The Effects of Maternal Obesity on Porcine Placental Efficiency and Proteome
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Study Design
2.2. Data Collection and Sampling
2.3. Plasma Analysis
2.4. Biochemical Analysis of Placental Tissue
2.5. Placental Protein Extraction for Proteomics
2.6. iTRAQ Labelling and SCX Fractionation
2.7. LC-ESI-MS/MS Analysis Based on Triple TOF 5600
2.8. Protein Identification and Quantification
2.9. Bioinformatics Analysis
2.10. RNA Extraction and RT-qPCR
2.11. Statistical Method
3. Results
3.1. Characteristics of the Studied Sows
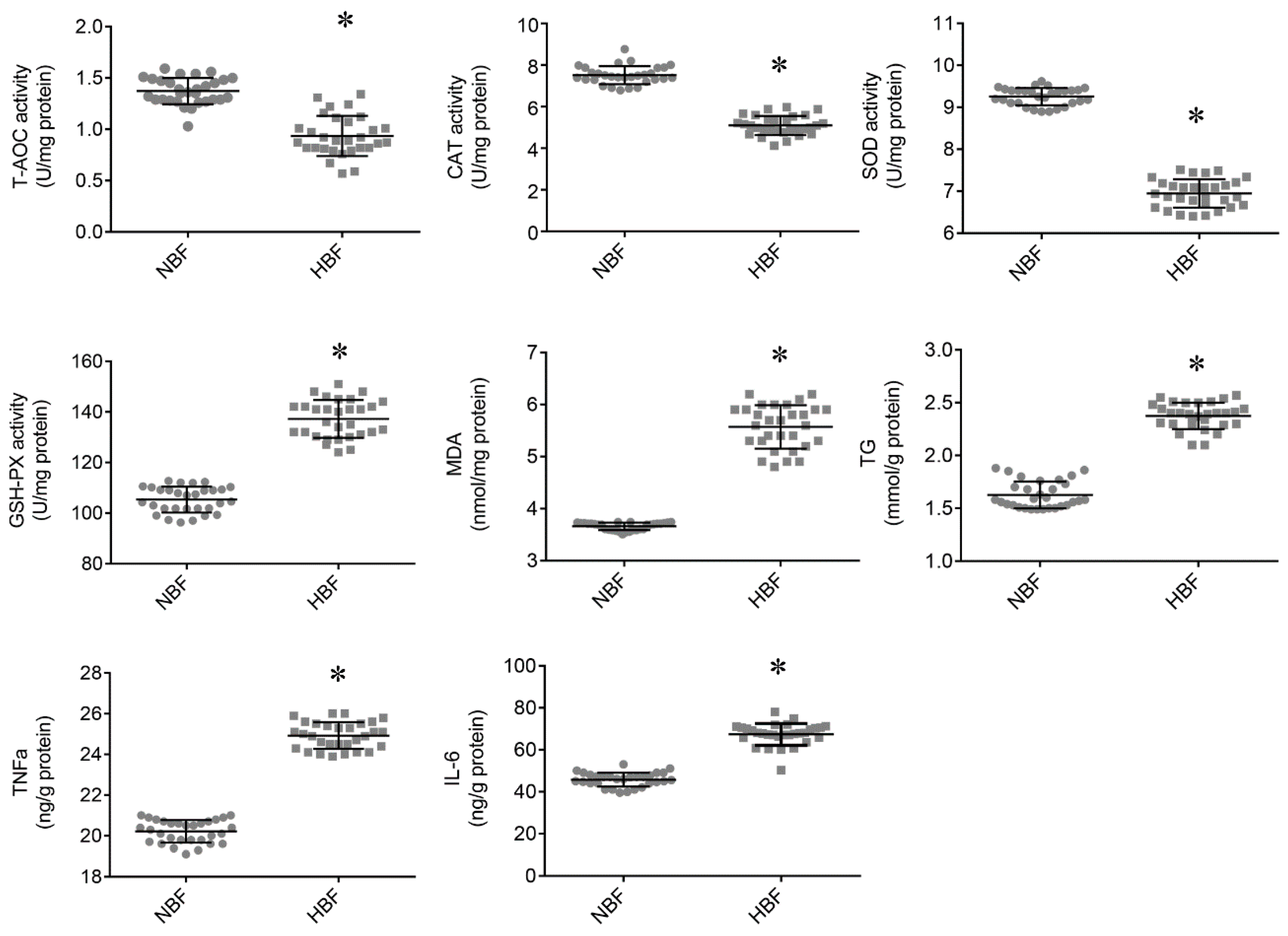
3.2. Antioxidant Status and TG and Inflammatory Cytokine Contents in the Placentas
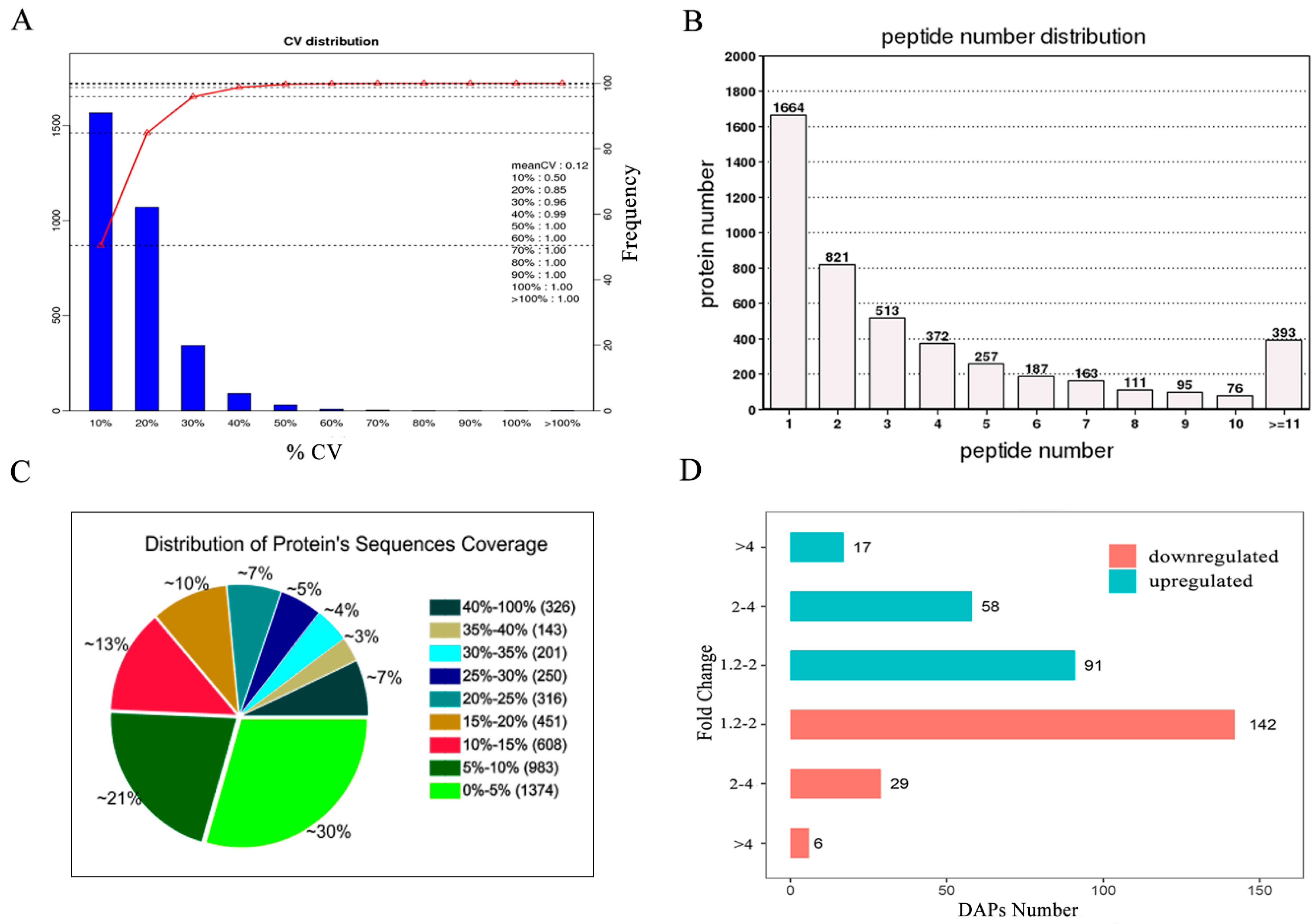
3.3. Identification and Quantification of the Placental Proteins
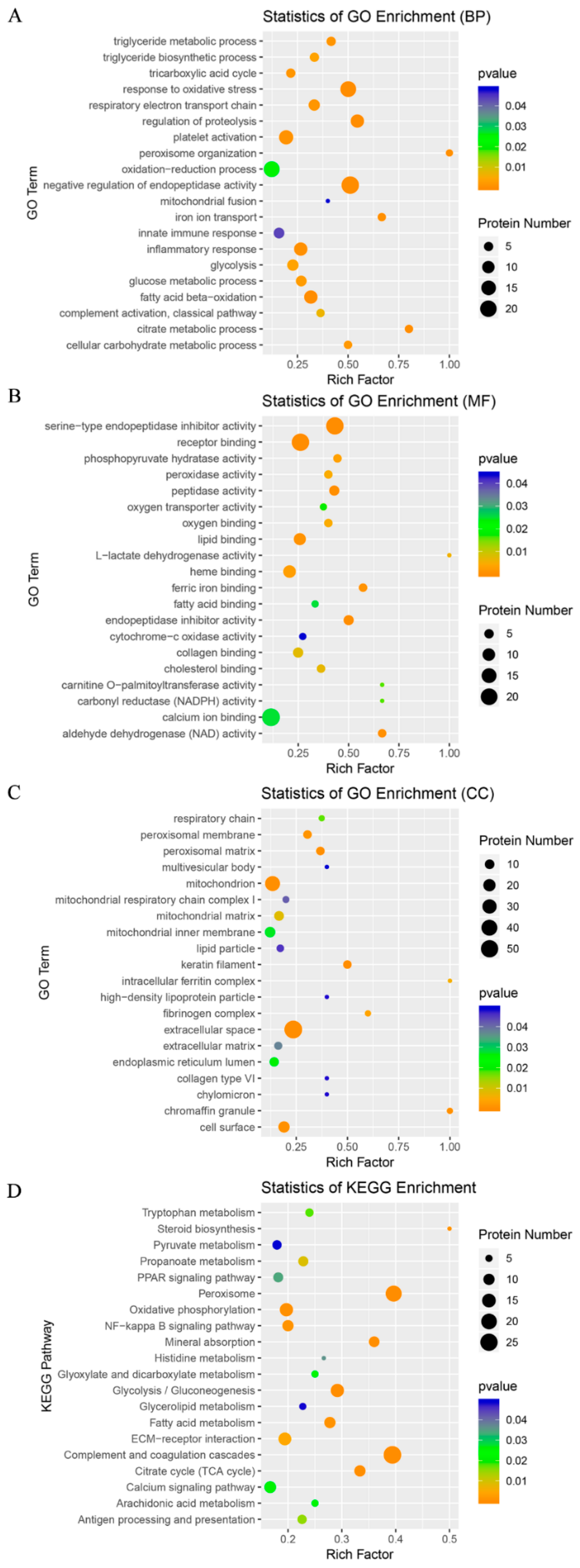
3.4. GO and KEGG Analyses of the DAPs
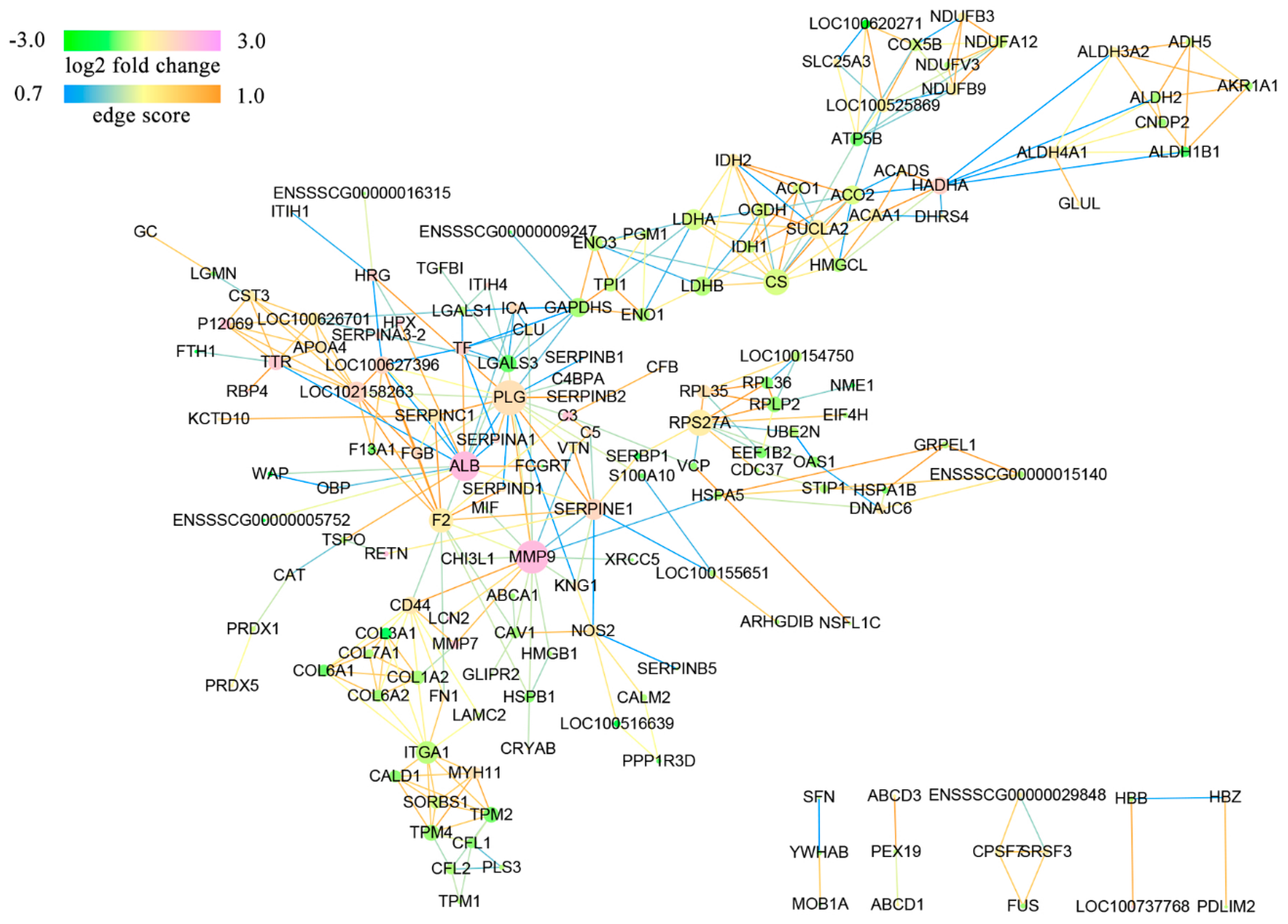
3.5. Protein Interaction Networks of the DAPs
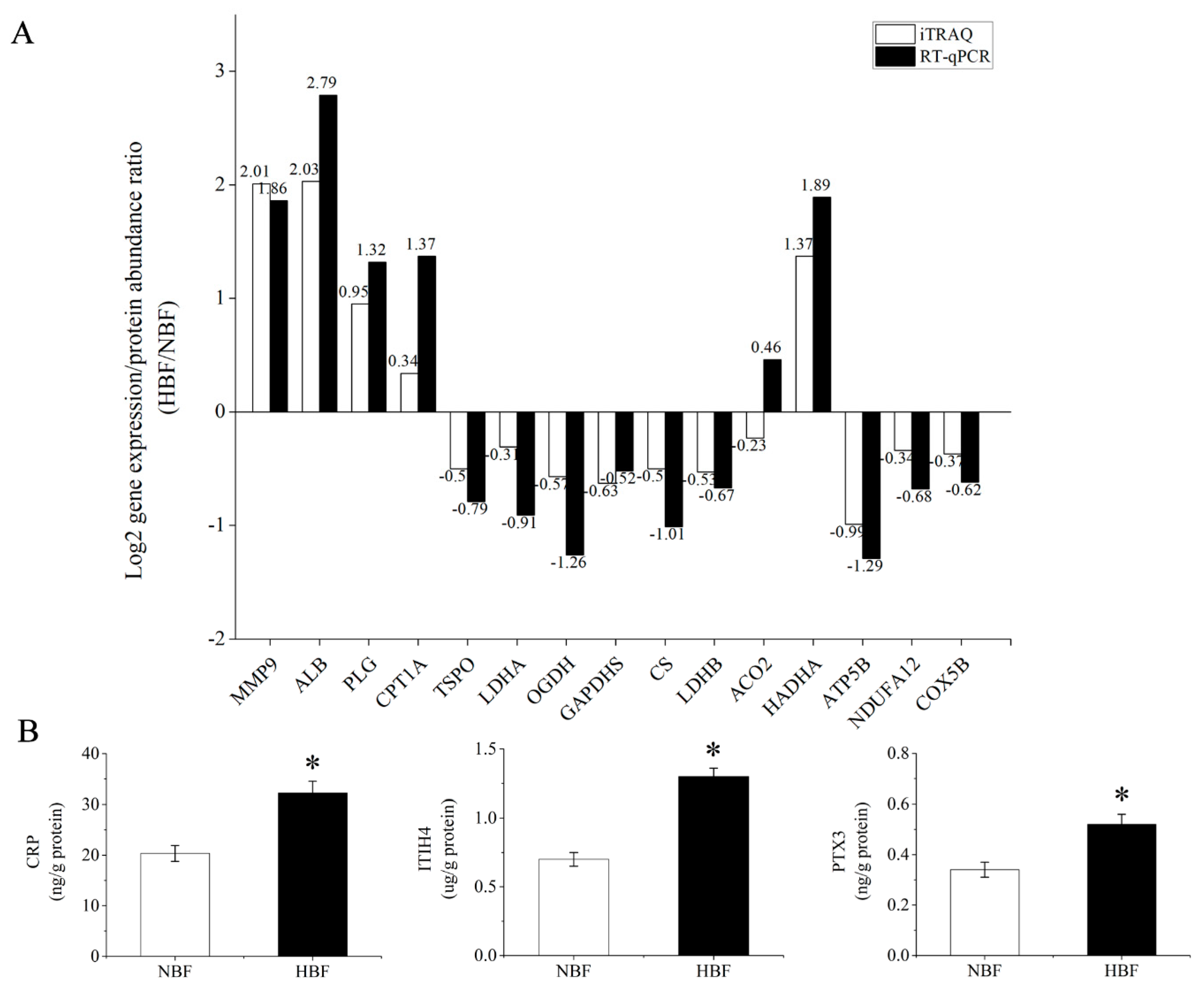
3.6. Validation of the DAPs Using RT-qPCR and ELISA Analysis
4. Discussion
4.1. Carbohydrate and Lipid Metabolism-Related DAPs
4.2. Mitochondrial Respiratory Chain and Mitochondrial Morphology-Related DAPs
4.3. Steroid Hormone Biosynthesis-Related DAPs
4.4. Oxidative Stress-Related DAPs
4.5. Inflammatory Response-Related DAPs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Flegal, K.M.; Carroll, M.D.; Kit, B.K.; Ogden, C.L. Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999–2010. J. Am. Med. Assoc. 2012, 307, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Howell, K.R.; Powell, T.L. Effects of maternal obesity on placental function and fetal development. Reproduction 2017, 153, R97–R108. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.F.; Xu, T.; Cai, A.L.; Wu, Y.H.; Wei, H.K.; Jiang, S.W.; Peng, J. Excessive backfat of sows at 109 d of gestation induces lipotoxic placental environment and is associated with declining reproductive performance. J. Anim. Sci. 2018, 96, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Lutsiv, O.; Mah, J.; Beyene, J.; Mcdonald, S.D. The effects of morbid obesity on maternal and neonatal health outcomes: A systematic review and meta-analyses. Obes. Rev. 2015, 16, 531–546. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.C.; Mahmood, T. Obesity in pregnancy. Best Pract. Res. Clin. Obstet. Gynaecol. 2015, 29, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Filha, W.S.A.; Bernardi, M.L.; Wentz, I.; Bortolozzo, F.P. Reproductive performance of gilts according to growth rate and backfat thickness at mating. Anim. Reprod. Sci. 2010, 121, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Houde, A.A.; Méthot, S.; Murphy, B.D.; Bordignon, V.; Palin, M.F. Relationships between backfat thickness and reproductive efficiency of sows: A two-year trial involving two commercial herds fixing backfat thickness at breeding. Can. J. Anim. Sci. 2010, 90, 429–436. [Google Scholar] [CrossRef]
- Saben, J.; Lindsey, F.; Zhong, Y.; Thakali, K.; Badger, T.M.; Andres, A.; Gomez-Acevedo, H.; Shankar, K. Maternal obesity is associated with a lipotoxic placental environment. Placenta 2014, 35, 171–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Challier, J.C.; Basu, S.; Bintein, T.; Hotmire, K.; Minium, J.; Catalano, P.M.; Mouzon, S.H. Obesity in pregnancy stimulates macrophage accumulation and inflammation in the placenta. Placenta 2008, 29, 274–281. [Google Scholar] [CrossRef]
- Roberts, V.H.J.; Smith, J.; Mclea, S.A.; Heizer, A.B.; Richardson, J.L.; Myatt, L. Effect of Increasing Maternal Body Mass Index on Oxidative and Nitrative Stress in the Human Placenta. Placenta 2009, 30, 169–175. [Google Scholar] [CrossRef]
- Malti, N.; Merzouk, H.; Merzouk, S.A.; Loukidi, B.; Karaouzene, N.; Malti, A.; Narce, M. Oxidative stress and maternal obesity: Feto-placental unit interaction. Placenta 2014, 35, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Farley, D.M.; Choi, J.; Dudley, D.J.; Li, C.; Jenkins, S.L.; Myatt, L.; Nathanielsz, P.W. Placental Amino Acid Transport and Placental Leptin Resistance in Pregnancies Complicated by Maternal Obesity. Placenta 2010, 31, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Dong, S.S.; Hu, J.; Yao, J.J.; Yan, P.S. The effect of maternal obesity on fatty acid transporter expression and lipid metabolism in the full-term placenta of lean breed swine. J. Anim. Physiol. Anim. Nutr. 2018, 102, e242–e253. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Lu, J.; Deng, Z.; Xu, T.; Yang, Y.; Wei, H.; Li, S.; Jiang, S.; Peng, J. Maternal obesity aggravates the abnormality of porcine placenta by increasing N⁶-methyladenosine. Int. J. Obes. 2018, 42, 1812–1820. [Google Scholar] [CrossRef] [PubMed]
- van Rens, B.; de Koning, G.; Bergsma, R.; van der Lende, T. Preweaning piglet mortality in relation to placental efficiency. J. Anim. Sci. 2005, 83, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.E.; Biensen, N.J.; Ford, S.P. Novel insight into the control of litter size in pigs, using placental efficiency as a selection tool. J. Anim. Sci. 1999, 77, 1654–1658. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shan, X.; Wu, Y.; Su, S.; Li, S.; Liu, H.; Han, J.; Xue, C.; Yuan, Y. iTRAQ-based quantitative proteomic analysis reveals new metabolic pathways responding to chilling stress in maize seedlings. J. Proteom. 2016, 146, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Datta, A.; Chai, Y.L.; Tan, J.M.; Lee, J.H.; Francis, P.T.; Chen, C.P.; Sze, S.K.; Lai, M.K.P. An iTRAQ-based proteomic analysis reveals dysregulation of neocortical synaptopodin in Lewy body dementias. Mol. Brain 2017, 10, 36. [Google Scholar] [CrossRef]
- Oliva, K.; Barker, G.; Riley, C.; Bailey, M.J.; Permezel, M.; Rice, G.E.; Lappas, M. The effect of pre-existing maternal obesity on the placental proteome: Two-dimensional difference gel electrophoresis coupled with mass spectrometry. J. Mol. Endocrinol. 2012, 48, 139–149. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, Y.; Pan, J.; Liu, X.; Chen, H.; Zhou, X.; Yuan, Z.; Wang, X.; Mo, D. iTRAQ-based quantitative proteomic analysis reveals the distinct early embryo myofiber type characteristics involved in landrace and miniature pig. BMC Genom. 2016, 17, 137. [Google Scholar] [CrossRef]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Lotia, S.; Pico, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods 2012, 9, 1069–1076. [Google Scholar] [CrossRef] [Green Version]
- Myatt, L.; Maloyan, A. Obesity and Placental Function. Semin. Reprod. Med. 2016, 34, 42–49. [Google Scholar] [CrossRef] [Green Version]
- Rebholz, S.L.; Burke, K.T.; Yang, Q.; Tso, P.; Woollett, L.A. Dietary fat impacts fetal growth and metabolism: Uptake of chylomicron remnant core lipids by the placenta. Am. J. Physiol. Endocrinol. Metab. 2011, 301, 416–425. [Google Scholar] [CrossRef]
- Dubé, E.; Gravel, A.; Martin, C.; Desparois, G.; Moussa, I.; Ethierchiasson, M.; Forest, J.C.; Giguère, Y.; Masse, A.; Lafond, J. Modulation of Fatty Acid Transport and Metabolism by Maternal Obesity in the Human Full-Term Placenta. Biol. Reprod. 2012, 87, 1–11. [Google Scholar] [CrossRef]
- Alvino, G.; Cozzi, V.; Radaelli, T.; Ortega, H.; Herrera, E.; Cetin, I. Maternal and fetal fatty acid profile in normal and intrauterine growth restriction pregnancies with and without preeclampsia. Pediatr. Res. 2008, 64, 615–620. [Google Scholar] [CrossRef]
- Aye, I.L.; Lager, S.; Ramirez, V.I.; Gaccioli, F.; Dudley, D.J.; Jansson, T.; Powell, T.L. Increasing maternal body mass index is associated with systemic inflammation in the mother and the activation of distinct placental inflammatory pathways. Biol. Reprod. 2014, 90, 129. [Google Scholar] [CrossRef]
- Cai, Q.; Zhao, M.; Liu, X.; Wang, X.; Nie, Y.; Li, P.; Liu, T.; Ge, R.; Han, F. Reduced expression of citrate synthase leads to excessive superoxide formation and cell apoptosis. Biochem. Biophys. Res. Commun. 2017, 485, 388–394. [Google Scholar] [CrossRef]
- Calabuig-Navarro, V.; Haghiac, M.; Minium, J.; Glazebrook, P.; Ranasinghe, G.C.; Hoppel, C.; de-Mouzon, S.H.; Catalano, P.; O’Tierney-Ginn, P. Effect of Maternal Obesity on Placental Lipid Metabolism. Endocrinology 2017, 158, 2543–2555. [Google Scholar] [CrossRef]
- Serra, D.; Mera, P.; Malandrino, M.I.; Mir, J.F.; Herrero, L. Mitochondrial fatty acid oxidation in obesity. Antioxid. Redox Signal. 2013, 19, 269–284. [Google Scholar] [CrossRef]
- Serviddio, G.; Giudetti, A.M.; Bellanti, F.; Priore, P.; Rollo, T.; Tamborra, R.; Siculella, L.; Vendemiale, G.; Altomare, E.; Gnoni, G.V. Oxidation of Hepatic Carnitine Palmitoyl Transferase-I (CPT-I) Impairs Fatty Acid Beta-Oxidation in Rats Fed a Methionine-Choline Deficient Diet. PLoS ONE 2011, 6, e24084. [Google Scholar] [CrossRef]
- Muralimanoharan, S.; Guo, C.; Myatt, L.; Maloyan, A. Sexual dimorphism in miR-210 expression and mitochondrial dysfunction in the placenta with maternal obesity. Int. J. Obes. 2015, 39, 1274–1281. [Google Scholar] [CrossRef] [Green Version]
- Schönfeld, P.; Wojtczak, L. Fatty acids as modulators of the cellular production of reactive oxygen species. Free Radical Biol. Med. 2008, 45, 231–241. [Google Scholar] [CrossRef]
- Nakamura, M.T.; Yudell, B.E.; Loor, J.J. Regulation of energy metabolism by long-chain fatty acids. Prog. Lipid Res. 2014, 53, 124–144. [Google Scholar] [CrossRef]
- Schneeberger, M.; Dietrich, M.O.; Sebastian, D.; Imbernon, M.; Castano, C.; Garcia, A.; Esteban, Y.; Gonzalez-Franquesa, A.; Rodriguez, I.C.; Bortolozzi, A.; et al. Mitofusin 2 in POMC neurons connects ER stress with leptin resistance and energy imbalance. Cell 2013, 155, 172–187. [Google Scholar] [CrossRef]
- Lassance, L.; Haghiac, M.; Minium, J.; Catalano, P.; Hauguel-De, M.S. Obesity-Induced Down-Regulation of the Mitochondrial Translocator Protein (TSPO) Impairs Placental Steroid Production. J. Clin. Endocrinol. Metab. 2015, 100, E11–E18. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.C.; Fischer, B.A.; Sridhar, V.; Blohowiak, S.E.; Coe, C.L.; Kling, P.J. Maternal obesity at delivery: A risk factor for newborn iron deficiency. Pediatr. Res. 2009, 66, 480. [Google Scholar]
- Kelner, G.S.; Lee, M.; Clark, M.E.; Maciejewski, D.; Mcgrath, D.; Rabizadeh, S.; Lyons, T.; Bredesen, D.; Jenner, P.; Maki, R.A. The copper transport protein Atox1 promotes neuronal survival. J. Biol. Chem. 2000, 275, 580–584. [Google Scholar] [CrossRef]
- Taira, T.; Saito, Y.; Niki, T.; Iguchiariga, S.M.M.; Takahashi, K.; Ariga, H. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 2004, 5, 213–218. [Google Scholar] [CrossRef]
- Almeida-Souza, L.; Goethals, S.; de Winter, V.; Dierick, I.; Gallardo, R.; Van Durme, J.; Irobi, J.; Gettemans, J.; Rousseau, F.; Schymkowitz, J.; et al. Increased Monomerization of Mutant HSPB1 Leads to Protein Hyperactivity in Charcot-Marie-Tooth Neuropathy. J. Biol. Chem. 2010, 285, 12778–12786. [Google Scholar] [CrossRef] [Green Version]
- Meunier, L.; Usherwood, Y.K.; Chung, K.T.; Hendershot, L.M. A Subset of Chaperones and Folding Enzymes Form Multiprotein Complexes in Endoplasmic Reticulum to Bind Nascent Proteins. Mol. Biol. Cell 2002, 13, 4456–4469. [Google Scholar] [CrossRef]
- Klos, A.; Tenner, A.J.; Johswich, K.O.; Ager, R.R.; Reis, E.S.; Köhl, J. The role of the anaphylatoxins in health and disease. Mol. Immunol. 2009, 46, 2753–2766. [Google Scholar] [CrossRef] [Green Version]
- Mold, C.; Rodriguez, W.; Rodic-Polic, B.; Du, C.T. C-reactive protein mediates protection from lipopolysaccharide through interactions with FcγR. J. Immunol. 2002, 169, 7019–7025. [Google Scholar] [CrossRef]
- Choimiura, N.H.; Takahashi, K.; Yoda, M.; Saito, K.; Hori, M.; Ozaki, H.; Mazda, T.; Tomita, M. The novel acute phase protein, IHRP, inhibits actin polymerization and phagocytosis of polymorphonuclear cells. Inflamm. Res. 2000, 49, 305–310. [Google Scholar] [CrossRef]
- Geisert, R.D.; Yelich, J.V.; Pratt, T.; Pomp, D. Expression of an inter-alpha-trypsin inhibitor heavy chain-like protein in the pig endometrium during the oestrous cycle and early pregnancy. J. Reprod. Fertil. 1998, 114, 35–43. [Google Scholar] [CrossRef]
- Llewellyn-Jones, C.G.; Lomas, D.A.; Carrell, R.W.; Stockley, R.A. The effect of the Z mutation on the ability of α1-antirtrypsin to prevent neutrophil mediated tissue damage. Biochim. Biophys. Acta 1994, 1227, 155–160. [Google Scholar] [CrossRef]
- Subramaniyam, D.; Virtala, R.; Pawłowski, K.; Clausen, I.G.; Warkentin, S.; Stevens, T.; Janciauskiene, S. TNF-α-induced self expression in human lung endothelial cells is inhibited by native and oxidized α1-antitrypsin. Int. J. Biochem. Cell Biol. 2008, 40, 258–271. [Google Scholar] [CrossRef]
- Hu, S.J.; Ren, G.; Liu, J.L.; Zhao, Z.A.; Yu, Y.S.; Su, R.W.; Ma, X.H.; Ni, H.; Lei, W.; Yang, Z.M. MicroRNA Expression and Regulation in Mouse Uterus during Embryo Implantation. J. Biol. Chem. 2008, 283, 23473–23484. [Google Scholar] [CrossRef] [Green Version]
- Lian, S.; Xia, Y.; Khoi, P.N.; Ung, T.T.; Yoon, H.J.; Kim, N.H.; Kim, K.K.; Jung, Y.D. Cadmium induces matrix metalloproteinase-9 expression via ROS-dependent EGFR, NF-кB, and AP-1 pathways in human endothelial cells. Toxicology 2015, 338, 104–116. [Google Scholar] [CrossRef]
| Reproductive performance parameters | NBF | HBF |
| Backfat at mating, mm | 18 ± 0.4 | 25 ± 0.5 # |
| Backfat at farrowing, mm | 20 ± 0.5 | 27 ± 0.6 # |
| Backfat gain during pregnancy, mm | 1.54 ± 0.32 | 1.61 ± 0.31 |
| Total born piglets, n | 423 | 421 |
| Live-born piglets, n | 371 | 362 |
| Litter size of live-born and stillborn piglets, n | 14.12 ± 0.47 | 14.04 ± 0.39 |
| Litter size of live-born piglets, n | 12.37 ± 0.31 | 12.08 ± 0.32 * |
| Litter weight of live-born piglets, kg | 17.15 ± 0.35 | 16.99 ± 0.64 |
| Litter weight of live-born and stillborn piglets, kg | 18.94 ± 0.51 | 18.78 ± 0.53 |
| Average weight of live-born piglets, kg | 1.51 ± 0.10 | 1.39 ± 0.09 * |
| Piglets with weights < 0.9 kg, n | 0.96 ± 0.08 | 1.31 ± 0.07 # |
| Rate of piglets with weight < 0.9 kg, % 1 | 7.17 ± 0.87 | 10.32 ± 0.93 * |
| CV for weights of live-born piglets, % 2 | 20.06 ± 1.69 | 23.38 ± 1.63 * |
| Placental weight, g | 286.29 ± 4.07 | 346.97 ± 5.66 * |
| Placental efficiency 3 | 4.96 ± 0.19 | 4.39 ± 0.24 # |
| Maternal plasma parameters | NBF | HBF |
| Glucose, mmol/L | 4.29 ± 0.23 | 4.35 ± 0.22 |
| HDL, mM | 1.27 ± 0.10 | 1.25 ± 0.08 |
| LDL, mM | 4.17 ± 0.27 | 4.20 ± 0.21 |
| CHOL, mM | 5.97 ± 0.54 | 6.11 ± 0.51 |
| TG, mM | 0.78 ± 0.15 | 0.89 ± 0.13 * |
| FFA, mM | 0.14 ± 0.07 | 0.22 ± 0.04 * |
| IL-6, ng/L | 237.69 ± 9.18 | 267.37 ± 9.29 * |
| TNF-α, ng/L | 64.87 ± 3.23 | 97.19 ± 4.04 # |
| NO. | Uniprot Accession | Score | Description | Gene | Ratio Average 1 | p-Value |
|---|---|---|---|---|---|---|
| DAPs involved in glycolysis | ||||||
| 1 | F1S814 | 699 | Phosphoglucomutase-1 | PGM1 | 0.73 | 0.001 |
| 2 | D0G7F6 | 1557 | Triosephosphate isomerase | TPI1 | 0.73 | 0.001 |
| 3 | F1RM74 | 281 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDHS | 0.65 | <0.001 |
| 4 | I3LK59 | 4047 | Enolase | ENO1 | 0.65 | 0.001 |
| 5 | Q1KYT0 | 1980 | Beta-enolase | ENO3 | 0.73 | 0.001 |
| 6 | P00336 | 1026 | l-lactate dehydrogenase B chain | LDHB | 0.69 | <0.001 |
| 7 | F1RII7 | 21595 | Hemoglobin subunit beta | HBB | 0.65 | 0.001 |
| 8 | P00339 | 1468 | l-lactate dehydrogenase A chain | LDHA | 0.81 | <0.001 |
| DAPs involved in tricarboxylic acid cycle | ||||||
| 1 | P00889 | 929 | Citrate synthase, mitochondrial | CS | 0.71 | <0.001 |
| 2 | I3LJW4 | 2855 | Aconitase 1 | ACO1 | 0.70 | <0.001 |
| 3 | F1SRC5 | 1602 | Aconitate hydratase, mitochondrial | ACO2 | 0.63 | 0.004 |
| 4 | Q0QF01 | 784 | Succinate dehydrogenase flavoprotein subunit | SDHA | 0.76 | 0.001 |
| 5 | F1SSH8 | 1077 | Oxoglutarate dehydrogenase | OGDH | 0.67 | <0.001 |
| DAPs involved in triglyceride biosynthetic process | ||||||
| 1 | A5YV76 | 6249 | Fatty acid synthase | FASN | 1.49 | <0.001 |
| 2 | G1FJ20 | 4204 | ATP-citrate synthase | ACLY | 1.48 | <0.001 |
| 3 | C0KEG5 | 98 | 1-acylglycerol-3-phosphate O-acyltransferase 7 | LPCAT4 | 1.51 | 0.001 |
| 4 | I3LLU0 | 495 | Glycerol-3-phosphate dehydrogenase | GPD1L | 1.57 | <0.001 |
| 5 | F1RWN0 | 159 | Uncharacterized protein | AGPAT2 | 1.32 | <0.001 |
| DAPs involved in fatty acid oxidation | ||||||
| 1 | A7J0B0 | 1777 | Acyl-coenzyme A oxidase | ACOX1 | 1.88 | 0.002 |
| 2 | Q28956 | 1346 | 17beta-estradiol dehydrogenase | HSD17B4 | 1.94 | <0.001 |
| 3 | D0G7F1 | 981 | Sterol carrier protein 2 | SCP2 | 1.49 | 0.001 |
| 4 | F1RRB7 | 1262 | Acetyl-CoA acyltransferase 1 | ACAA1 | 1.48 | <0.001 |
| 5 | F1S3F7 | 173 | ATP-binding cassette sub-family D member 4 | ABCD4 | 1.69 | <0.001 |
| 6 | I3LI20 | 242 | ATP-binding cassette sub-family D member 3 | ABCD3 | 1.58 | 0.001 |
| 7 | F1S2A1 | 71 | ATP binding cassette subfamily D member 1 | ABCD1 | 1.63 | <0.001 |
| 8 | F1SQH7 | 287 | Solute carrier family 27 member 2 | SLC27A2 | 1.57 | 0.005 |
| 9 | Q3Y5G5 | 447 | Peroxisomal enoyl coenzyme A hydratase 1 | ECH1 | 1.45 | 0.012 |
| 10 | F1SBY8 | 157 | Peroxisomal carnitine O-octanoyltransferase | CROT | 1.42 | 0.011 |
| 11 | F1RJH2 | 366 | Short-chain-specific acyl-CoA dehydrogenase | ACADS | 1.53 | 0.002 |
| 12 | Q29554 | 3820 | Trifunctional enzyme subunit alpha, mitochondrial | HADHA | 2.59 | 0.001 |
| 13 | Q95JG9 | 481 | Carnitine palmitoyltransferase I | CPT1A | 1.27 | <0.001 |
| 14 | B2ZF49 | 3810 | Hydroxyacyl-coenzyme A dehydrogenase | HADH | 0.16 | <0.001 |
| DAPs involved in mitochondrial respiratory chain and mitochondrial morphology | ||||||
| 1 | G8IFA6 | 228 | Mitochondrial NADH dehydrogenase Fe-S protein 4 | NDUFS4 | 0.55 | <0.001 |
| 2 | I3LRR4 | 389 | NADH dehydrogenase [ubiquinone] flavoprotein 3 | NDUFV3 | 0.67 | 0.002 |
| 3 | F1SQP4 | 264 | NADH dehydrogenase 1 alpha subcomplex subunit 12 | NDUFA12 | 0.66 | 0.001 |
| 4 | Q0QF01 | 784 | Succinate dehydrogenase flavoprotein subunit | SDHA | 0.76 | 0.001 |
| 5 | F1RPD4 | 340 | Cytochrome b-c1 complex subunit 2, mitochondrial | UQCRC2 | 0.17 | <0.001 |
| 6 | Q5S3G4 | 309 | Cytochrome c oxidase subunit 5B, mitochondrial | COX5B | 0.55 | 0.001 |
| 7 | F1SLA0 | 9014 | ATP synthase subunit beta | ATP5B | 0.50 | 0.001 |
| 8 | F1RF76 | 54 | Mitofusin 2 | MFN2 | 2.08 | 0.003 |
| 9 | F1SFG7 | 148 | mitochondrial dynamin like GTPase | OPA1 | 1.28 | <0.001 |
| DAPs involved in steroid hormone biosynthesis and regulation | ||||||
| 1 | F1SJR8 | 59 | Translocator protein | TSPO | 0.71 | <0.001 |
| 2 | F1RID6 | 57 | StAR related lipid transfer domain containing 5 | STARD5 | 0.60 | <0.001 |
| 3 | F1RSR5 | 28 | StAR related lipid transfer domain containing 13 | STARD13 | 0.68 | 0.003 |
| DAPs involved in oxidative stress | ||||||
| 1 | P79335 | 147 | Plasminogen activator inhibitor 1 | SERPINE1 | 2.30 | 0.005 |
| 2 | F1SGS9 | 1461 | Catalase | CAT | 0.75 | <0.001 |
| 3 | I3LDC7 | 2004 | Isocitrate dehydrogenase [NADP] | IDH1 | 0.73 | <0.001 |
| 4 | F1S131 | 165 | Caspase 6 | CASP6 | 0.51 | 0.006 |
| 5 | Q0R678 | 860 | DJ-1 protein | PARK7 | 0.59 | 0.001 |
| 6 | F1SJT7 | 182 | Apolipoprotein A-IV | APOA4 | 1.72 | 0.001 |
| 7 | P67937 | 1612 | Tropomyosin alpha-4 chain | TPM4 | 0.60 | <0.001 |
| 8 | F1SQP4 | 264 | NADH dehydrogenase 1 alpha subcomplex subunit 12 | NDUFA12 | 0.66 | 0.001 |
| 9 | F2Z5B6 | 1159 | Tropomyosin alpha-1 chain | TPM1 | 0.64 | <0.001 |
| 10 | Q6UJZ1 | 118 | Glutathione peroxidase | GPX3 | 2.89 | 0.003 |
| 11 | D0G0C7 | 172 | Antioxidant protein 1 homolog | ATOX1 | 0.53 | <0.001 |
| 12 | F1RIP3 | 647 | Ferritin | FTL | 0.28 | <0.001 |
| 13 | I3LR69 | 68 | Ferritin | FTH1 | 0.27 | <0.001 |
| 14 | F1S9Q3 | 3368 | Heat shock cognate 71 kDa protein | HSPA8 | 0.64 | <0.001 |
| 15 | F1RS36 | 5142 | 78 kDa glucose-regulated protein | HSPA5 | 0.62 | <0.001 |
| 16 | F1SC56 | 147 | Matrix metalloproteinase-9 | MMP9 | 4.02 | <0.001 |
| 17 | Q5S1U1 | 1057 | heat shock protein beta-1 | HSPB1 | 0.55 | <0.001 |
| 18 | F1RKG8 | 781 | Uncharacterized protein | PEBP1 | 0.65 | <0.001 |
| DAPs involved in inflammatory response | ||||||
| 1 | F1SME1 | 122 | Complement C5a anaphylatoxin | C5 | 2.21 | <0.001 |
| 2 | P79263 | 1833 | Inter-alpha-trypsin inhibitor heavy chain H4 | ITIH4 | 2.62 | 0.001 |
| 3 | F1S131 | 165 | Caspase 6 | CASP6 | 0.51 | 0.006 |
| 4 | A5D9N3 | 199 | Allograft inflammatory factor 1 | AIF1 | 0.59 | 0.001 |
| 5 | F1SCF0 | 2964 | Alpha-1-antitrypsin | SERPINA1 | 2.67 | <0.001 |
| 6 | A0MWC5 | 147 | Monocyte differentiation antigen CD14 | CD14 | 0.71 | <0.001 |
| 7 | F1RJ76 | 50 | C-reactive protein | CRP | 2.36 | 0.043 |
| 8 | F1SJM0 | 315 | Pentraxin-3 | PTX3 | 3.53 | 0.018 |
| 9 | F1SFI4 | 235 | Kininogen 1 | KNG1 | 2.01 | 0.003 |
| 10 | P80015 | 429 | Azurocidin | AZU1 | 5.18 | <0.001 |
| 11 | I3LL32 | 73 | Uncharacterized protein | CHIA | 1.75 | <0.001 |
| 12 | F1RUN2 | 7337 | Serum albumin | ALB | 4.08 | <0.001 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.-W.; Hu, J.; Wei, M.; Guo, Y.-Y.; Yan, P.-S. The Effects of Maternal Obesity on Porcine Placental Efficiency and Proteome. Animals 2019, 9, 546. https://doi.org/10.3390/ani9080546
Li J-W, Hu J, Wei M, Guo Y-Y, Yan P-S. The Effects of Maternal Obesity on Porcine Placental Efficiency and Proteome. Animals. 2019; 9(8):546. https://doi.org/10.3390/ani9080546
Chicago/Turabian StyleLi, Ji-Wei, Jian Hu, Ming Wei, Ying-Ying Guo, and Pei-Shi Yan. 2019. "The Effects of Maternal Obesity on Porcine Placental Efficiency and Proteome" Animals 9, no. 8: 546. https://doi.org/10.3390/ani9080546





