Proteomic Analyses of Mammary Glands Provide Insight into the Immunity and Metabolism Pathways Associated with Clinical Mastitis in Meat Sheep
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Tissue Collection
2.2. H&E Staining and Masson Staining
2.3. Protein Extraction
2.4. 2-DE and Image Analysis
2.5. In-Gel Digestion and MALDI-TOF Analysis
2.6. Protein Identification and Functional Enrichment Analysis
2.7. Total RNA Isolation and cDNA Synthesis
2.8. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.9. Western Blot
2.10. Statistical Analysis
3. Results
3.1. Morphological Comparisons of Mammary Tissues with Healthy and Clinical Mastitis
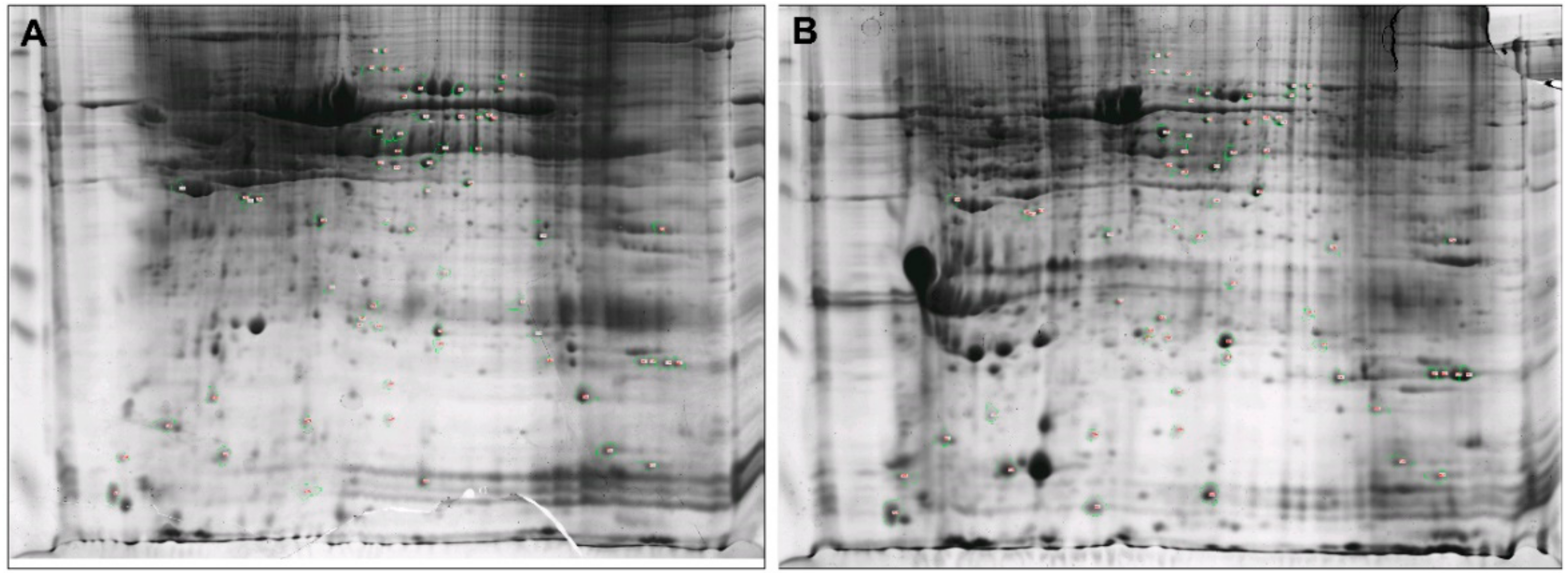
3.2. Distinct Protein Patterns in Mammary Glands of Healthy and Clinical Mastitis-Afflicted Sheep
3.3. Identification of Differentially Expressed Proteins Using MALDI-TOF/TOF-MS
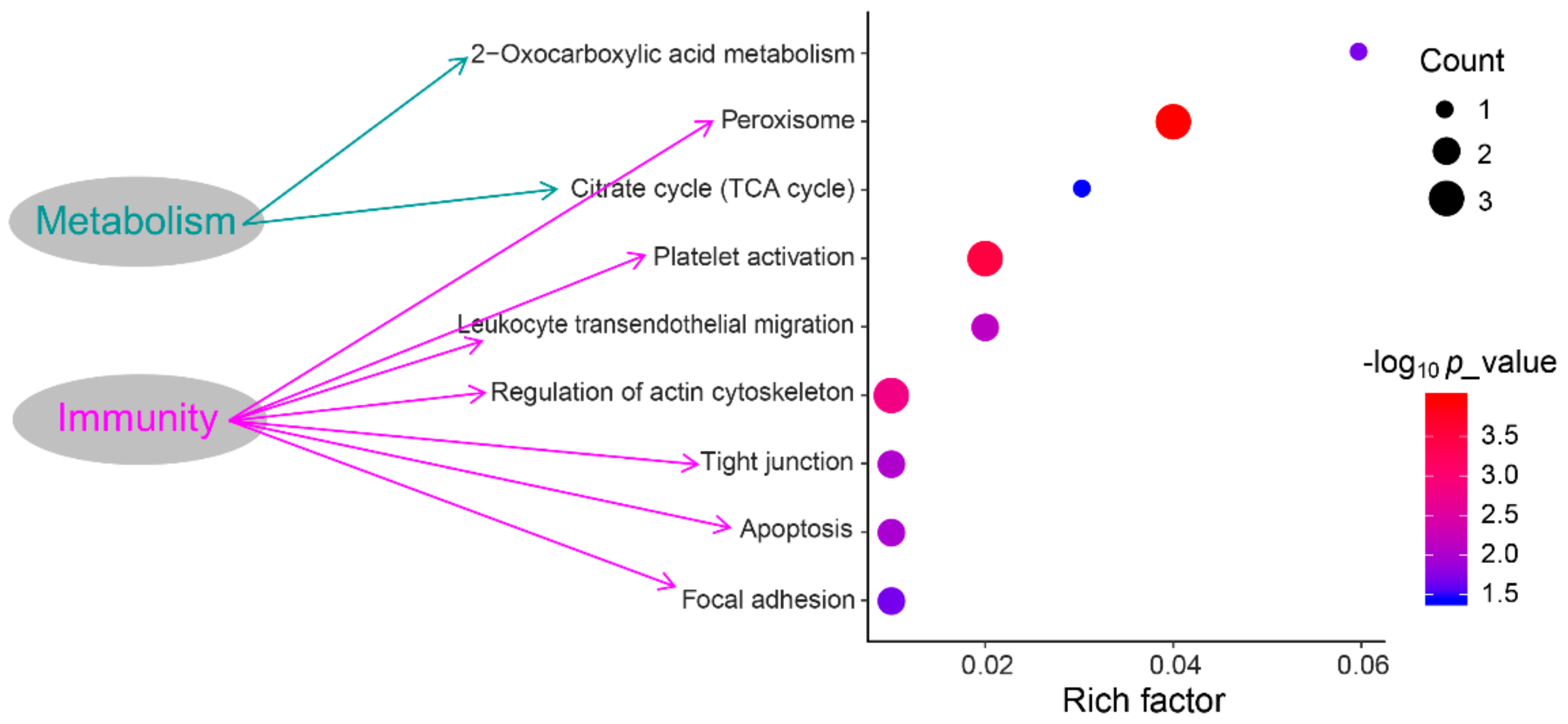
3.4. Functional Annotation and Pathway Enrichment Analysis of DEPs
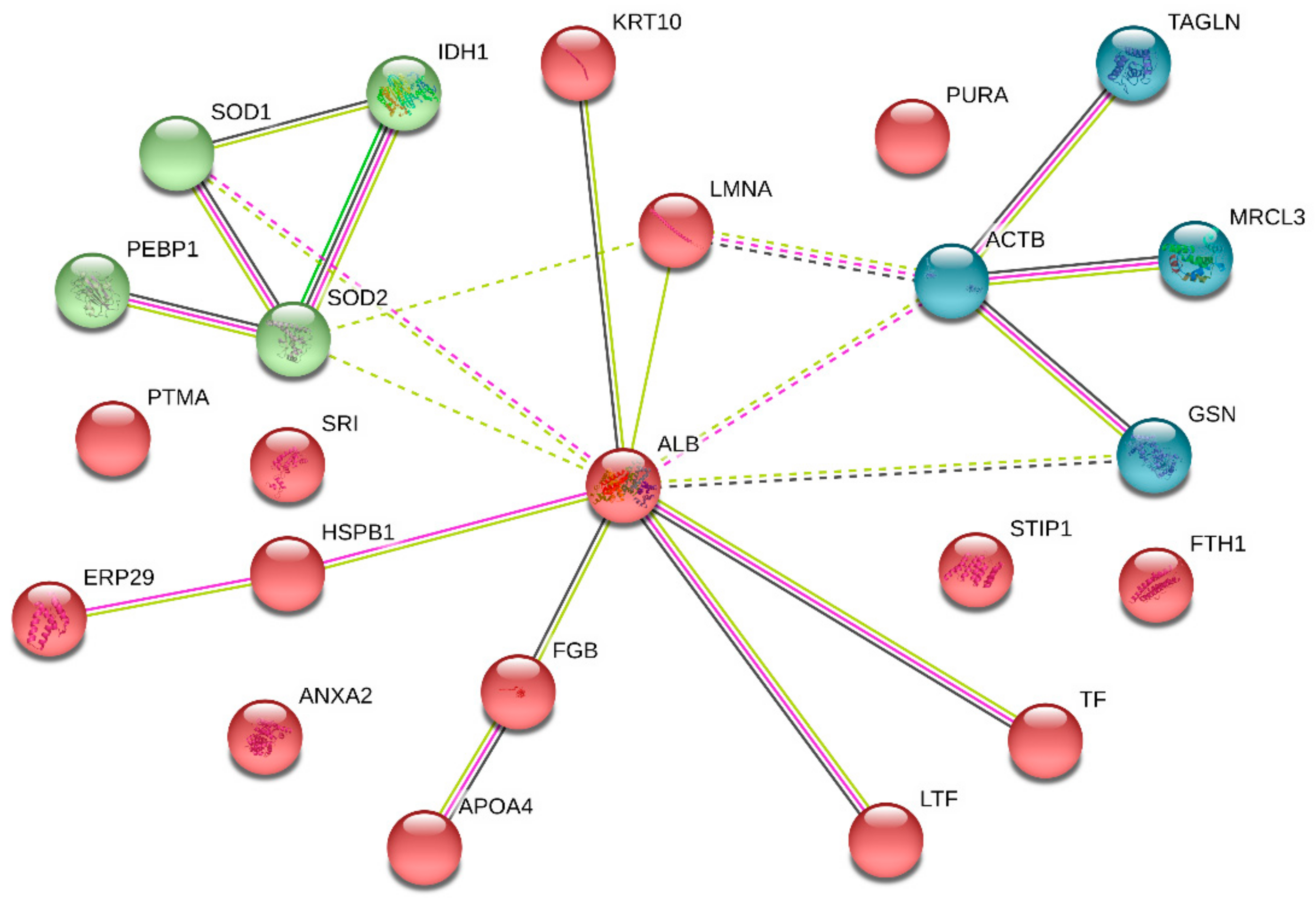
3.5. PPI Network Analysis
3.6. Verification of Differentially Expressed of Proteins
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gelasakis, A.I.; Mavrogianni, V.S.; Petridis, I.G.; Vasileiou, N.G.; Fthenakis, G.C. Mastitis in sheep-The last 10 years and the future of research. Vet. Microbiol. 2015, 181, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Varela-Ortiz, D.F.; Barboza-Corona, J.E.; González-Marrero, J.; León-Galván, M.F.; Valencia-Posadas, M.; Lechuga-Arana, A.A.; Sánchez-Felipe, C.G.; Ledezma-García, F.; Gutiérrez-Chávez, A.J. Antibiotic susceptibility of Staphylococcus aureus isolated from subclinical bovine mastitis cases and in vitro efficacy of bacteriophage. Vet. Res. Commun. 2018, 42, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Waage, S.; Vatn, S. Individual animal risk factors for clinical mastitis in meat sheep in Norway. Prev. Vet. Med. 2008, 87, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Munoz, C.; Campbell, A.; Barber, S.; Hemsworth, P.; Doyle, R. Using longitudinal assessment on extensively managed ewes to quantify welfare compromise and risks. Animals 2018, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Koop, G.; Rietman, J.F.; Pieterse, M.C. Staphylococcus aureus mastitis in Texel sheep associated with suckling twins. Vet. Rec. 2010, 167, 868–869. [Google Scholar] [CrossRef] [PubMed]
- Conington, J.; Cao, G.; Stott, A.; Bünger, L. Breeding for resistance to mastitis in United Kingdom sheep, a review and economic appraisal. Vet. Rec. 2008, 162, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Arsenault, J.; Dubreuil, P.; Higgins, R.; Bélanger, D. Risk factors and impacts of clinical and subclinical mastitis in commercial meat-producing sheep flocks in Quebec, Canada. Prev. Vet. Med. 2008, 87, 373–393. [Google Scholar] [CrossRef]
- Omaleki, L.; Browning, G.F.; Allen, J.L.; Markham, P.F.; Barber, S.R. Molecular epidemiology of an outbreak of clinical mastitis in sheep caused by Mannheimia haemolytica. Vet. Microbiol. 2016, 191, 82–87. [Google Scholar] [CrossRef]
- Huang, J.; Luo, G.; Zhang, Z.; Wang, X.; Ju, Z.; Qi, C.; Zhang, Y.; Wang, C.; Li, R.; Li, J. iTRAQ-proteomics and bioinformatics analyses of mammary tissue from cows with clinical mastitis due to natural infection with Staphylococci aureus. BMC Genom. 2014, 15, 839. [Google Scholar] [CrossRef]
- Zhang, H.; Jiang, H.; Fan, Y.; Chen, Z.; Li, M.; Mao, Y.; Karrow, N.A.; Loor, J.J.; Stephen, M.; Yang, Z. Transcriptomics and iTRAQ-proteomics analyses of bovine mammary tissue with Streptococcus agalactiae-induced mastitis. J. Agric. Food Chem. 2018, 66, 11188–11196. [Google Scholar] [CrossRef]
- Mudaliar, M.; Tassi, R.; Thomas, F.; Mcneilly, T.; Weidt, S.; Mclaughlin, M.; Wilson, D.; Burchmore, R.; Herzyk, P.; Eckersall, P.; et al. Mastitomics, the integrated omics of bovine milk in an experimental model of Streptococcus uberis mastitis: 2. Label-free relative quantitative proteomics. Mol. Biosyst. 2016, 12, 2748–2761. [Google Scholar] [CrossRef] [PubMed]
- Abdelmegid, S.; Murugaiyan, J.; Abo-Ismail, M.; Caswell, J.; Kelton, D.; Kirby, G. Mass spectrometry data from identification of host-defense related proteins using label-free quantitative proteomic analysis of milk whey from cows with Staphylococcus aureus subclinical mastitis. Data Brief 2019, 19, 909–913. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.X.; Cao, S.Z.; Zhang, Y.; Cheng, G.L.; Zhao, H.L.; Zhao, X.X. Differential proteomics analysis of plasma protein from Staphylococcus aureus mastitic and healthy dairy cows. J. Agric. Biotechnol. 2011, 19, 350–355. [Google Scholar] [CrossRef]
- Santana, A.M.; Thomas, F.C.; Silva, D.G.; Mcculloch, E.; Vidal, A.; Burchmore, R.; Fagliari, J.J.; Eckersall, P.D. Reference 1D and 2D electrophoresis maps for potential disease related proteins in milk whey from lactating buffaloes and blood serum from buffalo calves (Water buffalo, Bubalus bubalis). Res. Vet. Sci. 2018, 118, 449–465. [Google Scholar] [CrossRef] [PubMed]
- Chiaradia, E.; Valiani, A.; Tartaglia, M.; Scoppetta, F.; Renzone, G.; Arena, S.; Avellini, L.; Benda, S.; Gaiti, A.; Scaloni, A. Ovine subclinical mastitis: Proteomic analysis of whey and milk fat globules unveils putative diagnostic biomarkers in milk. J. Proteom. 2013, 83, 144–159. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.W.; Yang, Y.X.; Huang, D.W.; Cheng, G.L.; Zhao, H.L. Comparative proteomic analysis of proteins expression changes in the mammary tissue of cows infected with Escherichia coli mastitis. J. Vet. Sci. 2015, 16, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Hara, A.; Abe, T.; Hirao, A.; Sanbe, K.; Ayakawa, H.; Sarantonglaga, B.; Yamaguchi, M.; Sato, A.; Khurchabilig, A.; Ogata, K.; et al. Histochemical properties of bovine and ovine mammary glands during fetal development. J. Vet. Med. Sci. 2018, 80, 263–271. [Google Scholar] [CrossRef]
- Lu, Z.; Ma, Y.; Zhang, Q.; Zhao, X.; Zhang, Y.; Zhang, L. Proteomic analyses of ram (Ovis aries) testis during different developmental stages. Anim. Reprod. Sci. 2018, 189, 93–102. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yue, G.H. Reproductive characteristics of Chinese Hu sheep. Anim. Reprod. Sci. 1996, 44, 223–230. [Google Scholar] [CrossRef]
- Gong, Q.; Li, Y.; Ma, H.; Guo, W.; Kan, X.; Xu, D.; Liu, J.; Fu, S. Peiminine protects against lipopolysaccharide-induced mastitis by inhibiting the AKT/NF-κB, ERK1/2 and p38 signaling pathways. Int. J. Mol. Sci. 2018, 19, 2637. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, J.; Rutten, V.P.M.G.; van den Hout, M.; Spaninks, M.P.; Benedictus, L.; Koop, G. Differences between Staphylococcus aureus lineages isolated from ovine and caprine mastitis but not between isolates from clinical or subclinical mastitis. J. Dairy Sci. 2019, 102, 5430–5437. [Google Scholar] [CrossRef] [PubMed]
- Buitenhuis, B. In depth analysis of genes and pathways of the mammary gland involved in the pathogenesis of bovine Escherichia coli-mastitis. BMC Genom. 2011, 12, 130. [Google Scholar] [CrossRef] [PubMed]
- Cremonesi, P.; Capoferri, R.; Pisoni, G.; Corvo, M.D.; Strozzi, F.; Rupp, R.; Caillat, H.; Modesto, P.; Moroni, P.; Williams, J.L. Response of the goat mammary gland to infection with Staphylococcus aureus revealed by gene expression profiling in milk somatic and white blood cells. BMC Genom. 2012, 13, 540. [Google Scholar] [CrossRef] [PubMed]
- Rainard, P.; Riollet, C. Innate immunity of the bovine mammary gland. Vet. Res. 2015, 37, 369–400. [Google Scholar] [CrossRef] [PubMed]
- Bharathan, M.; Mullarky, I.K. Targeting mucosal immunity in the battle to develop a mastitis vaccine. J. Mammary Gland Biol. Neoplasia 2011, 16, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Kiku, Y.; Ozawa, T.; Takahashi, H.; Kushibiki, S.; Inumaru, S.; Shingu, H.; Nagasawa, Y.; Watanabe, A.; Hata, E.; Hayashi, T. Effect of intramammary infusion of recombinant bovine GM-CSF and IL-8 on CMT score, somatic cell count, and milk mononuclear cell populations in Holstein cows with Staphylococcus aureus subclinical mastitis. Vet. Res. Commun. 2017, 41, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Mabbott, N.A.; Kobayashi, A.; Sehgal, A.; Bradford, B.M.; Pattison, M.; Donaldson, D.S. Aging and the mucosal immune system in the intestine. Biogerontology 2015, 16, 133–145. [Google Scholar] [CrossRef]
- Brandtzaeg, P. The mucosal immune system and its integration with the mammary glands. J. Pediatr. 2010, 156, S8–S15. [Google Scholar] [CrossRef]
- Tokuhara, D.; Nochi, T.; Matsumura, A.; Mejima, M.; Takahashi, Y.; Kurokawa, S.; Kiyono, H.; Yuki, Y. Specific expression of apolipoprotein A-IV in the follicle-associated epithelium of the small intestine. Dig. Dis. Sci. 2014, 59, 2682–2692. [Google Scholar] [CrossRef]
- Wu, C.L.; Zhao, S.P.; Yu, B.L. Intracellular role of exchangeable apolipoproteins in energy homeostasis, obesity and non-alcoholic fatty liver disease. Biol. Rev. Camb. Philos. Soc. 2015, 90, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Sabherwal, M.; Duncan, E.; Stevens, S.; Stockwell, P.; McConnell, M.; Bekhit, A.-D.; Carne, A. In-depth characterization of sheep (Ovis aries) milk whey proteome and comparison with cow (Bos taurus). PLoS ONE 2015, 10, e0139774. [Google Scholar] [CrossRef] [PubMed]
- Soyeurt, H.; Bastin, C.; Colinet, F.G.; Arnould, V.R.; Berry, D.P.; Wall, E.; Dehareng, F.; Nguyen, H.N.; Dardenne, P.; Schefers, J.; et al. Mid-infrared prediction of lactoferrin content in bovine milk: Potential indicator of mastitis. Animal 2012, 6, 1830–1838. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, H.; Wang, C.; Li, J.; Li, Q.; Hou, M.; Zhong, J. Single nucleotide polymorphisms, haplotypes and combined genotypes of lactoferrin gene and their associations with mastitis in Chinese Holstein cattle. Mol. Biol. Rep. 2010, 37, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Kutila, T.; Suojala, L.; Lehtolainen, T.; Saloniemi, H.; Kaartinen, L.; Tähti, M.; Seppälä, K.; Pyörälä, S. The efficacy of bovine lactoferrin in the treatment of cows with experimentally induced Escherichia coli mastitis. J. Vet. Pharmacol. Ther. 2004, 27, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Ju, Z.H.; Li, Q.L.; Huang, J.M.; Hou, M.H.; Li, R.L.; Li, J.B.; Zhong, J.F.; Wang, C.F. Three novel SNPs of the bovine Tf gene in Chinese native cattle and their associations with milk production traits. Genet. Mol. Res. 2011, 10, 340–352. [Google Scholar] [CrossRef] [PubMed]
- Mitterhuemer, S.; Petzl, W.; Krebs, S.; Mehne, D.; Klanner, A.; Wolf, E.; Zerbe, H.; Blum, H. Escherichia coli infection induces distinct local and systemic transcriptome responses in the mammary gland. BMC Genom. 2010, 11, 138. [Google Scholar] [CrossRef]
- Kosciuczuk, E.M.; Lisowski, P.; Jarczak, J.; Majewska, A.; Rzewuska, M.; Zwierzchowski, L.; Bagnicka, E. Transcriptome profiling of Staphylococci-infected cow mammary gland parenchyma. BMC Vet. Res. 2017, 13, 161. [Google Scholar] [CrossRef]
- Che, M.; Wang, R.; Wang, H.Y.; Zheng, X.F. Expanding roles of superoxide dismutases in cell regulation and cancer. Drug Discov. Today 2016, 21, 143–149. [Google Scholar] [CrossRef]
- Fukai, T.; Ushio-Fukai, M. Superoxide dismutases: Role in redox signaling, vascular function, and diseases. Antioxid. Redox Signal. 2011, 15, 1583–1606. [Google Scholar] [CrossRef]
- Liu, W.; Degner, S.C.; Romagnolo, D.F. Trans-10, cis-12 conjugated linoleic acid inhibits prolactin-induced cytosolic NADP+-dependent isocitrate dehydrogenase expression in bovine mammary epithelial cells. J. Nutr. 2006, 136, 2743–2747. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Amankulor, N.M.; Kim, Y.; Arora, S.; Kargl, J.; Szulzewsky, F.; Hanke, M.; Margineantu, D.H.; Rao, A.; Bolouri, H.; Delrow, J. Mutant IDH1 regulates the tumor-associated immune system in gliomas. Genes Dev. 2017, 31, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Banos, G.; Bramis, G.; Bush, S.J.; Clark, E.L.; Mcculloch, M.E.B.; Smith, J.; Schulze, G.; Arsenos, G.; Hume, D.A.; Psifidi, A. The genomic architecture of mastitis resistance in dairy sheep. BMC Genom. 2017, 18, 624. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.L.; Mcgowen, M.R.; Weckle, A.; Pantham, P.; Caravas, J.; Agnew, D.; Benirschke, K.; Savage-Rumbaugh, S.; Nevo, E.; Kim, C.J. The core transcriptome of mammalian placentas and the divergence of expression with placental shape. Placenta 2017, 57, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Ji, N.Y.; Park, M.Y.; Kang, Y.H.; Lee, C.I.; Kim, D.G.; Yeom, Y.I.; Jang, Y.J.; Myung, P.K.; Kim, J.W.; Lee, H.G. Evaluation of annexin II as a potential serum marker for hepatocellular carcinoma using a developed sandwich ELISA method. Int. J. Mol. Med. 2009, 24, 765–771. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Jin, Y.; Yan, C.; Yu, Y.; Bai, J.; Chen, F.; Zhao, Y.; Fu, S. Involvement of Annexin A2 in p53 induced apoptosis in lung cancer. Mol. Cell. Biochem. 2008, 309, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Stukes, S.; Coelho, C.; Rivera, J.; Jedlicka, A.E.; Hajjar, K.A.; Casadevall, A. The membrane phospholipid binding protein annexin A2 promotes phagocytosis and nonlytic exocytosis of Cryptococcus neoformans and impacts survival in fungal infection. J. Immunol. 2016, 197, 1252–1261. [Google Scholar] [CrossRef]
- Chick, J.M.; Munger, S.C.; Simecek, P.; Huttlin, E.L.; Choi, K.; Gatti, D.M.; Raghupathy, N.; Svenson, K.L.; Churchill, G.A.; Gygi, S.P. Defining the consequences of genetic variation on a proteome–wide scale. Nature 2016, 534, 500–505. [Google Scholar] [CrossRef]
- Herman, A.B.; Autieri, M.V. Inflammation-regulated mRNA stability and the progression of vascular inflammatory diseases. Clin. Sci. 2017, 131, 2687–2699. [Google Scholar] [CrossRef]
- Liao, C.H.; Wang, Y.H.; Chang, W.W.; Yang, B.C.; Wu, T.J.; Liu, W.L.; Yu, A.L.; Yu, J. Leucine-rich repeat neuronal protein 1 regulates differentiation of embryonic stem cells by post-translational modifications of pluripotency factors. Stem Cells 2018, 36, 1514–1524. [Google Scholar] [CrossRef]
- Grindheim, A.K.; Saraste, J.; Vedeler, A. Protein phosphorylation and its role in the regulation of Annexin A2 function. Biochim. Biophys. Acta 2017, 1861, 2515–2529. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, I.; Takahashi, K.; Moll, I.; Moll, R. Expression of keratins in cutaneous epithelial tumors and related disorders—Distribution and clinical significance. Exp. Dermatol. 2011, 20, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.H.; Coulombe, P.A. Keratin function in skin epithelia: A broadening palette with surprising shades. Curr. Opin. Cell Biol. 2007, 19, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Magin, T.M.; Vijayaraj, P.; Leube, R.E. Structural and regulatory functions of keratins. Exp. Cell Res. 2007, 313, 2021–2032. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.Q. Profiles of Lymphocyte Subpopolations and Cytokines in Mastitis. Ph.D. Thesis, Northwest Agricultural Forest University, Yangling, China, 2013. [Google Scholar]
- Smolenski, G.A.; Cursons, R.T.; Hine, B.C.; Wheeler, T.T. Keratin and S100 calcium-binding proteins are major constituents of the bovine teat canal lining. Vet. Res. 2015, 46, 113. [Google Scholar] [CrossRef] [PubMed]




| Gene | Primer Sequences | Accession No. | Products Length (bp) |
|---|---|---|---|
| SOD2 | F: TAAACCGTCAGCCTTACACC | NM_001280703.1 | 198 |
| R: ACATTTTCAAACAGTTGCCTA | |||
| ANXA2 | F: CAAGCCCCTGTATTTCGCTGA | NM_001093788.1 | 194 |
| R: CTTTCTGGTAGTCGCCCTT | |||
| ERp29 | F: CCTTCCCCTGGATACAATCACT | EU596595.1 | 125 |
| R: AGTTTTCAGCCAGACGCTTG | |||
| KRT10 | F: TGCCCCAGGTGTTGATCTCACT | XM_015098774.1 | 100 |
| R: ATTGAACCATGCTTCGGCGTCT | |||
| GAPDH | F: GAAGGTCGGAGTGAACGGAT | NM_001190390.1 | 196 |
| R: GATGACGAGCTTCCCGTTCT |
| Spot No. | Protein Name | Gene Name | Fold Change | MW(Da) | pI |
|---|---|---|---|---|---|
| 791 | Lactoferrin | LTF | 1.78 | 77186.4 | 8.4 |
| 1040 | Lamin A/C | LMNA | −3.12 | 65082.6 | 6.54 |
| 844 | Transferrin | TF | −4.39 | 77290.5 | 6.41 |
| 1785 | Annexin A2 | ANXA2 | −1.83 | 39021 | 7.71 |
| 2244 | Keratin 10 | KRT10 | −4.47 | 57248.7 | 5.4 |
| 1043 | STIP1 | STIP1 | −1.76 | 68120 | 6.81 |
| 1260 | Actin, cytoplasmic 1 | ACTB | −1.79 | 41967.9 | 5.29 |
| 2234 | Cathelicidin | CATH1 | 1.58 | 17637 | 7.55 |
| 2182 | PEBP1 | PEBP1 | 2.72 | 21002.7 | 6.96 |
| 2459 | Prothymosin alpha | PTMA | 1.86 | 6228.8 | 4.1 |
| 1642 | Apolipoprotein A4 | APOA4 | −1.94 | 41454.5 | 5.55 |
| 2292 | Ferritin heavy chain | FTH1 | −2.58 | 21037.2 | 5.53 |
| 1709 | Albumin | ALB | −3.03 | 66269.8 | 5.58 |
| 2358 | Beta-lactoglobulin | BLG | 2.03 | 18139.4 | 5.26 |
| 741 | Gelsolin | GSN | −3.08 | 80652.5 | 5.49 |
| 2190 | Transgelin | TAGLN | 3.19 | 22866.5 | 8.92 |
| 2252 | Sorcin | SRI | −1.65 | 20317.8 | 5.11 |
| 1070 | Fibrinogen beta chain | FGB | −3.15 | 56604.7 | 7.84 |
| 2446 | Superoxide dismutase [Cu-Zn] | SOD1 | 4.85 | 15684.9 | 6.14 |
| 2364 | Superoxide dismutase [Mn] | SOD2 | 1.85 | 24135.4 | 8.67 |
| 1560 | 40S ribosomal protein SA | RPSA | −1.83 | 33538.8 | 4.8 |
| 1580 | Transcriptional activator protein Pur-alpha | PURA | −1.75 | 30258.4 | 7.05 |
| 2301 | Myosin regulatory light chain MRCL3 | MRCL3 | −1.85 | 19794.5 | 4.72 |
| 2485 | Heat shock protein family B member 1 | HSPB1 | 4.74 | 22437.3 | 5.77 |
| 1971 | Endoplasmic reticulum resident protein 29 | ERP29 | 1.58 | 29034.1 | 5.64 |
| 1358 | Isocitrate dehydrogenase [NADP] | IDH1 | −1.97 | 46703.5 | 6.34 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Li, T.; Lu, Z.; Wang, X.; Zhao, X.; Ma, Y. Proteomic Analyses of Mammary Glands Provide Insight into the Immunity and Metabolism Pathways Associated with Clinical Mastitis in Meat Sheep. Animals 2019, 9, 309. https://doi.org/10.3390/ani9060309
Gao J, Li T, Lu Z, Wang X, Zhao X, Ma Y. Proteomic Analyses of Mammary Glands Provide Insight into the Immunity and Metabolism Pathways Associated with Clinical Mastitis in Meat Sheep. Animals. 2019; 9(6):309. https://doi.org/10.3390/ani9060309
Chicago/Turabian StyleGao, Jianfeng, Taotao Li, Zengkui Lu, Xia Wang, Xingxu Zhao, and Youji Ma. 2019. "Proteomic Analyses of Mammary Glands Provide Insight into the Immunity and Metabolism Pathways Associated with Clinical Mastitis in Meat Sheep" Animals 9, no. 6: 309. https://doi.org/10.3390/ani9060309
APA StyleGao, J., Li, T., Lu, Z., Wang, X., Zhao, X., & Ma, Y. (2019). Proteomic Analyses of Mammary Glands Provide Insight into the Immunity and Metabolism Pathways Associated with Clinical Mastitis in Meat Sheep. Animals, 9(6), 309. https://doi.org/10.3390/ani9060309





