Chemical composition (%, DM) | ||
| Moister | 4.7 | 7 |
| Crude protein | 50.1 | 41.5 |
| Fat | 16.2 | 16.6 |
| Ash | 9.4 | 7.4 |
| Fiber | 1.9 | 1.6 |
| Gross energy (MJ/kg) | 20.7 | 20.5 |
| Experimental Diets | p Value | ||||
|---|---|---|---|---|---|
| MC/MP (15% Digestible Starch) | HC/LP (30% Digestible Starch) | ||||
| Initial weight, g/fish 1 | 103.8 | 0.5 | 105.5 | 1.5 | >0.05 |
| Final weight, g/fish 1 | 117.8 | 0.7 | 118.0 | 0.1 | >0.05 |
| Weight gain, g/fish b1 | 14.0 * | 1.2 | 11.7 | 0.2 | 0.033 |
| Feed intake as fed, g/fish c1 | 26.5 | 1.7 | 25.6 | 0.3 | >0.05 |
| FCR d1 | 1.5 | 0.1 | 1.9 * | 0.0 | 0.032 |
| DGC e1 | 1.1 * | 0.1 | 0.9 | 0.0 | 0.024 |
| PER f2 | 1.2 | 0.0 | 1.1 | 0.0 | <0.05 |
| Hepatosomatic Index (%) g1 | 1.2 | 0.1 | 1.5 | 0.2 | >0.05 |
| Viscerosomatic Index (%) h1 | 6.5 | 0.6 | 6.7 | 0.7 | >0.05 |
| Statistical Test | Test Statistic | p Value | |
|---|---|---|---|
| Unweighted UniFrac | PERMANOVA | 1.965 | 0.007 * |
| Weighted UniFrac | PERMANOVA | 1.238 | 0.219 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villasante, A.; Ramírez, C.; Catalán, N.; Opazo, R.; Dantagnan, P.; Romero, J. Effect of Dietary Carbohydrate-to-Protein Ratio on Gut Microbiota in Atlantic Salmon (Salmo salar). Animals 2019, 9, 89. https://doi.org/10.3390/ani9030089
Villasante A, Ramírez C, Catalán N, Opazo R, Dantagnan P, Romero J. Effect of Dietary Carbohydrate-to-Protein Ratio on Gut Microbiota in Atlantic Salmon (Salmo salar). Animals. 2019; 9(3):89. https://doi.org/10.3390/ani9030089
Chicago/Turabian StyleVillasante, Alejandro, Carolina Ramírez, Natalia Catalán, Rafael Opazo, Patricio Dantagnan, and Jaime Romero. 2019. "Effect of Dietary Carbohydrate-to-Protein Ratio on Gut Microbiota in Atlantic Salmon (Salmo salar)" Animals 9, no. 3: 89. https://doi.org/10.3390/ani9030089
APA StyleVillasante, A., Ramírez, C., Catalán, N., Opazo, R., Dantagnan, P., & Romero, J. (2019). Effect of Dietary Carbohydrate-to-Protein Ratio on Gut Microbiota in Atlantic Salmon (Salmo salar). Animals, 9(3), 89. https://doi.org/10.3390/ani9030089





