MicroRNA Sequencing Reveals the Effect of Different Levels of Non-Fibrous Carbohydrate/Neutral Detergent Fiber on Rumen Development in Calves
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Design
2.1.1. Ethics Statement
2.1.2. Experimental Animals and RNA Isolation
2.1.3. Preparation and Observation of Rumen Sections
2.2. MicroRNA Sequencing
2.2.1. miRNA library Construction and Illumina Deep Sequencing
2.2.2. Sequence Analyses
2.2.3. Differential Expression Analysis of miRNAs
2.2.4. MiRNA Target Prediction, Functional Annotation, and Interaction Networks
2.3. Verification of Sequencing Results
2.3.1. Validation of Relative Expression of miRNAs and mRNAs
2.3.2. Vector Construction
2.3.3. Cell Culture and Luciferase Reporter Assay
2.4. Statistical Analyses
3. Results and Discussion
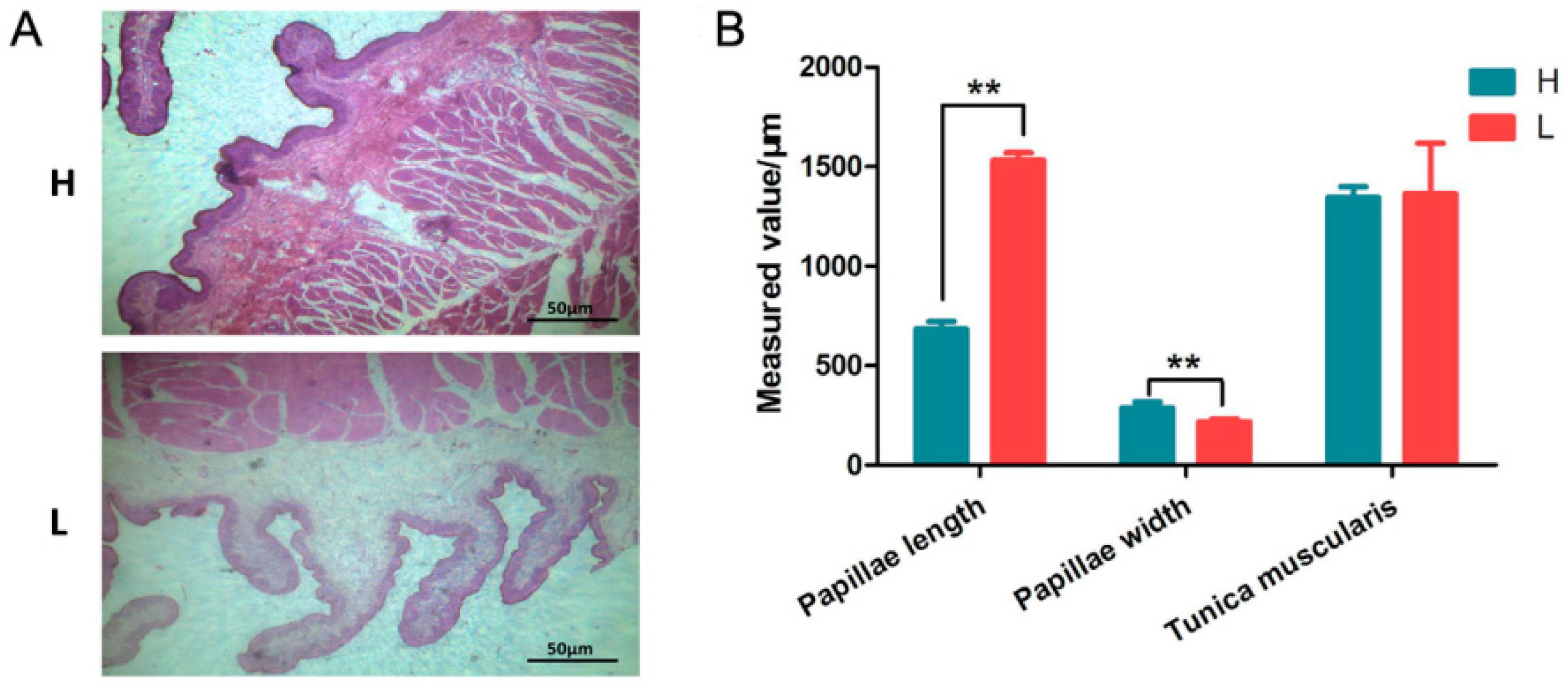
3.1. Effect of NFC/NDF Levels on Rumen Development of Calf
3.2. Overview of Small RNA Deep Sequencing Data
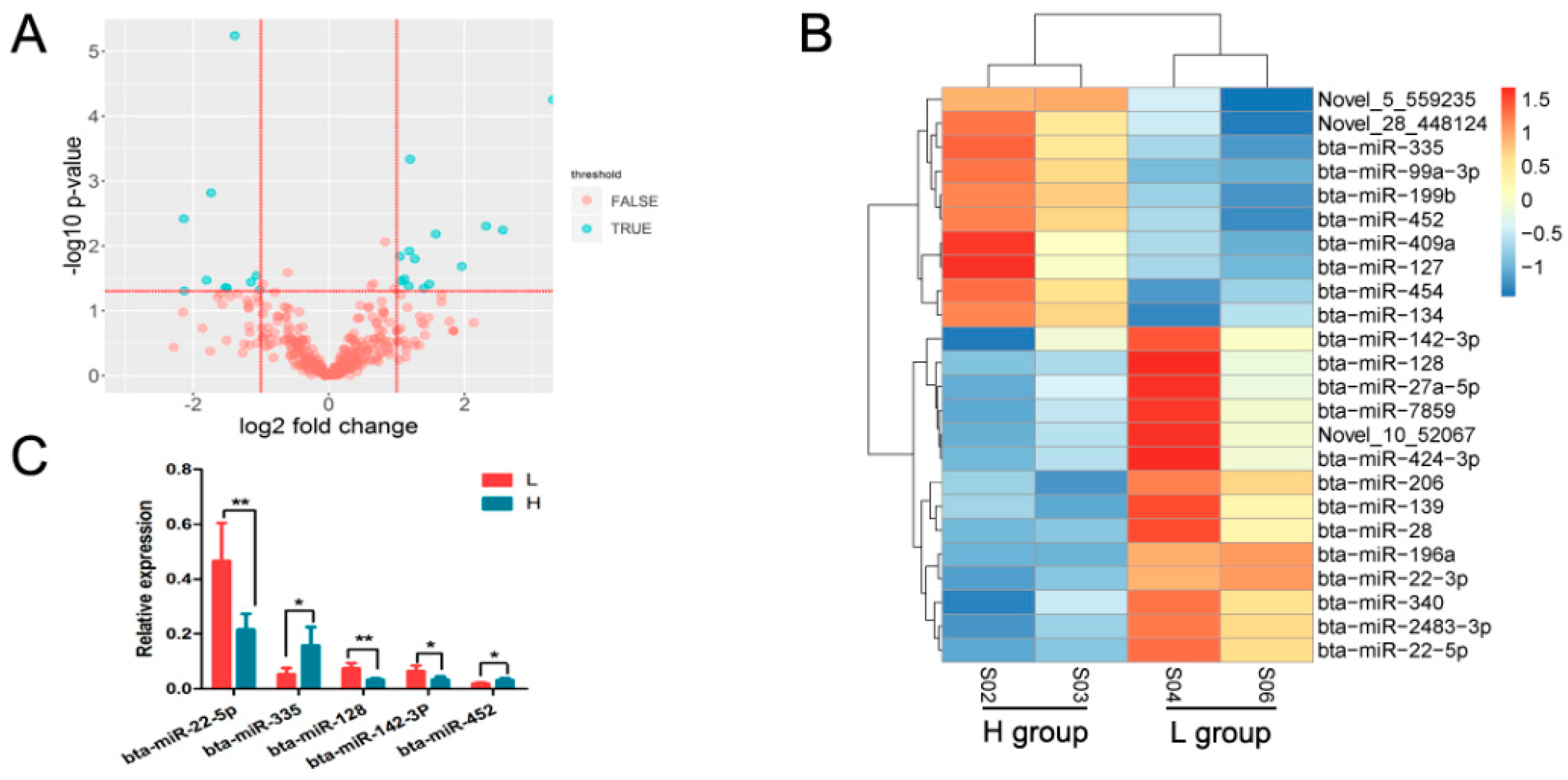
3.3. Identification of Differentially Expressed miRNAs
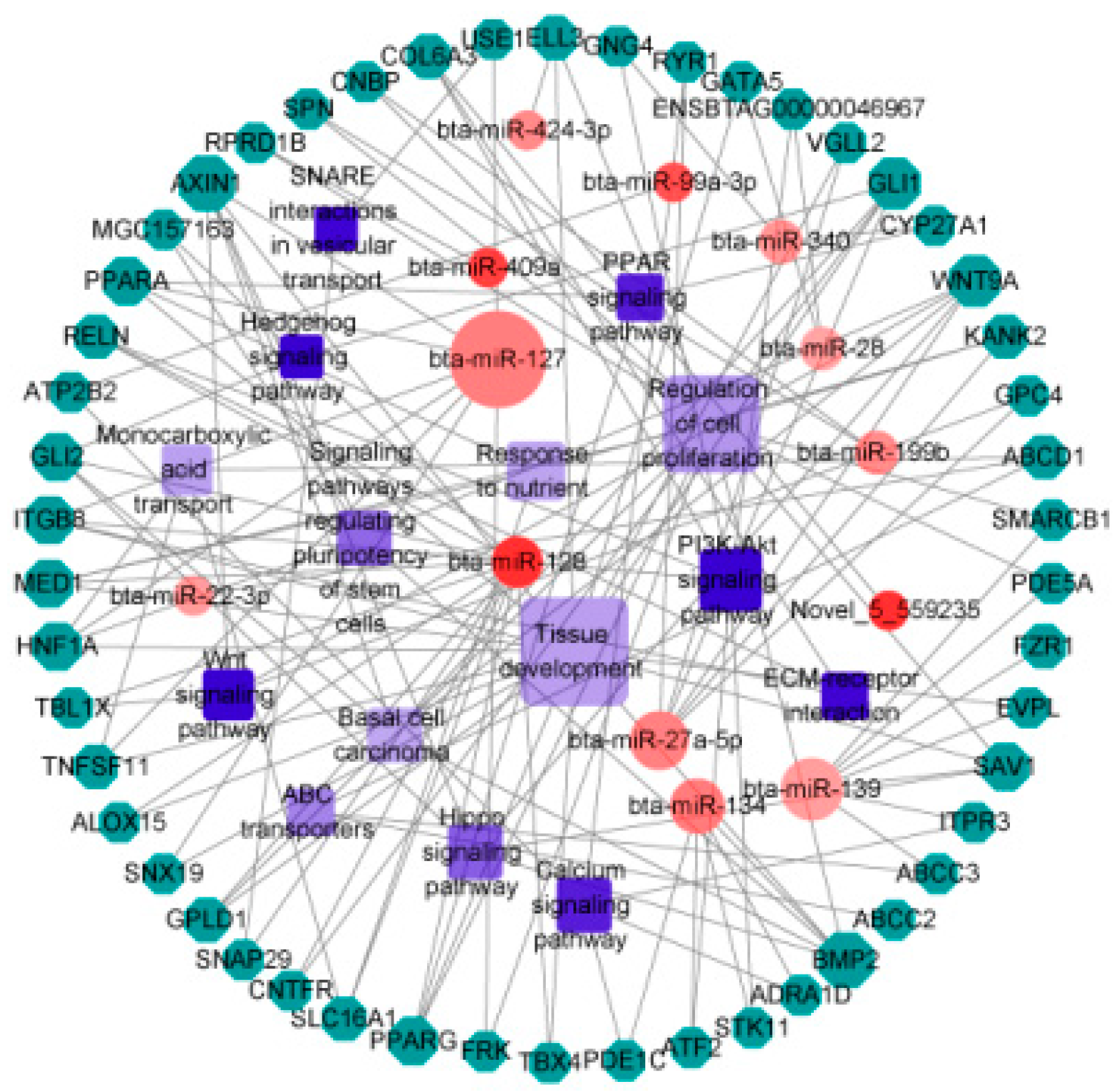
3.4. Prediction of DEmiRNA Target Genes
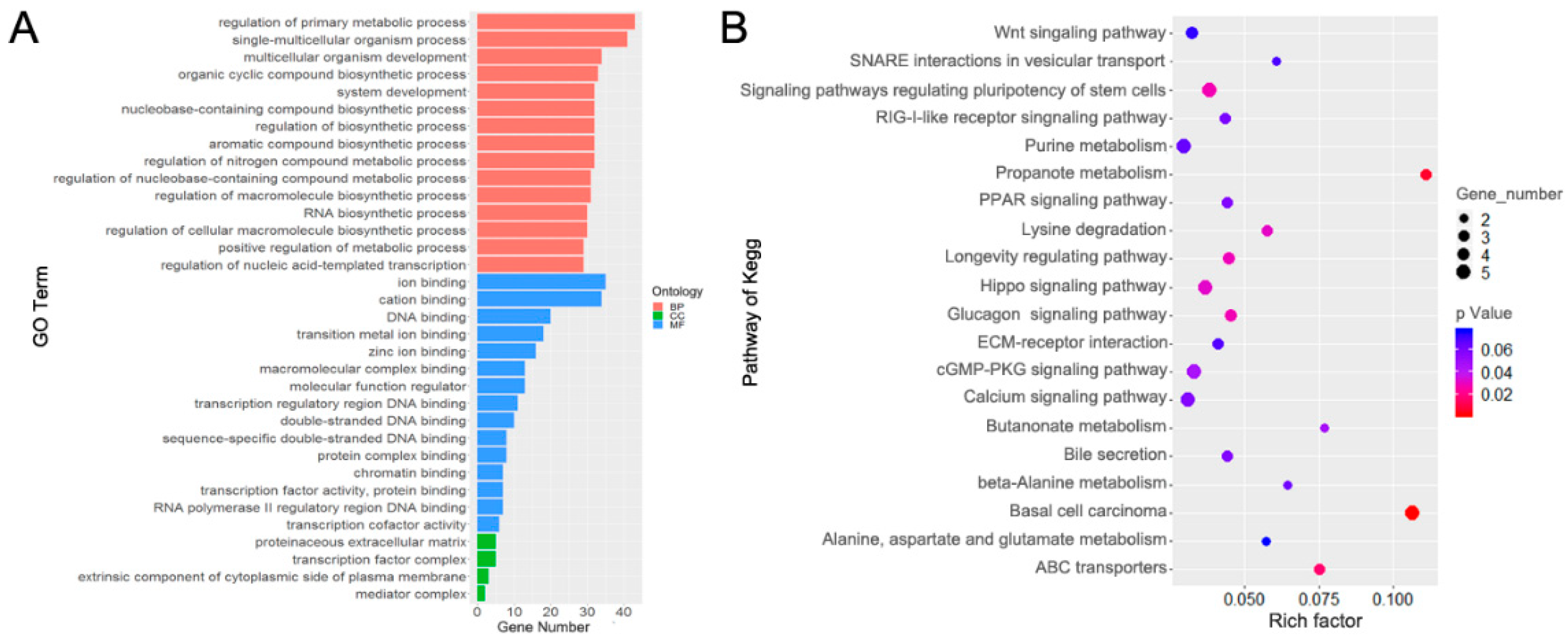
3.5. Functional Enrichment Analysis
3.6. Targeting Effect of Bta-miR-128 on PPARG and SLC16A1
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, B.; Wang, D.; Wu, X.; Cai, J.; Liu, M.; Huang, X.; Wu, J.; Liu, J.; Guan, L. Effects of dietary physical or nutritional factors on morphology of rumen papillae and transcriptome changes in lactating dairy cows based on three different forage-based diets. BMC Genom. 2017, 18, 353. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Carrillo, J.A.; Ding, Y.; He, Y.; Zhao, C.; Zan, L.; Song, J. Ruminal Transcriptomic Analysis of Grass-Fed and Grain-Fed Angus Beef Cattle. PLoS ONE 2015, 10, e0116437. [Google Scholar] [CrossRef] [PubMed]
- Zhong, T.; Hu, J.; Xiao, P.; Zhan, S.; Wang, L.; Guo, J.; Li, L.; Zhang, H.; Niu, L. Identification and Characterization of MicroRNAs in the Goat (Capra hircus) Rumen during Embryonic Development. Front. Genet. 2017, 8, 163. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Dudemaine, P.L.; Fomenky, B.E.; Ibeagha-Awemu, E.M. Integration of miRNA weighted gene co-expression network and miRNA-mRNA co-expression analyses reveals potential regulatory functions of miRNAs in calf rumen development. Genomics 2019, 111, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Steele, M.A.; Vandervoort, G.; AlZahal, O.; Hook, S.E.; Matthews, J.C.; McBride, B.W. Rumen epithelial adaptation to high-grain diets involves the coordinated regulation of genes involved in cholesterol homeostasis. Physiol. Genom. 2011, 43, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.Z.; Chen, Y.; Guan, L.L. MicroRNA expression profiles across blood and different tissues in cattle. Sci. Data 2019, 6, 190013. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, Y.; Wu, M.; Li, L.; Jin, C.; Zhang, Q.; Chen, C.; Song, W.; Wang, C. Small RNA Sequencing Reveals Differential miRNA Expression in the Early Development of Broccoli (Brassica oleracea var. italica) Pollen. Front. Plant Sci. 2017, 8, 404. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Hipfner, D.R.; Stark, A.; Russell, R.B.; Cohen, S.M. Bantam Encodes a Developmentally Regulated microRNA that Controls Cell Proliferation and Regulates the Proapoptotic Gene hid in Drosophila. Cell 2003, 113, 25–36. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, L.; Wen, G.; Zhao, H.; Luong, L.A.; Chen, Q.; Huang, Y.; Zhu, J.; Ye, S.; Xu, Q. Upregulated sirtuin 1 by miRNA-34a is required for smooth muscle cell differentiation from pluripotent stem cells. Cell Death Differ. 2015, 22, 1170–1180. [Google Scholar] [CrossRef]
- Ye, Q.; Zhao, X.; Xu, K.; Li, Q.; Cheng, J.; Gao, Y.; Du, J.; Shi, H.; Zhou, L. Polymorphisms in lipid metabolism related miRNA binding sites and risk of metabolic syndrome. Gene 2013, 528, 132–138. [Google Scholar] [CrossRef]
- Dragomir, M.P.; Knutsen, E.; Calin, G.A. SnapShot: Unconventional miRNA Functions. Cell 2018, 174, 1038–1038.e1031. [Google Scholar] [CrossRef]
- Lauressergues, D.; Couzigou, J.M.; Clemente, H.S.; Martinez, Y.; Dunand, C.; Bécard, G.; Combier, J.P. Primary transcripts of microRNAs encode regulatory peptides. Nature 2015, 520, 90–93. [Google Scholar] [CrossRef]
- Eiring, A.M.; Harb, J.G.; Neviani, P.; Garton, C.; Oaks, J.J.; Spizzo, R.; Liu, S.; Schwind, S.; Santhanam, R.; Hickey, C.J. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 2010, 140, 652–665. [Google Scholar] [CrossRef]
- Lehmann, S.M.; Krüger, C.; Park, B.; Derkow, K.; Rosenberger, K.; Baumgart, J.; Trimbuch, T.; Eom, G.; Hinz, M.; Kaul, D. An unconventional role for miRNA: Let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012, 15, 827. [Google Scholar] [CrossRef]
- Tang, R.; Li, L.; Zhu, D.; Hou, D.; Cao, T.; Gu, H.; Zhang, J.; Chen, J.; Zhang, C.Y.; Zen, K. Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: Evidence for a microRNA hierarchy system. Cell Res. 2012, 22, 504–515. [Google Scholar] [CrossRef]
- Lanjie, L.I.; Wang, A.; Lian, H.; Tong, F.U.; Wang, Z.; Diao, Q.; Yan, T.U.; Cheng, S. Effects of diets with different NFC/NDF levels on the growth performance and slaughter performance of meat calves. J. China Agric. Univ. 2017, 22, 101–109. [Google Scholar]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for Dietary Fiber, Neutral Detergent Fiber, and Nonstarch Polysaccharides in Relation to Animal Nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Mangos, J.A. American journal of physiology. Am. J. Physiol. 1917, 229, 553–559. [Google Scholar] [CrossRef]
- Carstens, G.E.; Min, B.R.; Pinchak, W.E. Effects of dietary tannin source on performance, feed efficiency, ruminal fermentation, and carcass and non-carcass traits in steers fed a high-grain diet. Anim. Feed Sci. Technol. 2010, 159, 1–9. [Google Scholar]
- Enright, A.J.; Saini, H.K.; van Dongen, S.; Griffiths-Jones, S. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2007, 36, D154–D158. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011, 40, 37–52. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, F.; Li, J.; Chen, J.P.; Zhang, H.M. Integrative Analysis of the microRNAome and Transcriptome Illuminates the Response of Susceptible Rice Plants to Rice Stripe Virus. PLoS ONE 2016, 11, e0146946. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010, 11, R90. [Google Scholar] [CrossRef]
- Jan, K.; Marc, R. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, 451–454. [Google Scholar]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Lotia, S.; Pico, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods 2012, 9, 1069–1076. [Google Scholar] [CrossRef]
- Guo, J.; Zhao, W.; Zhan, S.; Li, L.; Zhong, T.; Wang, L.; Dong, Y.; Zhang, H. Identification and Expression Profiling of miRNAome in Goat longissimus dorsi Muscle from Prenatal Stages to a Neonatal Stage. PLoS ONE 2016, 11, e0165764. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Wu, Z.; Sun, R.; Jin, H.; Ma, J.; Liu, L.; Ling, R.; Yi, J.; Wang, L.; et al. Three dysregulated microRNAs in serum as novel biomarkers for gastric cancer screening. Med. Oncol. 2014, 31, 298. [Google Scholar] [CrossRef]
- Khalili, M.; Sadeghizadeh, M.; Ghorbanian, K.; Malekzadeh, R.; Vasei, M.; Mowla, S.J. Down-regulation of miR-302b, an ESC-specific microRNA, in Gastric Adenocarcinoma. Cell J. 2012, 13, 251–258. [Google Scholar]
- Hou, L.; Gu, W.; Zhu, H.; Yao, W.; Wang, W.; Meng, Q. Spiroplasma eriocheiris induces mouse 3T6-Swiss albino cell apoptosis that associated with the infection mechanism. Mol. Immunol. 2017, 91, 75–85. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Thomas, P.; Smart, T.G. HEK293 cell line: A vehicle for the expression of recombinant proteins. J. Pharmacol. Toxicol. Methods 2005, 51, 187–200. [Google Scholar] [CrossRef]
- O’Shea, E.; Waters, S.M.; Keogh, K.; Kelly, A.K.; Kenny, D.A. Examination of the molecular control of ruminal epithelial function in response to dietary restriction and subsequent compensatory growth in cattle. J. Anim. Sci. Biotechnol. 2016, 7, 53. [Google Scholar] [CrossRef]
- Lesmeister, K.E.; Heinrichs, A.J. Effects of corn processing on growth characteristics, rumen development, and rumen parameters in neonatal dairy calves. J. Dairy Sci. 2004, 87, 3439–3450. [Google Scholar] [CrossRef]
- Steele, M.A.; Croom, J.; Kahler, M.; AlZahal, O.; Hook, S.E.; Plaizier, K.; McBride, B.W. Bovine rumen epithelium undergoes rapid structural adaptations during grain-induced subacute ruminal acidosis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 300, R1515–R1523. [Google Scholar] [CrossRef]
- Shi, Z.M.; Wang, J.; Yan, Z.; You, Y.P.; Li, C.Y.; Qian, X.; Yin, Y.; Zhao, P.; Wang, Y.Y.; Wang, X.F. MiR-128 inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS ONE 2012, 7, e32709. [Google Scholar] [CrossRef]
- Wang, D.; Tang, L.; Wu, H.; Wang, K.; Gu, D. MiR-127-3p inhibits cell growth and invasiveness by targeting ITGA6 in human osteosarcoma. IUBMB Life 2018, 70, 411–419. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, M.; Qian, J.; Bao, M.; Meng, X.; Zhang, S.; Zhang, L.; Zhao, R.; Li, S.; Cao, Q. miR-134 functions as a tumor suppressor in cell proliferation and epithelial-to-mesenchymal Transition by targeting KRAS in renal cell carcinoma cells. DNA Cell Biol. 2015, 34, 429–436. [Google Scholar] [CrossRef]
- Cui, Y.; Sun, X.; Jin, L.; Yu, G.; Li, Q.; Gao, X.; Ao, J.; Wang, C. MiR-139 suppresses β-casein synthesis and proliferation in bovine mammary epithelial cells by targeting the GHR and IGF1R signaling pathways. BMC Vet. Res. 2017, 13, 350. [Google Scholar] [CrossRef]
- Shveta, B.; John, B.; Shaun, H.; Katlin, M.; Janette, H.; Rachel, E.; Pasquinelli, A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 2005, 122, 553–563. [Google Scholar]
- Liu, X.G. Histogenesis of the Mongolia Sheep Embryonic Stomach Muscle Layer. Yinshan Acad. J. 2004, 18, 15–16. [Google Scholar]
- Steele, M.A.; Alzahal, O.; Hook, S.E.; Croom, J.; Mcbride, B.W. Ruminal acidosis and the rapid onset of ruminal parakeratosis in a mature dairy cow: A case report. Acta Vet. Scand. 2009, 51, 39. [Google Scholar] [CrossRef]
- Greenwood, R.H.; Morrill, J.L.; Titgemeyer, E.C.; Kennedy, G.A. A new method of measuring diet abrasion and its effect on the development of the forestomach. J. Dairy Sci. 1997, 80, 2534–2541. [Google Scholar] [CrossRef]
- Katoh, M.; Katoh, M. Transcriptional regulation of WNT2B based on the balance of Hedgehog, Notch, BMP and WNT signals. Int. J. Oncol. 2009, 34, 1411–1415. [Google Scholar] [CrossRef]
- Regl, G.; Neill, G.W.; Eichberger, T.; Kasper, M.; Ikram, M.S.; Koller, J.; Hintner, H.; Quinn, A.G.; Frischauf, A.M.; Aberger, F. Human GLI2 and GLI1 are part of a positive feedback mechanism in Basal Cell Carcinoma. Oncogene 2002, 21, 5529–5539. [Google Scholar] [CrossRef]
- Watt, K.I.; Harvey, K.F.; Gregorevic, P. Regulation of Tissue Growth by the Mammalian Hippo Signaling Pathway. Front. Physiol. 2017, 8, 942. [Google Scholar] [CrossRef]
- Oharazawa, H.; Ibaraki, N.; Lin, L.R.; Reddy, V.N. The effects of extracellular matrix on cell attachment, proliferation and migration in a human lens epithelial cell line. Exp. Eye Res. 1999, 69, 603–610. [Google Scholar] [CrossRef]
- Beharka, A.A.; Nagaraja, T.G.; Morrill, J.L.; Kennedy, G.; Klemm, R.D. Effects of Form of the Diet on Anatomical, Microbial, and Fermentative Development of the Rumen of Neonatal Calves. J. Dairy Sci. 1998, 81, 1946–1955. [Google Scholar] [CrossRef]
- Zhang, S.Q.; Zan, L.S.; Liang, D.Y.; Gui, L.S. Effect of different dietary concentrate to forage ratio on rumen morphological structure of Chinese Hostein bull. J. Northwest Agric. For. Univ. 2009, 37, 59–64. [Google Scholar]
- Hennings, H.; Michael, D.; Cheng, C.; Steinert, P.; Holbrook, K.; Yuspa, S.H. Calcium regulation of growth and differentiation of mouse epidermal cells in culture. Cell 1980, 19, 245–254. [Google Scholar] [CrossRef]
- Xie, Z.; Bikle, D.D. Phospholipase C-γ1 Is Required for Calcium-induced Keratinocyte Differentiation. J. Biol. Chem. 1999, 274, 20421–20424. [Google Scholar] [CrossRef]
- Weerachayaphorn, J.; Amaya, M.J.; Spirli, C.; Chansela, P.; Mitchellrichards, K.A.; Ananthanarayanan, M.; Nathanson, M.H. Nuclear Factor, Erythroid 2-Like 2 Regulates Expression of Type 3 Inositol 1,4,5-Trisphosphate Receptor and Calcium Signaling in Cholangiocytes. Gastroenterology 2015, 149, 211–222.e210. [Google Scholar] [CrossRef]
- Minich, R.R.; Li, J.; Tempel, B.L. Early growth response protein 1 regulates promoter activity ofα-plasma membrane calcium ATPase 2, a major calcium pump in the brain and auditory system. BMC Mol. Biol. 2017, 18, 14. [Google Scholar] [CrossRef]
- Hurne, A.; O’Brien, J.; Wingrove, D.; Cherednichenko, G.; Allen, P.; Beam, K.; Pessah, I. Ryanodine receptor type 1 (RyR1) mutations C4958S and C4961S reveal excitation-coupled calcium entry (ECCE) is independent of sarcoplasmic reticulum store depletion. J. Biol. Chem. 2005, 280, 36994–37004. [Google Scholar] [CrossRef]
- Mikami, Y.; Kanemaru, K.; Okubo, Y.; Nakaune, T.; Suzuki, J.; Shibata, K.; Sugiyama, H.; Koyama, R.; Murayama, T.; Ito, A. Nitric Oxide-induced Activation of the Type 1 Ryanodine Receptor Is Critical for Epileptic Seizure-induced Neuronal Cell Death. Ebiomedicine 2016, 11, 253–261. [Google Scholar] [CrossRef]
- Pachera, N.; Papin, J.; Zummo, F.P.; Rahier, J.; Mast, J.; Meyerovich, K.; Cardozo, A.K.; Herchuelz, A. Heterozygous inactivation of plasma membrane Ca2+-ATPase in mice increases glucose-induced insulin release and beta cell proliferation, mass and viability. Diabetologia 2015, 58, 2843–2850. [Google Scholar] [CrossRef]
- Blackshaw, S.; Sawa, A.; Sharp, A.H.; Ross, C.A.; Snyder, S.H.; Khan, A.A. Type 3 inositol 1,4,5-trisphosphate receptor modulates cell death. FASEB J. 2000, 14, 1375–1379. [Google Scholar] [CrossRef]
- Rowther, F.B.; Wei, W.; Dawson, T.P.; Ashton, K.; Singh, A.; Madiessetimchou, M.P.; Thomas, D.G.; Darling, J.L.; Warr, T. Cyclic nucleotide phosphodiesterase-1C (PDE1C) drives cell proliferation, migration and invasion in glioblastoma multiforme cells in vitro. Mol. Carcinog. 2016, 55, 268–279. [Google Scholar] [CrossRef]
- Burdick, A.D.; Kim, D.J.; Peraza, M.A.; Gonzalez, F.J.; Peters, J.M. The role of peroxisome proliferator-activated receptor-β/δ in epithelial cell growth and differentiation. Cell. Signal. 2006, 18, 9–20. [Google Scholar] [CrossRef]
- Liliane, M.; Béatrice, D.; Walter, W. Peroxisome-proliferator-activated receptors and cancers: Complex stories. Nat. Rev. Cancer 2004, 4, 61–70. [Google Scholar]
- Norouzian, M.A.; Valizadeh, R. Effect of forage inclusion and particle size in diets of neonatal lambs on performance and rumen development. J. Anim. Physiol. Anim. Nutr. 2015, 98, 1095–1101. [Google Scholar] [CrossRef]
- Doaa, K.; Junji, M.; Hideaki, H.; Hidetomo, I.; Hiroshi, Y.; Hiroyuki, T.; Seiyu, K. Monocarboxylate transporter 1 (MCT1) plays a direct role in short-chain fatty acids absorption in caprine rumen. J. Physiol. 2010, 576, 635–647. [Google Scholar]
- Kirat, D.; Inoue, H.; Iwano, H.; Hirayama, K.; Yokota, H.; Taniyama, H.; Kato, S. Expression and distribution of monocarboxylate transporter 1 (MCT1) in the gastrointestinal tract of calves. Res. Vet. Sci. 2005, 79, 45–50. [Google Scholar] [CrossRef]
- Halestrap, A.P.; David, M. The SLC16 gene family-from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflügers Arch. Eur. J. Physiol. 2004, 447, 619–628. [Google Scholar] [CrossRef]
- Graham, C.; Gatherar, I.; Haslam, I.; Glanville, M.; Simmons, N.L. Expression and localization of monocarboxylate transporters and sodium/proton exchangers in bovine rumen epithelium. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 292, R997–R1007. [Google Scholar] [CrossRef]
- Frank, M.; Korinna, H.; Helga, P.; Aschenbach, J.R.R.; Gerhard, B.; Gotthold, G.B. Transport of ketone bodies and lactate in the sheep ruminal epithelium by monocarboxylate transporter 1. Am. J. Physiol. Gastrointest. Liver Physiol. 2002, 283, G1139. [Google Scholar]
- Ippolito, L.; Morandi, A.; Giannoni, E.; Chiarugi, P. Lactate: A Metabolic Driver in the Tumour Landscape. Trends Biochem. Sci. 2019, 44, 153–166. [Google Scholar] [CrossRef]
- Payen, V.L.; Hsu, M.Y.; Rädecke, K.S.; Wyart, E.; Vazeille, T.; Bouzin, C.; Porporato, P.E.; Sonveaux, P. Monocarboxylate Transporter MCT1 Promotes Tumor Metastasis Independently of Its Activity as a Lactate Transporter. Cancer Res. 2017, 77, 5591–5601. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, M.; Wang, K.; Wang, A.; Li, R.; Wang, Y.; Sun, S.; Yan, D.; Song, G.; Xu, H.; Sun, G.; et al. MicroRNA Sequencing Reveals the Effect of Different Levels of Non-Fibrous Carbohydrate/Neutral Detergent Fiber on Rumen Development in Calves. Animals 2019, 9, 496. https://doi.org/10.3390/ani9080496
Xue M, Wang K, Wang A, Li R, Wang Y, Sun S, Yan D, Song G, Xu H, Sun G, et al. MicroRNA Sequencing Reveals the Effect of Different Levels of Non-Fibrous Carbohydrate/Neutral Detergent Fiber on Rumen Development in Calves. Animals. 2019; 9(8):496. https://doi.org/10.3390/ani9080496
Chicago/Turabian StyleXue, Mingming, Kejun Wang, Ansi Wang, Ruiting Li, Yadong Wang, Shuaijie Sun, Duo Yan, Guohua Song, Huifen Xu, Guirong Sun, and et al. 2019. "MicroRNA Sequencing Reveals the Effect of Different Levels of Non-Fibrous Carbohydrate/Neutral Detergent Fiber on Rumen Development in Calves" Animals 9, no. 8: 496. https://doi.org/10.3390/ani9080496
APA StyleXue, M., Wang, K., Wang, A., Li, R., Wang, Y., Sun, S., Yan, D., Song, G., Xu, H., Sun, G., & Li, M. (2019). MicroRNA Sequencing Reveals the Effect of Different Levels of Non-Fibrous Carbohydrate/Neutral Detergent Fiber on Rumen Development in Calves. Animals, 9(8), 496. https://doi.org/10.3390/ani9080496




