Effects of Land Transport Stress on Variations in Ruminal Microbe Diversity and Immune Functions in Different Breeds of Cattle
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Care and Study Design
2.2. Blood and Rumen Fluid Sample Collection and Storage
2.3. DNA Extraction
2.4. PCR Amplification, Library Construction and Illumina Sequencing
2.5. Library Preparation and Sequencing
2.6. Bioinformatics and Statistical Analysis
2.7. Correlation Between Rumen Microbiota and Cattle Physiological Variables
3. Results
3.1. Serum Hormones
3.2. Rumen Fluid Characteristics
3.3. Immunity Levels
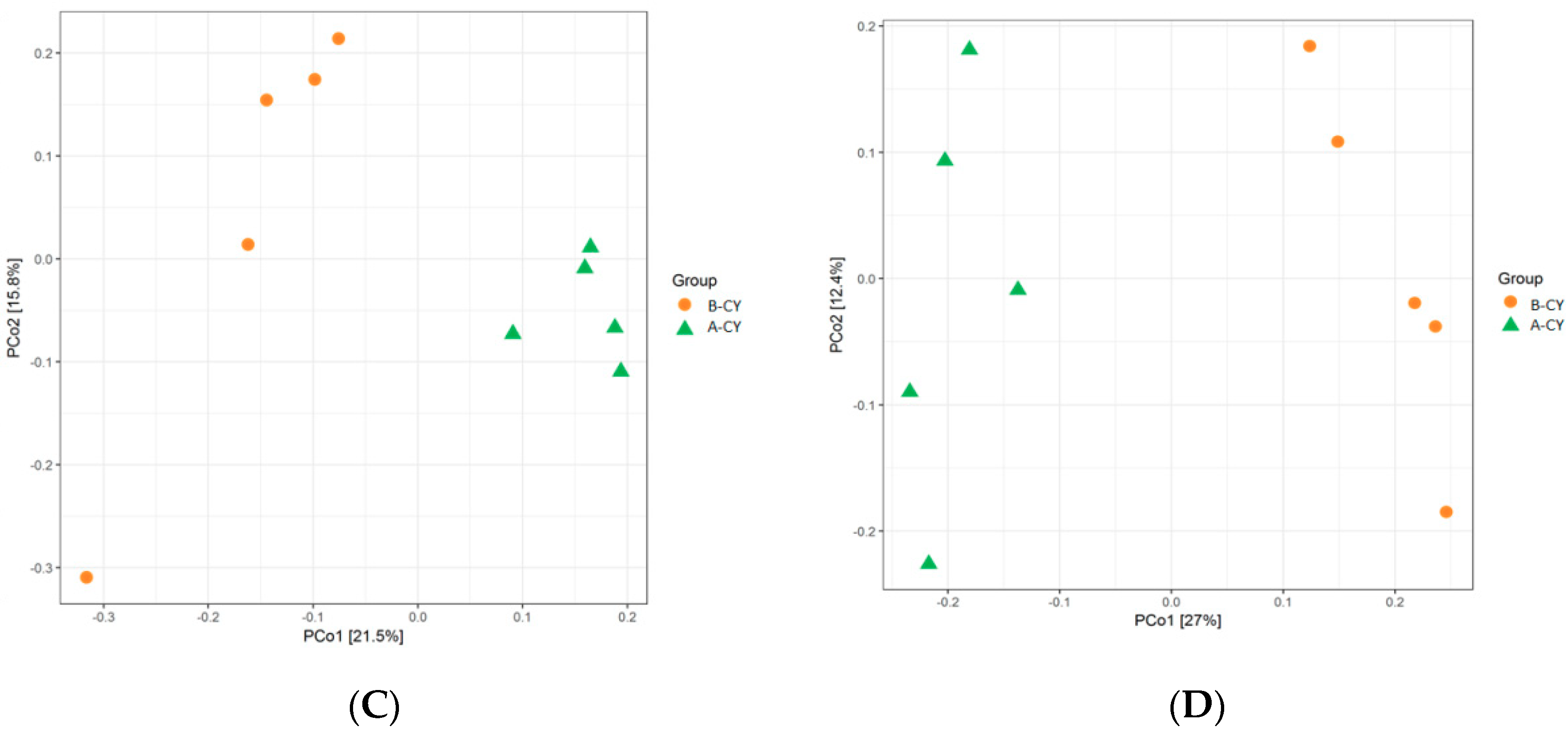
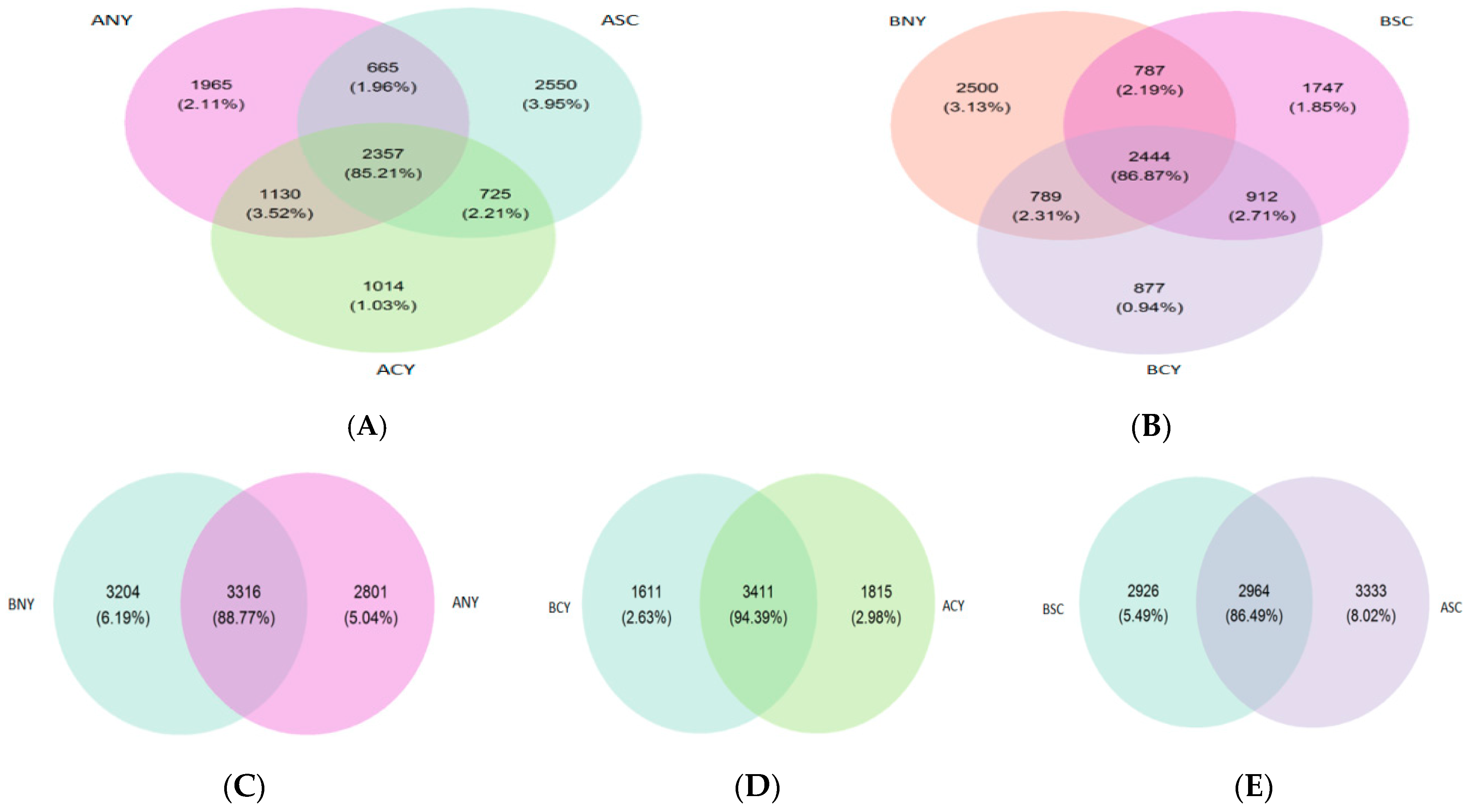
3.4. Alpha-Diversity Measures and OTU Analysis
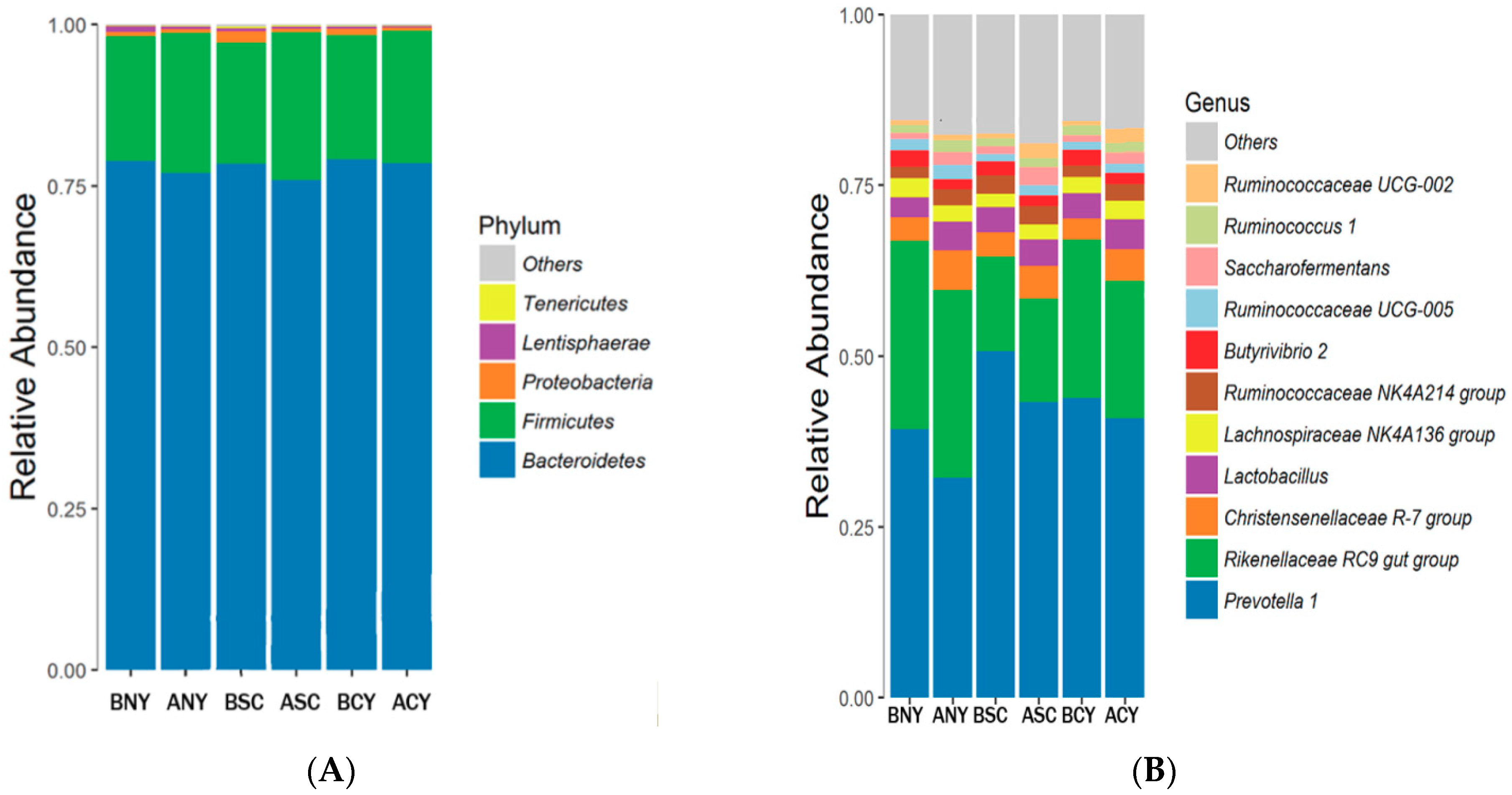
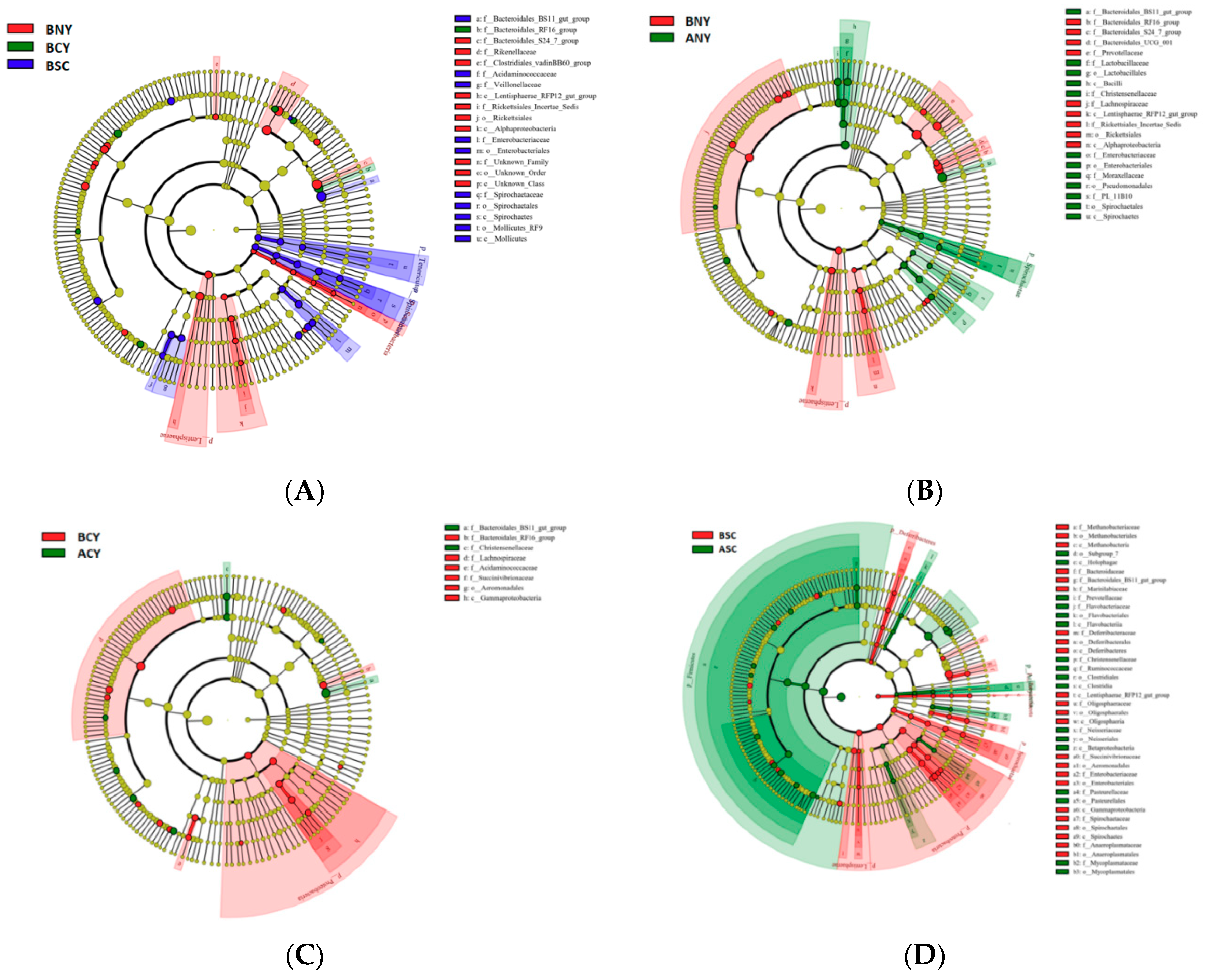
Microbiota Composition of Rumen
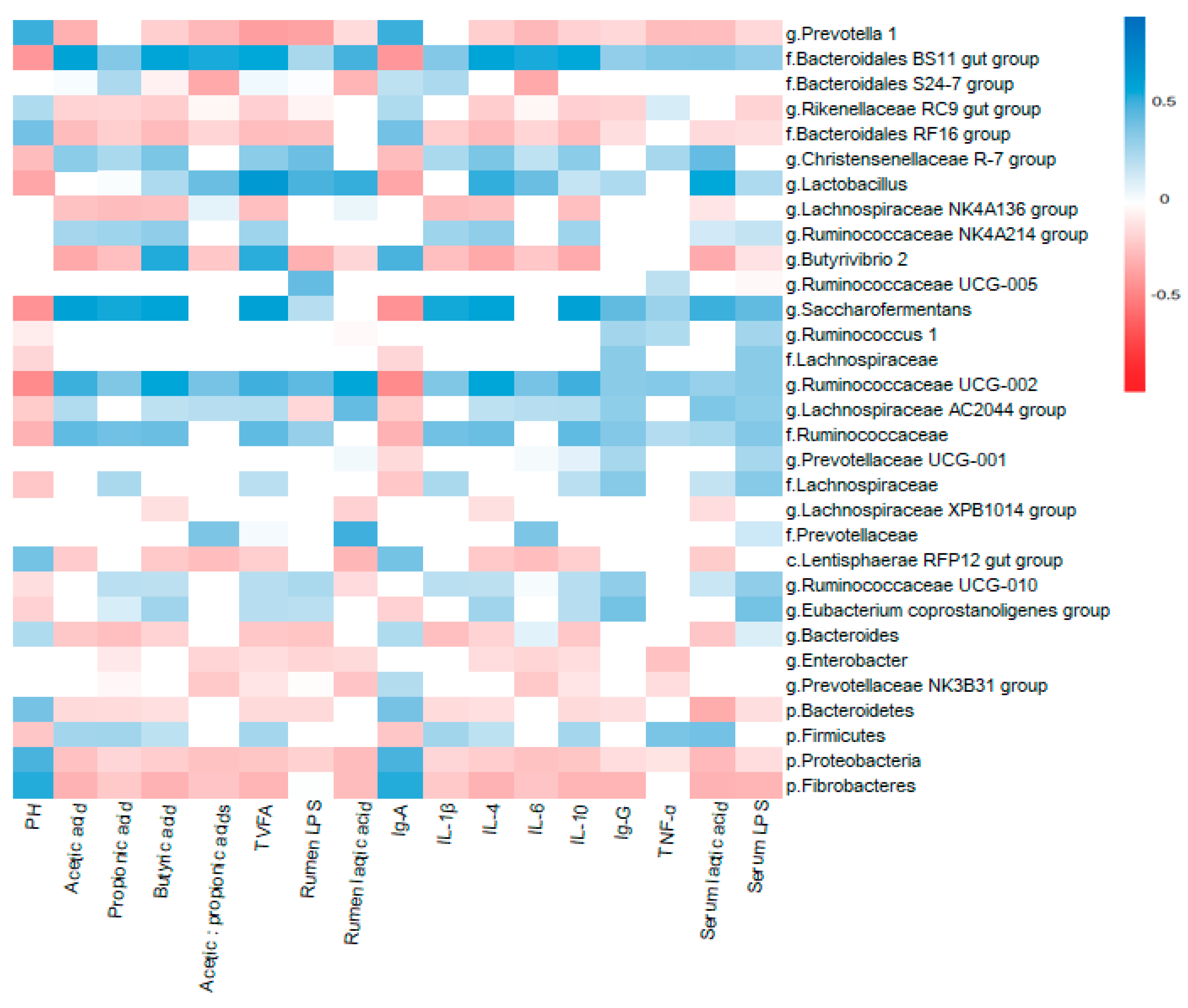
3.5. Correlation Between Rumen Microbiota and Physiological Variables
4. Discussion
4.1. Effect of Hormone Balance in Serum
4.2. Effects of Transport Stress on Rumen Fermentation Characteristics
4.3. Effect of Transport Stress on Rumen Microorganisms
4.4. Effect of Transportation Stress on Immunity
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- González, L.; Schwartzkopf-Genswein, K.; Bryan, M.; Silasi, R.; Brown, F. Relationships between transport conditions and welfare outcomes during commercial long haul transport of cattle in North America. J. Anim. Sci. 2012, 90, 3640–3651. [Google Scholar] [CrossRef] [PubMed]
- Earley, B.; McDonnell, B.; Murray, M.; Prendiville, D.; Crowe, M. The effect of sea transport from Ireland to the Lebanon on inflammatory, adrenocortical, metabolic and behavioural responses of bulls. Res. Vet. Sci. 2011, 91, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Earley, B.; Murray, M.; Prendiville, D.; Pintado, B.; Borque, C.; Canali, E. The effect of transport by road and sea on physiology, immunity and behaviour of beef cattle. Res. Vet. Sci. 2012, 92, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Silanikove, N. Effects of heat stress on the welfare of extensively managed domestic ruminants. Livest. Prod. Sci. 2000, 67, 1–18. [Google Scholar] [CrossRef]
- Silanikove, N. The physiological basis of adaptation in goats to harsh environments. Small Rumin. Res. 2000, 35, 181–193. [Google Scholar] [CrossRef]
- Zhou, G.; Liu, L.; Xiu, X.; Jian, H.; Wang, L.; Sun, B.; Tong, B. Productivity and carcass characteristics of pure and crossbred Chinese Yellow Cattle. Meat Sci. 2001, 58, 359–362. [Google Scholar] [CrossRef]
- Freestone, P.; Lyte, M. Stress and microbial endocrinology: Prospects for ruminant nutrition. Animal 2010, 4, 1248–1257. [Google Scholar] [CrossRef]
- Ingvartsen, K.L.; Andersen, J.B. Integration of metabolism and intake regulation: A review focusing on periparturient animals. J. Dairy Sci. 2000, 83, 1573–1597. [Google Scholar] [CrossRef]
- Deng, L.; He, C.; Zhou, Y.; Xu, L.; Xiong, H. Ground transport stress affects bacteria in the rumen of beef cattle: A real-time PCR analysis. Anim. Sci. J. 2017, 88, 790–797. [Google Scholar] [CrossRef]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Dethlefsen, L.; McFall-Ngai, M.; Relman, D.A. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 2007, 449, 811. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Lozupone, C.A.; Hamady, M.; Knight, R.; Gordon, J.I. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 2008, 6, 776. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Littman, D.R. The microbiome in infectious disease and inflammation. Annu. Rev. Immunol. 2012, 30, 759–795. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef] [PubMed]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2009, 26, 266–267. [Google Scholar] [CrossRef] [PubMed]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; O’hara, R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community ecology package. R package version 1.17–4. Acesso Em 2010, 23, 2010. [Google Scholar]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Huzzey, J.; Nydam, D.; Grant, R.; Overton, T. The effects of overstocking Holstein dairy cattle during the dry period on cortisol secretion and energy metabolism. J. Dairy Sci. 2012, 95, 4421–4433. [Google Scholar] [CrossRef] [PubMed]
- Minka, N.; Ayo, J. Physiological responses of food animals to road transportation stress. Afr. J. Biotechnol. 2009, 8, 6601–6613. [Google Scholar]
- Dijkstra, J.; Ellis, J.; Kebreab, E.; Strathe, A.; López, S.; France, J.; Bannink, A. Ruminal pH regulation and nutritional consequences of low pH. Anim. Feed Sci. Technol. 2012, 172, 22–33. [Google Scholar] [CrossRef]
- Nafikov, R.A.; Beitz, D.C. Carbohydrate and lipid metabolism in farm animals. J. Nutr. 2007, 137, 702–705. [Google Scholar] [CrossRef]
- Abeni, F.; Calamari, L.; Stefanini, L. Metabolic conditions of lactating Friesian cows during the hot season in the Po valley. 1. Blood indicators of heat stress. Int. J. Biometeorol. 2007, 52, 87–96. [Google Scholar] [CrossRef]
- Wheelock, J.; Rhoads, R.; VanBaale, M.; Sanders, S.; Baumgard, L. Effects of heat stress on energetic metabolism in lactating Holstein cows. J. Dairy Sci. 2010, 93, 644–655. [Google Scholar] [CrossRef]
- Decuypere, J.; Dierick, N. The combined use of triacylglycerols containing medium-chain fatty acids and exogenous lipolytic enzymes as an alternative to in-feed antibiotics in piglets: Concept, possibilities and limitations. An overview. Nutr. Res. Rev. 2003, 16, 193–210. [Google Scholar] [CrossRef] [PubMed]
- Pryde, S.E.; Duncan, S.H.; Hold, G.L.; Stewart, C.S.; Flint, H.J. The microbiology of butyrate formation in the human colon. Fems Microbiol. Lett. 2002, 217, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.A.; Withington, S.; Newland, A.C.; Kelsey, S.M. Monocyte anergy in septic shock is associated with a predilection to apoptosis and is reversed by granulocyte-macrophage colony-stimulating factor ex vivo. J. Infect. Dis. 1998, 178, 1421–1433. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wells, J.E.; Russell, J.B. The effect of growth and starvation on the lysis of the ruminal cellulolytic bacterium Fibrobacter succinogenes. Appl. Environ. Microbiol. 1996, 62, 1342–1346. [Google Scholar] [PubMed]
- Meale, S.J.; Li, S.; Azevedo, P.; Derakhshani, H.; DeVries, T.; Plaizier, J.; Steele, M.; Khafipour, E. Weaning age influences the severity of gastrointestinal microbiome shifts in dairy calves. Sci. Rep. 2017, 7, 198. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.; He, Y.; Xia, C.; Rahman, M.A.U.; Qiu, Q.; Shao, T.; Liang, Y.; Ji, L.; Wang, H.; Cao, B. Effects of replacing Leymus chinensis with whole-crop wheat hay on Holstein bull apparent digestibility, plasma parameters, rumen fermentation, and microbiota. Sci. Rep. 2017, 7, 2114. [Google Scholar] [CrossRef] [PubMed]
- Paz, H.A.; Anderson, C.L.; Muller, M.J.; Kononoff, P.J.; Fernando, S.C. Rumen bacterial community composition in Holstein and Jersey cows is different under same dietary condition and is not affected by sampling method. Front. Microbiol. 2016, 7, 1206. [Google Scholar] [CrossRef] [PubMed]
- Walsh, K.; O’Kiely, P.; Taweel, H.; McGee, M.; Moloney, A.; Boland, T. Intake, digestibility and rumen characteristics in cattle offered whole-crop wheat or barley silages of contrasting grain to straw ratios. Anim. Feed Sci. Technol. 2009, 148, 192–213. [Google Scholar] [CrossRef]
- Jolazadeh, A.; Dehghan-Banadaky, M.; Rezayazdi, K. Effects of soybean meal treated with tannins extracted from pistachio hulls on performance, ruminal fermentation, blood metabolites and nutrient digestion of Holstein bulls. Anim. Feed Sci. Technol. 2015, 203, 33–40. [Google Scholar] [CrossRef]
- France, J.; Dijkstra, J. Volatile fatty acid production. Quant. Asp. Rumin. Dig. Metab. 2005, 2, 157–175. [Google Scholar]
- Wetzels, S.; Mann, E.; Metzler-Zebeli, B.; Wagner, M.; Klevenhusen, F.; Zebeli, Q.; Schmitz-Esser, S. Pyrosequencing reveals shifts in the bacterial epimural community relative to dietary concentrate amount in goats. J. Dairy Sci. 2015, 98, 5572–5587. [Google Scholar] [CrossRef] [PubMed]
- Jami, E.; Mizrahi, I. Composition and similarity of bovine rumen microbiota across individual animals. PLoS ONE 2012, 7, e33306. [Google Scholar] [CrossRef] [PubMed]
- Pope, P.B.; Mackenzie, A.K.; Gregor, I.; Smith, W.; Sundset, M.A.; McHardy, A.C.; Morrison, M.; Eijsink, V.G. Metagenomics of the Svalbard reindeer rumen microbiome reveals abundance of polysaccharide utilization loci. PLoS ONE 2012, 7, e38571. [Google Scholar] [CrossRef] [PubMed]
- Naas, A.E.; Mackenzie, A.; Mravec, J.; Schückel, J.; Willats, W.G.T.; Eijsink, V.; Pope, P. Do rumen Bacteroidetes utilize an alternative mechanism for cellulose degradation? MBio 2014, 5, e01401–e01414. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Laursen, M.F.; Bahl, M.I.; Michaelsen, K.F.; Licht, T.R. First foods and gut microbes. Front. Microbiol. 2017, 8, 356. [Google Scholar] [CrossRef] [PubMed]
- Broderick, G.A. Altering ruminal nitrogen metabolism to improve protein utilization. Introduction. J. Nutr. 1996, 126, 1324S. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- Russell, J.B.; Hespell, R.B. Microbial rumen fermentation. J. Dairy Sci. 1981, 64, 1153–1169. [Google Scholar] [CrossRef]
- Wood, T.M.; Wilson, C.A.; Stewart, C.S. Preparation of the cellulase from the cellulolytic anaerobic rumen bacterium Ruminococcus albus and its release from the bacterial cell wall. Biochem. J. 1982, 205, 129–137. [Google Scholar] [CrossRef]
- Petersen, C.; Round, J.L. Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef] [PubMed]
- Kiecolt-Glaser, J.K.; Preacher, K.J.; MacCallum, R.C.; Atkinson, C.; Malarkey, W.B.; Glaser, R. Chronic stress and age-related increases in the proinflammatory cytokine IL-6. Proc. Natl. Acad. Sci. USA 2003, 100, 9090–9095. [Google Scholar] [CrossRef] [PubMed]
- Alesci, S.; Martinez, P.E.; Kelkar, S.; Ilias, I.; Ronsaville, D.S.; Listwak, S.J.; Ayala, A.R.; Licinio, J.; Gold, H.K.; Kling, M.A. Major depression is associated with significant diurnal elevations in plasma interleukin-6 levels, a shift of its circadian rhythm, and loss of physiological complexity in its secretion: Clinical implications. J. Clin. Endocrinol. Metab. 2005, 90, 2522–2530. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59. [Google Scholar] [CrossRef] [PubMed]
- Vgontzas, A.N.; Chrousos, G.P. Sleep, the hypothalamic–pituitary–adrenal axis, and cytokines: Multiple interactions and disturbances in sleep disorders. Endocrinol. Metab. Clin. 2002, 31, 15–36. [Google Scholar] [CrossRef]
- Yudkin, J.S.; Kumari, M.; Humphries, S.E.; Mohamed-Ali, V. Inflammation, obesity, stress and coronary heart disease: Is interleukin-6 the link? Atherosclerosis 2000, 148, 209–214. [Google Scholar] [CrossRef]
- Andrews, R.C.; Walker, B.R. Glucocorticoids and insulin resistance: Old hormones, new targets. Clin. Sci. 1999, 96, 513–523. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446. [Google Scholar] [CrossRef]
- Obata, T.; Fernie, A.R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef]
- Xiang, W.; Lam, J.; Shen, J. Stability analysis and L1-gain characterization for switched positive systems under dwell-time constraint. Automatica 2017, 85, 1–8. [Google Scholar] [CrossRef]
- Frank, D.N.; Amand, A.L.S.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; Rudensky, A.Y. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451. [Google Scholar] [CrossRef] [PubMed]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313. [Google Scholar] [CrossRef]
- Fukuda, S.; Ohno, H. Gut Microbiome and Metabolic Diseases; Seminars in Immunopathology; Springer: Berlin, Germany, 2014; pp. 103–114. [Google Scholar]
- Vijay-Kumar, M.; Aitken, J.D.; Carvalho, F.A.; Cullender, T.C.; Mwangi, S.; Srinivasan, S.; Sitaraman, S.V.; Knight, R.; Ley, R.E.; Gewirtz, A.T. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 2010, 328, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Rubtsov, Y.P.; Rasmussen, J.P.; Chi, E.Y.; Fontenot, J.; Castelli, L.; Ye, X.; Treuting, P.; Siewe, L.; Roers, A.; Henderson Jr, W.R. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 2008, 28, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Sawa, S.; Lochner, M.; Satoh-Takayama, N.; Dulauroy, S.; Bérard, M.; Kleinschek, M.; Cua, D.; Di Santo, J.P.; Eberl, G. RORγt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat. Immunol. 2011, 12, 320. [Google Scholar] [CrossRef] [PubMed]
- Tien, M.-T.; Girardin, S.E.; Regnault, B.; Le Bourhis, L.; Dillies, M.-A.; Coppée, J.-Y.; Bourdet-Sicard, R.; Sansonetti, P.J.; Pédron, T. Anti-inflammatory effect of Lactobacillus casei on Shigella-infected human intestinal epithelial cells. J. Immunol. 2006, 176, 1228–1237. [Google Scholar] [CrossRef]
- Fleming, L.; Floch, M. Digestion and absorption of fiber carbohydrate in the colon. Am. J. Gastroenterol. 1986, 81, 507–511. [Google Scholar]
- Segain, J.; De La Blétiere, D.R.; Bourreille, A.; Leray, V.; Gervois, N.; Rosales, C.; Ferrier, L.; Bonnet, C.; Blottiere, H.; Galmiche, J. Butyrate inhibits inflammatory responses through NFκB inhibition: Implications for Crohn’s disease. Gut 2000, 47, 397–403. [Google Scholar] [CrossRef]
- Vanhoutvin, S.A.; Troost, F.J.; Hamer, H.M.; Lindsey, P.J.; Koek, G.H.; Jonkers, D.M.; Kodde, A.; Venema, K.; Brummer, R.J. Butyrate-induced transcriptional changes in human colonic mucosa. PLoS ONE 2009, 4, e6759. [Google Scholar] [CrossRef]

| Item | Treatment | NY | SC | CY |
|---|---|---|---|---|
| COR | B | 130.77 ± 34.45 B | 133.06 ± 23.78 B | 125.57 ± 12.37 B |
| (ng/mL) | A | 158.37 ± 22.26 A | 160.54 ± 25.17 A | 156.89 ± 18.71 A |
| ACTH | B | 338.86 ± 70.49 B | 340.58 ± 58.31 B | 295.15 ± 48.59 B |
| (pg/mL) | A | 422.71 ± 61.02 Aa | 437.84 ± 70.92 Aa | 357.32 ± 66.26 Ab |
| T 3 | B | 6.89 ± 0.778 a | 6.91 ± 0.885 Aa | 5.37 ± 0.776 b |
| (ng/mL) | A | 7.18 ± 0.386 a | 5.14 ± 0.684 Bb | 5.51 ± 0.618 b |
| T 4 | B | 243.89 ± 35.286 a | 231.14 ± 35.594 a | 184.32 ± 70.257 b |
| (ng/mL) | A | 253.56 ± 37.249 a | 206.31 ± 43.599 b | 186.63 ± 47.442 b |
| Item | Treatment | NY | SC | CY |
|---|---|---|---|---|
| Serum LPS | B | 13.63 ± 3.144 | 14.61 ± 2.022 | 13.25 ± 2.199 B |
| (ng/mL) | A | 15.77 ± 2.08 | 17.56 ± 2.186 | 17.54 ± 1.69 A |
| Rumen LPS | B | 13.03 ± 2.589 | 10.93 ± 2.557 B | 10.65 ± 2.55 B |
| (ng/mL) | A | 15.26 ± 2.323 a | 14.86 ± 2.383 Ab | 14.22 ± 1.923 Ac |
| Serum lactic acid | B | 0.91 ± 0.349 B | 0.99 ± 0.464 B | 1.27 ± 0.479 |
| (mmol/L) | A | 1.74 ± 0.192 A | 1.85 ± 0.363 A | 1.7 ± 0.31 |
| Rumen lactic acid | B | 0.26 ± 0.049 c | 0.78 ± 0.641 Ab | 1.97 ± 0.373 Aa |
| (mmol/L) | A | 0.25 ± 0.066 b | 0.43 ± 0.099 Ba | 0.54 ± 0.168 Ba |
| Acetic acid | B | 21.52 ± 0.986 | 23.51 ± 7.56 B | 21.69 ± 4.495 B |
| (mmol/L) | A | 33.36 ± 5.628 c | 45.22 ± 7.21 Aab | 36.01 ± 2.486 Ab |
| Propionic acid | B | 6.03 ± 1.35 | 6.17 ± 2.2 B | 5.17 ± 0.883 B |
| (mmol/L) | A | 7.2 ± 1.608 b | 11.43 ± 2.93 Aa | 7 ± 1.06 Ac |
| Butyric acid | B | 2.3 ± 0.411 B | 3.2 ± 1.73 B | 2.46 ± 0.181 B |
| (mmol/L) | A | 5.06 ± 1.557 Ac | 7.7 ± 1.406 Aa | 6.85 ± 1.969 Ab |
| Acetic : propionic | B | 3.72 ± 0.691 B | 3.84 ± 0.093 | 4.16 ± 0.218 B |
| acids | A | 4.59 ± 0.311 Aab | 4.05 ± 0.372 b | 5.13 ± 0.267 Aa |
| TVFA | B | 29.85 ± 1.49 B | 32.87 ± 3.32 B | 29.31 ± 5.55 B |
| (mmol/L) | A | 45.62 ± 4.34 Ab | 64.345 ± 5.52 Aa | 47.93 ± 3.48 Ab |
| PH | B | 7.27 ± 0.09 | 7.3 ± 0.06 A | 7.22 ± 0.11 A |
| A | 6.98 ± 0.03 a | 6.75 ± 0.09 Bb | 6.79 ± 0.12 Bb |
| Item | Treatment | NY | SC | CY |
|---|---|---|---|---|
| IgG | B | 1.03 ± 0.05 A | 1.13 ± 0.01 A | 1.32 ± 0.06 A |
| (mg/mL) | A | 0.75 ± 0.08 B | 1.05 ± 0.02 B | 0.95 ± 0.02 B |
| IgA | B | 14.98 ± 5.31 | 19.22 ± 2.19 | 15.21 ± 3.39 |
| (ug/mL) | A | 13.82 ± 5.04 ab | 18.03 ± 3.16 a | 13.64 ± 2.34 b |
| TNF-α | B | 12.65 ± 1.86 Ba | 9.77 ± 1.48 Ab | 11.58 ± 1.03 Ba |
| (pg/mL) | A | 15.61 ± 1.24 A | 16.02 ± 1.31 B | 15.38 ± 2.81 A |
| IL-1β | B | 2000.61 ± 455.54 Bb | 1716.15 ± 239.57 Bb | 2225.56 ± 391.49 Ba |
| (pg/mL) | A | 2682.02 ± 511.69 Ab | 2834.96 ± 310.28 Ab | 3265.82 ± 335.59 Aa |
| IL-6 | B | 695.02 ± 135.81 Ba | 688.45 ± 57.96 Ba | 594.1 ± 45.63 Bb |
| (pg/mL) | A | 845.49 ± 68.06 A | 802.35 ± 82.38 A | 857.58 ± 45.92 A |
| IL-10 | B | 783.66 ± 97.12 a | 621.33 ± 70.11 b | 621.03 ± 127.17 b |
| (pg/mL) | A | 806.59 ± 78.53 a | 689.08 ± 111.145 ab | 648.91 ± 171.76 b |
| IL-4 | B | 498.49 ± 113.535 a | 444.01 ± 50.79 ab | 361.82 ± 64.257 Bb |
| (pg/mL) | A | 504.49 ± 204.821 | 501.25 ± 47.275 | 477.85 ± 50.824 A |
| Item | Treatment | NY | SC | CY |
|---|---|---|---|---|
| OTUs | B | 2610.75 ± 100.24 a | 2255.8 ± 372.33 Bb | 2093.6 ± 146.02 b |
| A | 2410.25 ± 387.46 b | 2755.6 ± 177 Aa | 2184 ± 113.74 b | |
| Chao1 | B | 4219.42 ± 340.24 | 3678.18 ± 1014.29 B | 3343.85 ± 375.36 |
| A | 3947.86 ± 849.05 a | 4315.88 ± 606.76 Aa | 3688.9 ± 426.59 b | |
| Shannon | B | 6.5 ± 0.14 | 6.38 ± 0.19 B | 6.15 ± 0.18 |
| A | 6.41 ± 0.3 b | 6.67 ± 0.07 Aa | 6.2 ± 0.08 b |
| Item | Transport | Breeds | Transport × Breeds | |||
|---|---|---|---|---|---|---|
| F | p | F | p | F | p | |
| COR | 13.856 | <0.001 | 0.262 | 0.77 | 0.192 | 0.826 |
| ACTH | 10.057 | 0.0812 | 0.817 | 0.445 | 14.668 | <0.001 |
| Serum LPS | 21.157 | 0.029 | 24.027 | <0.001 | 0.593 | <0.001 |
| Rumen LPS | 9.094 | 0.006 | 0.654 | 0.529 | 0.149 | 0.862 |
| Rumen lactic acid | 24.048 | <0.001 | 1.634 | 0.216 | 9.769 | 0.001 |
| Serum lactic acid | 21.823 | <0.001 | 20.274 | <0.001 | 0.82 | 0.452 |
| pH | 33.873 | <0.001 | 22.273 | <0.001 | 4.392 | <0.001 |
| Acetic acid | 42.38 | <0.001 | 14.98 | <0.001 | 1.684 | 0.207 |
| Propionic acid | 16.37 | <0.001 | 3.349 | 0.052 | 3.73 | 0.039 |
| Butyric acid | 47.527 | <0.001 | 16.458 | <0.001 | 0.89 | 0.424 |
| Acetic: propionic | 22.877 | <0.001 | 3.658 | 0.041 | 2.733 | 0.085 |
| TVFA | 39.951 | <0.001 | 8.164 | 0.002 | 1.934 | 0.166 |
| IgA | 0.365 | 0.547 | 10.874 | <0.001 | 0.138 | 0.871 |
| IgG | 26.078 | <0.001 | 0.109 | 0.897 | 0.308 | 0.738 |
| TNF-α | 10.252 | 0.0617 | 16.084 | <0.001 | 10.169 | <0.001 |
| IL-1β | 15.095 | <0.001 | 7.98 | 0.044 | 14.218 | <0.001 |
| IL-6 | 1.78 | 0.186 | 15.228 | <0.001 | 1.025 | 0.363 |
| IL-10 | 1.878 | 0.174 | 1.917 | 0.153 | 10.522 | <0.001 |
| IL-4 | 13.701 | <0.001 | 1.112 | 0.334 | 2.305 | 0.106 |
| OTUs | 2.351 | 0.138 | 4.388 | 0.024 | 2.765 | <0.001 |
| Chao1 | 1.529 | 0.228 | 1.297 | 0.292 | 0.414 | 0.665 |
| Shannon | 1.621 | 0.215 | 7.464 | 0.003 | 1.905 | 0.171 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Shah, A.M.; Wang, Z.; Peng, Q.; Hu, R.; Zou, H.; Tan, C.; Zhang, X.; Liao, Y.; Wang, Y.; et al. Effects of Land Transport Stress on Variations in Ruminal Microbe Diversity and Immune Functions in Different Breeds of Cattle. Animals 2019, 9, 599. https://doi.org/10.3390/ani9090599
Li F, Shah AM, Wang Z, Peng Q, Hu R, Zou H, Tan C, Zhang X, Liao Y, Wang Y, et al. Effects of Land Transport Stress on Variations in Ruminal Microbe Diversity and Immune Functions in Different Breeds of Cattle. Animals. 2019; 9(9):599. https://doi.org/10.3390/ani9090599
Chicago/Turabian StyleLi, Fengpeng, Ali Mujtaba Shah, Zhisheng Wang, Quanhui Peng, Rui Hu, Huawei Zou, Cui Tan, Xiangfei Zhang, Yupeng Liao, Yongjie Wang, and et al. 2019. "Effects of Land Transport Stress on Variations in Ruminal Microbe Diversity and Immune Functions in Different Breeds of Cattle" Animals 9, no. 9: 599. https://doi.org/10.3390/ani9090599
APA StyleLi, F., Shah, A. M., Wang, Z., Peng, Q., Hu, R., Zou, H., Tan, C., Zhang, X., Liao, Y., Wang, Y., Wang, X., Zeng, L., Xue, B., & Wang, L. (2019). Effects of Land Transport Stress on Variations in Ruminal Microbe Diversity and Immune Functions in Different Breeds of Cattle. Animals, 9(9), 599. https://doi.org/10.3390/ani9090599








